Targeting the epigenome in malignant pleural mesothelioma
Introduction
Malignant pleural mesotheliomas (MPM) are rare and highly lethal neoplasms attributable primarily to environmental or occupational exposure to asbestos and related fibers, influenced in some cases by predisposing germline mutations (1). Recent advances in “omics” technologies have enabled comprehensive gene and transcriptome analyses that have provided considerable insight regarding the pathogenesis, prognosis, and treatment of MPM (2,3). Presently, less information is available regarding mechanisms and clinical relevance of epigenetic derangements in MPM (4,5). This review will focus on recent advances pertaining to the epigenetics of MPM, and potential implications regarding the development of epigenetic therapies for these neoplasms.
Epigenetic regulation of gene expression
Epigenetic regulation of gene expression occurs in the context of chromatin, the basic unit of which is the nucleosome. Each nucleosome is composed of 147 bp of DNA wrapped twice around an octamer of core histones (H2A, H2B, H3, and H4). Lysine rich tails of core histones (particularly histone H3) extend out from the nucleosome, providing sites for reversible, covalent modifications such as acetylation, methylation, ubiquitination, phosphorylation, and SUMOylation that facilitate activation or repression of gene expression (6,7).
DNA methylation is the major epigenetic mechanism mediating dynamic changes in gene expression during normal cellular homeostasis and tissue differentiation, as well as long-term repression of imprinted alleles, germ cell restricted genes, repetitive DNA, and endogenous retroviral sequences (8,9). Three major DNA methyltransferases (DNMT1, 3A, and 3B) have been identified in normal somatic cells, all of which mediate transfer of a methyl group from S-adenosyl-methionine to the 5' position of cytosine in the context of CpG (10). Clusters of CpG dinucleotides (CpG islands) are located in promoters of approximately 60% of genes; most of these islands are not methylated, thereby allowing a relaxed, transcriptionally active (euchromatin) structure (11). Additional CpG dinucleotides and CpG islands are dispersed throughout the genome; these CpGs are typically hypermethylated in normal cells (11). Whereas considerable overlap exists, DNMT1 binds preferentially to hemimethylated DNA, and functions mostly as a maintenance methyltransferase, restoring DNA methylation patterns during DNA replication or repair. In contrast, DNMT3A and 3B recognize unmethylated or hemimethylated DNA and mediate de-novo DNA methylation (10). DNA methylation inhibits binding of methylation-sensitive transcription factors (12), and induces recruitment of methyl CpG binding domain (MBD) and related proteins such as UHRF1, as well as co-repressor complexes containing sin3a, NCoR and histone deacetylases (HDACs) resulting in formation of compact, transcriptionally silent heterochromatin (13,14). During malignant transformation, over-expression or aberrant targeting of components of the DNA methylation machinery results in epigenetic silencing of differentiation-related genes, many of which are tumor suppressors (11). Additionally, DNA methylation can inactivate tumor suppressor genes by transition mutations resulting from deamination of 5-methylcytosine (5-MC) (11), or adduct formation with environmental carcinogens such as benzo(a) pyrene (15).
DNA demethylation occurs passively during DNA replication (16). Furthermore, DNA is actively demethylated by ten-eleven translocations (TET) enzymes which catalyze oxidation of 5-MC to 5-hydroxymethylcytosine, 5-formylcytosine, and 5-carboxylcytosine (17). During malignant transformation, the total amount of methylated CpGs except for those in promoter CpG islands becomes markedly reduced (up to 50%) (9). Whereas genome wide DNA demethylation may be attributable to deficient DNA repair (18,19), decreased DNA methyltransferase 1 expression (20,21), glycosylase-mediated excision of 5-MC (16), and aberrant expression/targeting of TET proteins (22), the mechanisms mediating this phenomenon have not been fully elucidated. Genome-wide DNA demethylation facilitates de-repression of imprinted alleles, endogenous retroviruses, and transposable elements, thereby inducing genomic instability (11,23). Global DNA demethylation also results in de-repression of a variety of cancer-germline (CG) genes that are silent in normal somatic cells, yet exhibit stage-specific expression during germ cell development in testes or ovary (24). Simultaneous hypermethylation of tumor suppressors and hypomethylation of other genomic regions in cancer cells coincide with alterations in nucleosomal positioning, as well as modifications of core histones reflecting activated, repressed or bivalent chromatin structure within the respective loci (25).
Acetylation/deacetylation and methylation/demethylation have been the most extensively characterized histone modifications in normal and cancer cells (25,26). Histone acetylation is mediated by a variety of histone acetyltransferases (HATs) whereas histone deacetylation is mediated by HDACs that are divided into four classes [reviewed in ref (26-28)]. Histone acetylation increases net negative charge leading to repulsion of DNA, relaxation of chromatin, and activation of gene expression (7). Many non-histone proteins including Hsp90, SP1, p53, and HDAC1 are targets for HATs and HDACs (27-29).
Histone lysine methylation is mediated by a variety of histone lysine methyltransferases (KMTs) that mediate mono- di- and trimethylation of specific residues, whereas histone demethylation is mediated by histone demethylases (KDMs) (26,30-32). These histone modifications are highly dynamic in response to environmental signals (33,34). Unlike histone acetylation, histone lysine methylation does not alter charge of core histones. Furthermore, in contrast to histone acetylation which is always a histone activation mark, histone lysine methylation may facilitate or inhibit gene expression depending on the site. For example, methylation of histone H3K9 and H3K27 coincides with transcriptional repression; in contrast, H3K4, H3K36 or H3K79 methylation is associated with gene activation. A variety of non-histone proteins including NFκB, p53, and E2F1 are targets of KMTs and KDM (30,31).
Recently, ATP-dependent chromatin remodeling complexes (CRC) have emerged as critical mediators of epigenetic regulation of gene expression in normal and malignant cells (35,36). To date four families have been characterized including switch/sucrose nonfermentable (SWI/SNF), imitation SWI (ISWI), chromodomain helicase DNA-binding (CHD), and INO80, named for its ability to regulate inositol-responsive gene expression. These complexes have multiple subunits with various isoforms, and exhibit pleiotropic functions including regulation of gene expression, maintenance of chromatin structure, replication of pericentrometic heterochromatin, ribosomal RNA repression, and DNA double strand break repair (37). The mechanisms by which these complexes remodel chromatin vary among and within different families. For instance, SWI/SNF complexes disassemble nucleosomes to expose DNA, whereas ISWI, INO80 and CDH family members reposition (slide) nucleosomes and stretch out the intervening DNA, thereby increasing accessibility to transcription factors; these latter complexes can also assemble nucleosomes, and are thus important for maintaining chromatin structure and genomic stability (35-37).
Advances in transcriptome analysis have revealed that >90% of the genome is transcribed as noncoding RNAs (38). Particularly relevant to this discussion are recent observations that long noncoding RNAs (lncRNA) are critical mediators of chromatin structure and gene expression during normal cellular homeostasis and malignant transformation (39). In addition to other highly diverse activities (40), lncRNAs function as scaffolds to recruit DNMTs and histone methyltransferases to chromatin (41), thereby adding another layer of epigenetic regulation in normal cells that is perturbed in malignancies.
Epigenetic alterations in MPM
Methylation-mediated silencing of tumor suppressors
Although initially thought not to contribute to the pathogenesis of mesotheliomas (42), it is become clear that epigenetic alterations are common events in this disease. Indeed, given the relatively low mutational burden of mesotheliomas (3), epigenetic perturbations may be critical determinants of malignant transformation of pleural mesothelial cells following exposure to asbestos and related fibers. Christensen et al. (43) examined promoter DNA methylation status of six genes regulating cell cycle progression in 70 MPM. Extent of methylation of these genes correlated with lung asbestos burden as well as overall survival. Goto et al. (44) used micro-array and quantitative methylation specific PCR techniques to examine methylation status of over 6,000 CpG islands in 20 MPM relative to 20 pulmonary adenocarcinomas. An average of 387 genes (6.3%) were hypermethylated in mesotheliomas compared to 544 genes (8.8%) in lung adenocarcinomas. Higher levels of DNA methylation correlated with decreased patient survival. Three genes (TMEM30B, KAZALD1, and MAPK13) were specifically hypermethylated in MPM. Numerous other reports have documented DNA methylation-mediated silencing of tumor suppressor genes in MPM (Table 1); hypermethylation of some these genes may impact survival of mesothelioma patients [reviewed in ref (4)].
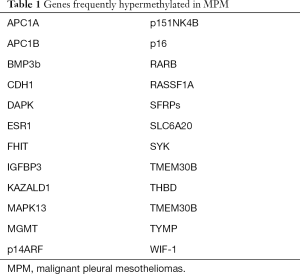
Full table
Whereas accumulating evidence indicates recurrent hypermethylation of tumor suppressor genes in MPM, the mechanisms underlying this phenomenon as yet have not been elucidated. Cytokine signaling can modulate DNMT expression and mediate hypermethylation of target genes in colorectal carcinoma and erythroleukemia cells (45,46); conceivably, cytokines induced by high mobility group box 1 (HMGB1) or the NLRP3 inflammasome in response to asbestos exposure dysregulate expression and/or targeting of DNMTs and other components of the DNA methylation machinery during evolution of MPM (47-50). Recently we performed qRT-PCR analysis of a panel of genes encoding epigenetic regulators in a panel of cultured cell lines derived from asbestos associated MPM relative to either LP-9 (a commercially available normal mesothelial cell line) or a normal mesothelial line established in our laboratory. DNMT1, DNMT3A and DNMT3B appeared to be over-expressed in the majority of MPM lines (Table 2). Consistent with these findings, TCGA data demonstrate a spectrum of DNMT expression in MPM, and suggest that over-expression of DNMT1, DNMT3A and DNMT3B correlates with shorter survival of pleural mesothelioma patients (Figure 1).
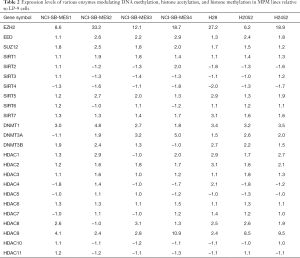
Full table

In recent studies, Kim et al. (51) used RNA-seq and methylated DNA immunoprecipitation techniques to comprehensively characterize methylation and gene expression profiles in pluripotent side populations (SP) and non-SP fractions of a human mesothelioma line. Six thousand and four hundred genes were hypermethylated, while 3,483 were hypomethylated in SP compared to non-SP fractions. Seven hundred and ninety-five genes were upregulated whereas 335 were significantly repressed in SP fractions relative to non-SP. Concomitant changes in DNA methylation and expression levels were noted for 122 genes; 118 were hypermethylated and downregulated, whereas 4 were hypomethylated and upregulated. Ten genes exhibited hypermethylation of promoter CpG islands in association with repression. Gene ontology analysis revealed significant enrichment for stem cell maintenance, stem cell development, and stem cell differentiation.
In additional studies Kim et al. (52) used micro-array techniques to comprehensively examine microRNA (miR) expression profiles in SP and non-SP fractions of the human mesothelioma cell line used for the aforementioned DNA methylation analysis. Ninety-five miRs were differentially expressed in the SP fraction. Gene ontology analysis demonstrated enrichment for stem cell maintenance, programmed cell death, cell proliferation, cell migration, and cellular response to stress. ErbB2 receptor tyrosine kinase signaling was the most represented pathway, suggesting that similar to what has been observed in other malignancies (53,54), ErbB2 signaling is critical for maintaining pluripotency and possibly treatment resistance in mesothelioma cells.
Loss of imprinting (LOI) and de-repression of CG genes
DNA hypomethylation has been implicated in LOI, de-repression of endogenous retroviral sequences, and activation of CG genes which may enhance proliferation, genomic instability, and resistance to apoptosis during malignant transformation (55-57). Presently, limited information is available regarding mechanisms and clinical relevance of DNA hypomethylation in MPM. For instance, with the exception of a single report of LOI of IGF-II in a pleural mesothelioma inducing hypoglycemia (58), this phenomenon has not been described in MPM. Similarly, expression of endogenous retroviral sequences has not been evaluated in MPM.
Aberrant activation of CG genes [also referred to as cancer-testis (CT) genes] in somatic cells during malignant transformation results in expression of highly restricted tumor antigens that induce serologic as well as cell-mediated immune responses in cancer patients; as such, cancer-testis antigens (CTAs) have emerged as attractive targets for cancer immunotherapy (59). To date, more than 270 CG genes have been registered in the CT database (http://www.cta.lncc.br); 75% of these genes are expressed only in normal testis and malignancies, whereas the remainder exhibit high level expression in testis and variable expression in other normal tissues, and cancers (24). Approximately half of the CG genes are encoded on the X chromosome. These cancer-testis-X chromosome (CT-X) genes frequently comprise extended families with inverted DNA repeats. In contrast, the non-X CT genes are not associated with extended families or inverted repetitive DNA sequences (60,61). Relative to autosomal CT genes, CT-X genes tend to be more frequently activated in cancers, and particular gene families are coordinately up-regulated in a tumor-specific manner suggesting transcriptional co-regulation, and functional relatedness of the respective gene products.
In general, the magnitude of CG gene de-repression in human cancers coincides with advanced stage of disease, and there is mounting evidence that activation of these genes enhances the malignant phenotype of cancer cells. For example, BORIS/CTCFL up-regulates h-TERT (62), and through poorly defined mechanisms inhibits apoptosis in cancer cells (63). MAGE-A11 inhibits function of the RBL1/p107 tumor suppressor (64), and MAGE-B2 enhances E2F activity to promote cell cycle progression (65). MAGE-A2, and MAGE-C2 impair p53 function by directly inhibiting binding of p53 to target promoters, promoting deacetylation (inactivation) of p53, or by enhancing ubiquitin-mediated degradation of this tumor suppressor (66-68). Recent reports demonstrating high level CT-X gene expression in cancer stem cells (69-71), raise the possibility that CT-X antigens function to enhance pluripotency.
Although associated with genomic hypomethylation, de-repression of CG genes does not appear to be simply a manifestation of pluripotency. Loriot et al. (72) observed no up-regulation of 18 different CG genes in human ESC, mesenchymal stem cells or adipose derived stem cells. Consistent with these findings, we observed that CG genes such as NY-ESO-1, MAGE-A1, and MAGE-A3 that are commonly upregulated in thoracic malignancies, remain transcriptionally repressed in induced pluripotent stem cells (iPSC) derived from normal small airway epithelial cells (Shukla et al.; submitted). Although these findings could suggest incomplete reprogramming in iPSC, up-regulation of CG genes in cancer cells may require more extensive DNA hypomethylation, as well as tissue-specific activation of transcription factors.
Several studies have been performed to examine the mechanisms regulating CG gene expression in cancer cells. Cartron et al. (73) observed that epigenetic repression of NY-ESO-1 in mesothelioma cells was mediated by sequential recruitment of HDAC1-mSin3A-NCOR, DNMT3b- HDAC1-Egr1, and DNMT1-PCNA-UHRF1-G9a complexes to the NY-ESO-1 promoter. Consistent with these findings, de-novo activation of NY-ESO-1 requires genetic depletion or pharmacologic inactivation of both DNMT1 and DNMT3B (74). Hong et al. (74) reported that spontaneous or pharmacologic induction of NY-ESO-1 coincides with up-regulation and recruitment of BORIS/CTCFL with displacement of CTCF from the NY-ESO-1 promoter. Kang et al. (75) demonstrated that the transcription factor specificity protein 1 (SP1) directly interacts with BORIS/CTCFL but not CTCF, and that SP1 is required for BORIS-mediated activation of NY-ESO1.
Recently, Cannuyer et al. (76) sought to examine mechanisms regulating activation of CG genes in melanoma cells. Analysis of transcriptomic data revealed that CG gene de-repression was not associated with differential expression of gametogenic regulators. Instead, CG gene activation coincided with repression of a set of genes regulating mitosis/cell division. This gene expression signature was similar to one previously observed in epithelial cells following depletion of DNMT1 (77). CG gene activation and downregulation of inversely correlated mitosis/cell division genes in melanoma samples was associated with a modest, but statistically significant decrease in expression of DNMT1, but did not correlate with alterations in DNMT3A, DNMT3B, TET1, TET2, TET3, or UHRF1 expression.
Presently, limited information is available regarding expression of CG genes in MPM. Sigalotti et al. (78) used RT-PCR techniques to examine expression of MAGE1-4, NY-ESO-1, GAGE1-2, GAGE1-6, SSX2, SSX1-6 and RAGE-1 in five MPM lines relative to normal mesothelial cells. Consistent with what we have previously reported for lung cancers (79), heterogeneous CG gene expression was observed in these MPM lines, with each line expressing a unique profile. Normal mesothelial cells did not express any of these genes. Consistent with our previously published data (80), CG genes including NY-ESO-1 were readily up-regulated in MPM lines by the DNA demethylating agent 5-aza-CdR (78).
To more comprehensively examine CG gene expression in MPM, we have analyzed the TCGA database focusing primarily on those genes that are normally expressed only in germ cells, and aberrantly activated in cancers, and have been shown to be regulated by DNA methylation mechanisms (24). Table 3 depicts results of 15 of the 87 mesothelioma samples in the data base. Virtually all MPM exhibited de-repression of CG genes, although the patterns and magnitude of activation of these genes were quite variable. MAGE family members were the most consistently up-regulated CT-X genes, whereas BAGE2 and CAGE1 were the most commonly up-regulated autosomal CG genes activated in MPM. Several tumors exhibited extensive de-repression of CG genes, which did not appear to coincide with relative expression levels of genes encoding DNMTs, HDACs, SP1, sirtuins, or TET proteins (data not shown).
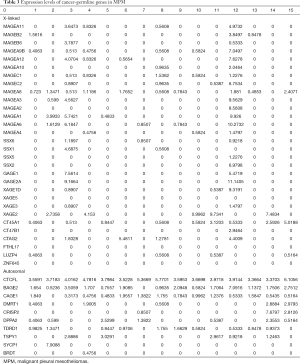
Full table
Polycomb mediated gene silencing
Polycomb group proteins (PcG) proteins are critical determinants of pluripotency and differentiation of stem cells (81), as well as aberrant gene expression during malignant transformation (82,83). Two major polycomb repressor complexes (PRC) have been identified in mammals (83). The initiation complex, PRC-2, contains EZH1/EZH2, SUZ12, EED and RBAP46/48 subunits, and mediates trimethylation of histone 3 lysine 27 (H3K27Me3). The maintenance complex, PRC-1, containing PCAF, PHC, RING1, CBX, and BMI1 subunits, mediates ubiquitination of H2AK119 (H2AK119Ub) (82,83). These histone marks coincide with recruitment of CRC, formation of heterochromatin - frequently in the context of DNA hypermethylation, and repression of gene expression (82,83). Several proteins such as JARID2 and additional sex comb-like (ASXL) family members physically interact with EZH2 and SUZ12 to target PRC-2 to polycomb response elements (PRE) throughout the genome (84,85). Although often associated with promoter hypermethylation, polycomb-mediated gene silencing may occur independent of DNA methylation (86,87), typically in the context of bivalent chromatin, exhibiting occupancy of PcG proteins and simultaneous activation (H3K4Me3) and repressive (H3K27Me3) histone marks. Frequently observed in stem cells, bivalent chromatin maintains differentiation-related genes in a repressed, but poised state for rapid activation or permanent silencing depending on the differentiation signal (88,89).
Observations by Goto et al. (44) that a subset of genes repressed in MPM exhibited H3K27Me3 without DNA hypermethylation suggested that perturbations of polycomb gene expression might contribute to the pathogenesis of these neoplasms. To examine this issue, we used microarray, qRT-PCR, immunoblot and immunofluorescence (IHC) techniques to examine polycomb group (PcG) gene/protein expression in a panel of cultured MPM lines and normal mesothelial cells (90). This analysis demonstrated over-expression of EZH2 (both splice variants), and to a lesser extent, EED and SUZ12 in MPM cells relative to cultured normal mesothelial cells (LP3, LP9). Immunoblot experiments demonstrated over-expression of EZH2 with concomitant increases in global H3K27Me3 levels in MPM cells relative to normal mesothelial cells. qRT-PCR, immunoblot, and IHC experiments demonstrated over-expression of EZH2 in approximately 80% of primary MPMs (the vast majority of which were epithelioid histology). shRNA-mediated knock-down of EZH2 (both variants) or EED [which is critical for maintaining stability of PRC-2, and histone methyltransferase activity of EZH2 (83)] decreased global H3K27Me3 levels and significantly inhibited proliferation, migration, clonogenicity and tumorigenicity of MPM cells. The S-adenosylhomocysteine hydrolase (SAH) inhibitor, DZNep, which is known to deplete PRC-2 components (91), recapitulated the effects of EZH2/EED depletion in MPM cells in-vitro and in-vivo. Microarray with Gene Set Enrichment Analysis confirmed enrichment of polycomb targets in MPM xenografts from DZNep treated mice. Additional analysis revealed that increased intratumoral expression of either of the two EZH2 splice variants detected by Illumina array techniques correlated with decreased overall survival of MPM patients undergoing potentially curative resections (90).
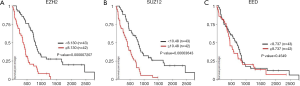
Collectively, these experiments were the first demonstration that EZH2 is over-expressed in MPM, and that PRC-2 is a potential therapeutic target in these neoplasms. Subsequent analysis of TCGA has confirmed up-regulation of EZH2 in MPM (92), as well as the significant association between EZH2 over-expression and decreased survival of MPM patients (Figure 2A). Further analysis of TCGA demonstrates that SUZ12 over-expression also portends poor survival in MPM patients (Figure 2B). In contrast, there does not appear to be any association between EED expression and survival of MPM patients (Figure 2C).
The aforementioned findings are of particular relevance given recent observations that rare familial MPMs, as well as ~60% of sporadic MPMs exhibit inactivating mutations involving BRCA-1 associated protein-1 (BAP1), which encodes a nuclear ubiquitin hydrolase with diverse activities including de-ubiquitination of H2AK119Ub (93-95). BAP1 directly interacts with ASXL1, but not EZH2 or SUZ12 (85). As previously mentioned, ASXL1 interacts with EZH2 and SUZ12 to recruit PRC-2 to DNA (85). As such, the ASXL1-BAP1 complex may function to mitigate the repressive activities of ASXL1-PRC-2. Collectively, these findings suggest that perturbations of the BAP1-ASXL1-polycomb repressor (BAPR) axis are central themes of mesothelioma development. Whereas the effects of BAP1 mutations on gene expression in MPM have been described (93), the epigenomic effects of BAPR dysregulation in these neoplasms have not been evaluated in a comprehensive manner. Conceivably, global and promoter-specific PRC-2 marks, DNA methylation, micro-RNA and gene expression profiles, as well as responses to biochemical or pharmacologic inhibition of PRC-2 activity may be contingent on BAP1 mutation status as well as magnitude of EZH2 over-expression in MPM.
In a series of elegant experiments, LaFave et al. (92) observed that BAP1 mutations, which typically result in loss of protein expression, increased EZH2 as well as SUZ12 expression in MPM cells. Up-regulation of EZH2 in BAP1 mutant cells was associated with reduced levels of H4K2Me1, as well as decreased occupancy of L3MBTL2 (an atypical polycomb protein which recognizes this repressive histone mark) within the EZH2 promoter (96,97). Additional experiments demonstrated that BAP1 deubiquitinates and thereby stabilizes and co-localizes with L3MBTL2 within the EZH2 promoter. BAP1 mutant MPM cells exhibited decreased H4K20Me1 in the EZH2 promoter; presently it is not known if this was due to a decrease in expression/activity of SETD8, the KMT that catalyzes mono-methylation of H4K20 (30), or up-regulation of an unidentified histone demethylase. BAP1 mutations decreased occupancy of BAP1 as well as L3MBTL2 within the EZH2 promoter. Relative to MPM cells expressing BAP1, MPM cells with BAP1 mutations were markedly more sensitive to pharmacologic inhibitors of EZH2 in-vitro and in-vivo (92), suggesting that BAP1 mutations render MPM cells addicted to PRC-2.
Despite strong association between BAP1 mutations and repression of stem cell polycomb targets (93), there does not appear to be a unique clinical phenotype of BAP1 mutant MPM. Interestingly, current or former smokers with MPM appear more likely to have somatic BAP1 mutations, although nucleotide substitutions do not suggest a direct causal role of smoker exposure and BAP1 mutations (98). Additionally, in contrast to EZH2 or SUZ12 over-expression which appears to be associated with decreased patient survival, BAP1 mutations appear be associated with improved patient survival despite up-regulation of both of these PRC-2 components (99,100). This paradox may be related to deficient BRCA-1-mediated DNA repair, which may render BAP1 mutant MPM cells more sensitive to DNA damaging agents. To date however, there have been no published studies examining mutational loads in BAP1 mutant vs BAP1 wild type MPM.
Additional studies have been performed to examine mechanisms contributing to EZH2 over-expression that potentially could be targeted therapeutically in MPM. In silico analysis of the EZH2 promoter revealed numerous recognition elements for SP1, a zinc-finger transcription factor over-expressed in a variety of human malignancies (101). qRT-PCR, immunoblot and IHC experiments demonstrated markedly higher SP1 expression levels in MPM lines and primary MPMs relative to cultured normal mesothelia or normal pleura. Over-expression or knock-down of SP1 significantly increased or decreased EZH2 expression, respectively; furthermore, knock-down of SP1 diminished proliferation of MPM cells (102).
SWI/SNF
CRC: SWI/SNF complexes- mammalian homologs of yeast trithorax, function to antagonize the repressive activities of PRC-2 in part by disrupting DNA-nucleosome contacts, and facilitating movement, ejection, or substitution of nuclosomes to enhance transcription factor accessibility to DNA (103,104). SWI/SNF complexes are composed of 10–15 subunit multimers encoded by 29 genes. Genes encoding SWI/SNF complexes rank among the most frequently mutated genes in human cancers, with specific subunit mutations being associated with unique cancer histologies (103,104). Yoshikawa et al. (105) performed whole exome sequencing on short-term lines established from 8 MPM, and noted significant enrichment for mutations in genes involved in SWI/SNF pathways, including homozygous mutations of SMARCA4, ARID2, and PBRM1; one patient had homozygous germline mutations in SMARCC1 and SETD2, a histone methyltransferase that catalyzes formation of H3K36ME3 (activation mark). More recently, Yoshikawa et al. (2) used high-density array comparative genomic hybridization (a-CGH) and targeted next-generation sequencing (NGS) techniques to examine somatic copy loss in the 3p21 region (approximately 10.7 Mb containing 251 genes) in 33 MPM. Minute (<3 Kb) bi-allelic deletions were detected in 46 genes; 4 of which have been associated with malignancies, including two SWI/SNF related genes [PBRM1 (15%) and SMARCC1 (6%)], BAP1 (48%) and SETD2 (27%). In a recent comprehensive genomic analysis of over 200 MPM, Bueno et al. (3) reported no mutations involving genes encoding SWI/SNF components, but did observe SETD2 mutations in 8% of specimens, as well as mutations involving two additional histone methyltransferases (SETDB1 and SETD5) in approximately 3% of specimens. Discrepancies between results reported by Yoshikawa and colleagues (2) and Bueno et al. (3) may be attributable to identification of minute deletions by high-density a-CGH and targeted NGS that are not detectable by conventional NGS techniques. Additional studies are required to more fully interrogate the frequency and clinical implications of SWI/SNF mutations in MPM.
Epigenetic strategies for mesothelioma therapy
Aforementioned data demonstrate that similar to other cancers (11), MPM exhibit silencing of tumor suppressor genes via site specific DNA hypermethylation and/or polycomb repressive complexes in the context of genome wide hypomethylation that facilitates LOI and de-repression of CG genes. This “DNA methylation paradox”, recapitulates epigenomic states in normal germ cells, and provides the rationale for the development of epigenetic regimens that induce growth arrest/apoptosis via restoration of tumor suppressor gene expression (106-108), while simultaneously augmenting antitumor immunity by up-regulation of CTAs (79), induction of viral mimicry by de-repression of endogenous retroviruses (109,110), and modulation of the tumor microenvironment (111-113).
Given their direct roles in silencing tumor suppressor genes and maintaining pluripotency (114,115), DNMTs are attractive targets for MPM therapy. However, previous clinical efforts to inhibit DNMT activity in MPM have been disappointing. Yogelzang et al. (116) reported a 17% objective response rate in 41 MPM patients receiving continuous 120 h dihydro-5-azacytidine infusions; interestingly, the one complete responder was free of disease six years following treatment. Schrump et al. (117) observed transient stabilization of disease in 2 of 6 MPM patients receiving continuous 72 h decitabine infusions.
The lack of efficacy of DNA hypomethyating agents in solid tumors to date may be related to the fact that these agents have been dosed to maximum tolerated levels resulting in myelosuppression rather than administered chronically at lower doses to achieve pharmacodynamic effects without systemic toxicities. Data from our phase I decitabine (DAC) trial clearly demonstrate that chronic exposures are required to achieve maximal gene induction effects in cancer tissues (117). Furthermore, the short half-lives (<5 min) and poor biodistribution of 5-AZA and DAC administered by IV, SQ or PO routes limits their potential use in patients with solid tumors. These compounds are rapidly inactivated by cytidine deaminase (CDA) which is present in virtually all organs-particularly those in the GI tract (118). Recent studies in nonhuman primates (119) as well as a phase I trial in patients with sickle cell disease (Saunthararajah et al.; submitted) have demonstrated that oral tetrahydrouridine [an inhibitor of CDA that has been administered intravenously to thousands of cancer patients without documented toxicities (120,121)] significantly increases Cmax and t1/2 (>50 nM and 4 h, respectively) and increases biodistribution of oral decitabine thereby significantly decreasing interpatient variability regarding drug levels; oral DAC-THU mediated systemic DNA hypomethylation evidenced by significant increases in fetal hemoglobin, without neutropenia, thrombocytopenia or lymphopenia. A phase II trial (NCT02664181) is presently underway at the Cleveland Clinic and NCI to examine if DAC/THU can improve responses to nivolumab when administered as second line therapy to patients with non-small cell lung cancers (NSCLC). A phase I/II dose escalation trial will commence at the NCI in the very near future to ascertain if oral DAC/THU can enhance responses to pembrolizumab when administered as first line therapy for NSCLC with high PD-L1 expression. If positive, results of these trials would support evaluation of similar regimens in mesothelioma patients (see below).
Despite encouraging preclinical data (122), efforts to target HDACs in MPM have been discouraging as well. Krug et al. (123) randomized 661 MPM patients to receive the HDAC inhibitor, vorinostat or placebo as 2nd or 3rd line therapy. Primary endpoints were overall survival (OS) as well as safety and tolerability of vorinostat. Median OS for vorinostat treated patients was 30.7 weeks (95% CI: 26.7–36.1) compared to 27.1 weeks (95% CI: 23.1–31.9) for patients receiving placebo. The lack of efficacy of single agent vorinostat in MPM patients is not surprising given little evidence of over-expression of HDACs in MPM (Table 2), and rather limited antitumor effects of HDAC inhibitors alone in preclinical experiments (124). Combination strategies such as the use of HDAC inhibitors to sensitize cells to TRAIL mediated apoptosis (125), or the use of flavopiridol to enhance romidepsin mediated growth arrest and apoptosis (124) might be appropriate to evaluate in future clinical trials.
Given the frequency and negative prognostic impact of EZH2 over-expression in MPM (90,92), PRC-2 has emerged as a major therapeutic target in these neoplasms- particularly those with BAP1 mutations. Whereas DZNep is not available for clinical trials, several potent and specific inhibitors of EZH2 activity are in early clinical development. A multicenter phase II trial (NCT02860286) is underway to examine response rates in patients with inoperable, BAP1 mutant MPM treated with oral tazemetostat (800 mg BID). A two arm phase II trial will commence in the near future at the NCI to examine response rates in patients with wild type vs mutant BAP1 MPM receiving GSK126 as induction therapy prior to pleurectomy/decortication; a variety of translational endpoints will be assessed in this trial. Additionally, mithramycin, which depletes EZH2 as well as several other PRC-2 associated proteins (102), is being evaluated in patients with inoperable thoracic malignancies (including MPM) at the NCI (NCT01624090, NCT02859415).
It may be possible to further exploit BAP1 mutations for MPM therapy. BAP1 functions to stabilize BRCA-1 and promote poly(ADP-ribose)-dependent recruitment of polycomb deubiquitylase complex PR-DUB to DNA damage sites (126,127). This activity is dependent on deubiquitinase activity as well as phosphorylation of BAP1 (128). BAP1 mutations, which always appear to be manifested as loss of function, decrease BRCA-1 levels (129), and inhibit double strand DNA repair (126-128). Parotta et al (130) observed that a BAP1 isoform lacking part of the catalytic domain sensitized MPM cells to the PARP1 inhibitor, olaparid; and this sensitivity could be augmented by concomitant treatment with the dual PI3K-mTOR inhibitor, GDC0980, which downregulates BRCA-1. Such strategies might enhance responses to cisplatin/pemetrexed in patients with BAP1 mutant MPM, and should be evaluated in future clinical trials.
Given extensive preclinical studies demonstrating that DNA demethylating agents, HDAC inhibitors and KMT inhibitors mediate potentially significant immunomodulatory effects (111), there is considerable interest in utilizing chromatin remodeling agents in conjunction with either adoptive cell transfer or immune checkpoint inhibitors for cancer therapy. Previously we demonstrated that a cancer testis antigen upregulated in-vivo by decitabine could be targeted by cytolytic T cells to eradicate metastatic cancer in a syngeneic murine tumor model; these experiments established the preclinical rationale for combining gene induction regimens with adoptive immunotherapy for cancer (131). Recently, Corve et al. (132) evaluated the potential efficacy of combining a gene induction regimen with anti-CTLA 4 therapy in MPM. Consistent with results of a recent phase II double blind, placebo controlled trial demonstrating no efficacy of Tremelimumad in MPM patients (133), the murine anti-CTLA4 Mab 9H10 did not significantly inhibit growth of MPM xenografts. 5-azacytidine (5-AZA) induced a slight but insignificant reduction in growth of MPM xenografts. In contrast, combined 5-AZA/9H10 treatment mediated an 81% inhibition of MPM xenograft growth (P<0.05). This phenomenon coincided with up-regulation of the murine CTA, P1A, as well as increased class I HLA expression. Collectively, these data support evaluation of DNA demethylating agents in combination with immune checkpoint inhibitors in MPM. Despite evidence that 5-AZA augmented responses to anti-CTLA therapy in preclinical models, a better translational strategy might be to combine DNA demethylating agents with pembrolizumab given recent observations that this PD-L1 inhibitor mediated a 17% objective response rate in MPM patients (134), and high levels of PD-L1 expression in MPM-particularly sarcomatoid subtypes (3). Observations that combined decitabine/GSK126 or 5-AZA/entinostat treatment markedly augment efficacy of adoptively transferred CTL or anti-PD-L1 via up-regulation of Th1 signaling and inhibition of immunosuppressive myeloid derived suppressor cells within the tumor microenvironment in murine cancer models (113,135) support evaluation of such combinatorial regimens in clinical settings. These findings, together with recent observations that the immune microenvironment impacts outcome of patients with MPM (3,136) provide compelling rationale for combined epigenetic-immunotherapies for these neoplams.
Conclusions
Due to their recalcitrance to conventional treatment modalities, MPMs continue to challenge clinicians. Recent insights into epigenetic mechanisms which dysregulate gene expression in MPM, together with novel potent, and potentially efficacious regimens targeting DNMTs, EZH2 and PARP1, as well as immune checkpoints provide new opportunities to target the epigenome for the treatment and possible prevention of MPM.
Acknowledgements
None.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Bononi A, Napolitano A, Pass HI, et al. Latest developments in our understanding of the pathogenesis of mesothelioma and the design of targeted therapies. Expert Rev Respir Med 2015;9:633-54. [Crossref] [PubMed]
- Yoshikawa Y, Emi M, Hashimoto-Tamaoki T, et al. High-density array-CGH with targeted NGS unmask multiple noncontiguous minute deletions on chromosome 3p21 in mesothelioma. Proc Natl Acad Sci U S A 2016;113:13432-7. [Crossref] [PubMed]
- Bueno R, Stawiski EW, Goldstein LD, et al. Comprehensive genomic analysis of malignant pleural mesothelioma identifies recurrent mutations, gene fusions and splicing alterations. Nat Genet 2016;48:407-16. [Crossref] [PubMed]
- Zhang X, Tang N, Rishi AK, et al. Methylation profile landscape in mesothelioma: possible implications in early detection, disease progression, and therapeutic options. Methods Mol Biol 2015;1238:235-47. [Crossref] [PubMed]
- Vandermeers F, Neelature SS, Costa C, et al. The role of epigenetics in malignant pleural mesothelioma. Lung Cancer 2013;81:311-8. [Crossref] [PubMed]
- Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell 2012;150:12-27. [Crossref] [PubMed]
- Tessarz P, Kouzarides T. Histone core modifications regulating nucleosome structure and dynamics. Nat Rev Mol Cell Biol 2014;15:703-8. [Crossref] [PubMed]
- Li E, Zhang Y. DNA methylation in mammals. Cold Spring Harb Perspect Biol 2014;6:a019133. [Crossref] [PubMed]
- Baylin SB, Jones PA. A decade of exploring the cancer epigenome - biological and translational implications. Nat Rev Cancer 2011;11:726-34. [Crossref] [PubMed]
- Jin B, Robertson KD. DNA methyltransferases, DNA damage repair, and cancer. Adv Exp Med Biol 2013;754:3-29. [Crossref] [PubMed]
- Baylin SB, Jones PA. Epigenetic Determinants of Cancer. Cold Spring Harb Perspect Biol 2016;8:a019505. [Crossref] [PubMed]
- Yin Y, Morgunova E, Jolma A, et al. Impact of cytosine methylation on DNA binding specificities of human transcription factors. Science 2017;356:eaaj2239. [Crossref] [PubMed]
- Zhu H, Wang G, Qian J. Transcription factors as readers and effectors of DNA methylation. Nat Rev Genet 2016;17:551-65. [Crossref] [PubMed]
- Shimbo T, Wade PA. Proteins That Read DNA Methylation. Adv Exp Med Biol 2016;945:303-20. [Crossref] [PubMed]
- Yoon JH, Smith LE, Feng Z, et al. Methylated CpG dinucleotides are the preferential targets for G-to-T transversion mutations induced by benzo[a]pyrene diol epoxide in mammalian cells: similarities with the p53 mutation spectrum in smoking-associated lung cancers. Cancer Res 2001;61:7110-7. [PubMed]
- Bochtler M, Kolano A, Xu GL. DNA demethylation pathways: Additional players and regulators. Bioessays 2017;39:1-13. [Crossref] [PubMed]
- Yin X, Xu Y. Structure and Function of TET Enzymes. Adv Exp Med Biol 2016;945:275-302. [Crossref] [PubMed]
- Schomacher L, Niehrs C. DNA repair and erasure of 5-methylcytosine in vertebrates. Bioessays 2017.39. [PubMed]
- Schuermann D, Weber AR, Schar P. Active DNA demethylation by DNA repair: Facts and uncertainties. DNA Repair (Amst) 2016;44:92-102. [Crossref] [PubMed]
- Ha K, Lee GE, Palii SS, et al. Rapid and transient recruitment of DNMT1 to DNA double-strand breaks is mediated by its interaction with multiple components of the DNA damage response machinery. Hum Mol Genet 2011;20:126-40. [Crossref] [PubMed]
- Cai Y, Tsai HC, Yen RC, et al. Critical threshold levels of DNA methyltransferase 1 are required to maintain DNA methylation across the genome in human cancer cells. Genome Res 2017;27:533-44. [Crossref] [PubMed]
- An J, Rao A, Ko M. TET family dioxygenases and DNA demethylation in stem cells and cancers. Exp Mol Med 2017;49:e323. [Crossref] [PubMed]
- Zoghbi HY, Beaudet AL. Epigenetics and Human Disease. Cold Spring Harb Perspect Biol 2016;8:a019497. [Crossref] [PubMed]
- Van Tongelen A, Loriot A, De Smet C. Oncogenic roles of DNA hypomethylation through the activation of cancer-germline genes. Cancer Lett 2017;396:130-7. [Crossref] [PubMed]
- Audia JE, Campbell RM. Histone Modifications and Cancer. Cold Spring Harb Perspect Biol 2016;8:a019521. [Crossref] [PubMed]
- Zhao Y, Garcia BA. Comprehensive Catalog of Currently Documented Histone Modifications. Cold Spring Harb Perspect Biol 2015;7:a025064. [Crossref] [PubMed]
- Wapenaar H, Dekker FJ. Histone acetyltransferases: challenges in targeting bi-substrate enzymes. Clin Epigenetics 2016;8:59. [Crossref] [PubMed]
- Li Y, Seto E. HDACs and HDAC Inhibitors in Cancer Development and Therapy. Cold Spring Harb Perspect Med 2016.6. [PubMed]
- Yu X, Guo ZS, Marcu MG, et al. Modulation of p53, ErbB1, ErbB2, and Raf-1 expression in lung cancer cells by depsipeptide FR901228. J Natl Cancer Inst 2002;94:504-13. [Crossref] [PubMed]
- Hyun K, Jeon J, Park K, et al. Writing, erasing and reading histone lysine methylations. Exp Mol Med 2017;49:e324. [Crossref] [PubMed]
- Liu Q, Wang MW. Histone lysine methyltransferases as anti-cancer targets for drug discovery. Acta Pharmacol Sin 2016;37:1273-80. [Crossref] [PubMed]
- Yi X, Jiang XJ, Li XY, et al. Histone methyltransferases: novel targets for tumor and developmental defects. Am J Transl Res 2015;7:2159-75. [PubMed]
- Liu F, Killian JK, Yang M, et al. Epigenomic alterations and gene expression profiles in respiratory epithelia exposed to cigarette smoke condensate. Oncogene 2010;29:3650-64. [Crossref] [PubMed]
- Filipp FV. Crosstalk between epigenetics and metabolism-Yin and Yang of histone demethylases and methyltransferases in cancer. Brief Funct Genomics 2017. [Epub ahead of print]. [Crossref] [PubMed]
- Kumar R, Li DQ, Muller S, et al. Epigenomic regulation of oncogenesis by chromatin remodeling. Oncogene 2016;35:4423-36. [Crossref] [PubMed]
- Längst G, Manelyte L. Chromatin Remodelers: From Function to Dysfunction. Genes (Basel) 2015;6:299-324. [Crossref] [PubMed]
- Witkowski L, Foulkes WD. In Brief: Picturing the complex world of chromatin remodelling families. J Pathol 2015;237:403-6. [Crossref] [PubMed]
- Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem 2012;81:145-66. [Crossref] [PubMed]
- Rinn JL. lncRNAs: linking RNA to chromatin. Cold Spring Harb Perspect Biol 2014.6. [PubMed]
- Meseure D, Drak Alsibai K, Nicolas A, et al. Long Noncoding RNAs as New Architects in Cancer Epigenetics, Prognostic Biomarkers, and Potential Therapeutic Targets. Biomed Res Int 2015;2015:320214.
- Davidovich C, Cech TR. The recruitment of chromatin modifiers by long noncoding RNAs: lessons from PRC2. Rna 2015;21:2007-22. [Crossref] [PubMed]
- Bagwe AN, Kay PH, Spagnolo DV. Evidence that DNA methylation imbalance is not involved in the development of malignant mesothelioma. Anticancer Res 1997;17:3341-3. [PubMed]
- Christensen BC, Godleski JJ, Marsit CJ, et al. Asbestos exposure predicts cell cycle control gene promoter methylation in pleural mesothelioma. Carcinogenesis 2008;29:1555-9. [Crossref] [PubMed]
- Goto Y, Shinjo K, Kondo Y, et al. Epigenetic profiles distinguish malignant pleural mesothelioma from lung adenocarcinoma. Cancer Res 2009;69:9073-82. [Crossref] [PubMed]
- Foran E, Garrity-Park MM, Mureau C, et al. Upregulation of DNA methyltransferase-mediated gene silencing, anchorage-independent growth, and migration of colon cancer cells by interleukin-6. Mol Cancer Res 2010;8:471-81. [Crossref] [PubMed]
- Hodge DR, Xiao W, Clausen PA, et al. Interleukin-6 regulation of the human DNA methyltransferase (HDNMT) gene in human erythroleukemia cells. J Biol Chem 2001;276:39508-11. [Crossref] [PubMed]
- Yang H, Rivera Z, Jube S, et al. Programmed necrosis induced by asbestos in human mesothelial cells causes high-mobility group box 1 protein release and resultant inflammation. Proc Natl Acad Sci U S A 2010;107:12611-6. [Crossref] [PubMed]
- Thompson JK, MacPherson MB, Beuschel SL, et al. Asbestos-Induced Mesothelial to Fibroblastic Transition Is Modulated by the Inflammasome. Am J Pathol 2017;187:665-78. [Crossref] [PubMed]
- Hillegass JM, Miller JM, MacPherson MB, et al. Asbestos and erionite prime and activate the NLRP3 inflammasome that stimulates autocrine cytokine release in human mesothelial cells. Part Fibre Toxicol 2013;10:39. [Crossref] [PubMed]
- Sayan M, Mossman BT. The NLRP3 inflammasome in pathogenic particle and fibre-associated lung inflammation and diseases. Part Fibre Toxicol 2016;13:51. [Crossref] [PubMed]
- Kim MC, Kim NY, Seo YR, et al. An Integrated Analysis of the Genome-Wide Profiles of DNA Methylation and mRNA Expression Defining the Side Population of a Human Malignant Mesothelioma Cell Line. J Cancer 2016;7:1668-79. [Crossref] [PubMed]
- Kim MC, Kim NY, Seo YR, et al. A subset of microRNAs defining the side population of a human malignant mesothelioma cell line. Oncotarget 2017. [Epub ahead of print]. [Crossref] [PubMed]
- Schneider MR, Yarden Y. The EGFR-HER2 module: a stem cell approach to understanding a prime target and driver of solid tumors. Oncogene 2016;35:2949-60. [Crossref] [PubMed]
- Nami B, Wang Z. HER2 in Breast Cancer Stemness: A Negative Feedback Loop towards Trastuzumab Resistance. Cancers (Basel) 2017.9. [PubMed]
- Gaudet F, Hodgson JG, Eden A, et al. Induction of tumors in mice by genomic hypomethylation. Science 2003;300:489-92. [Crossref] [PubMed]
- Howard G, Eiges R, Gaudet F, et al. Activation and transposition of endogenous retroviral elements in hypomethylation induced tumors in mice. Oncogene 2008;27:404-8. [Crossref] [PubMed]
- Salmaninejad A, Zamani MR, Pourvahedi M, et al. Cancer/Testis Antigens: Expression, Regulation, Tumor Invasion, and Use in Immunotherapy of Cancers. Immunol Invest 2016;45:619-40. [Crossref] [PubMed]
- Hodzic D, Delacroix L, Willemsen P, et al. Characterization of the IGF system and analysis of the possible molecular mechanisms leading to IGF-II overexpression in a mesothelioma. Horm Metab Res 1997;29:549-55. [Crossref] [PubMed]
- De Smet C, Loriot A. DNA hypomethylation and activation of germline-specific genes in cancer. Adv Exp Med Biol 2013;754:149-66. [Crossref] [PubMed]
- Simpson AJ, Caballero OL, Jungbluth A, et al. Cancer/testis antigens, gametogenesis and cancer. Nat Rev Cancer 2005;5:615-25. [Crossref] [PubMed]
- Zendman AJ, Ruiter DJ, Van Muijen GN. Cancer/testis-associated genes: identification, expression profile, and putative function. J Cell Physiol 2003;194:272-88. [Crossref] [PubMed]
- Renaud S, Loukinov D, Alberti L, et al. BORIS/CTCFL-mediated transcriptional regulation of the hTERT telomerase gene in testicular and ovarian tumor cells. Nucleic Acids Res 2011;39:862-73. [Crossref] [PubMed]
- Dougherty CJ, Ichim TE, Liu L, et al. Selective apoptosis of breast cancer cells by siRNA targeting of BORIS. Biochem Biophys Res Commun 2008;370:109-12. [Crossref] [PubMed]
- Su S, Minges JT, Grossman G, et al. Proto-oncogene activity of melanoma antigen-A11 (MAGE-A11) regulates retinoblastoma-related p107 and E2F1 proteins. J Biol Chem 2013;288:24809-24. [Crossref] [PubMed]
- Peche LY, Ladelfa MF, Toledo MF, et al. Human MageB2 Protein Expression Enhances E2F Transcriptional Activity, Cell Proliferation, and Resistance to Ribotoxic Stress. J Biol Chem 2015;290:29652-62. [Crossref] [PubMed]
- Marcar L, Maclaine NJ, Hupp TR, et al. Mage-A cancer/testis antigens inhibit p53 function by blocking its interaction with chromatin. Cancer Res 2010;70:10362-70. [Crossref] [PubMed]
- Monte M, Simonatto M, Peche LY, et al. MAGE-A tumor antigens target p53 transactivation function through histone deacetylase recruitment and confer resistance to chemotherapeutic agents. Proc Natl Acad Sci U S A 2006;103:11160-5. [Crossref] [PubMed]
- Peche LY, Scolz M, Ladelfa MF, et al. MageA2 restrains cellular senescence by targeting the function of PMLIV/p53 axis at the PML-NBs. Cell Death Differ 2012;19:926-36. [Crossref] [PubMed]
- Podberezin M, Wen J, Chang CC. Cancer stem cells: a review of potential clinical applications. Arch Pathol Lab Med 2013;137:1111-6. [Crossref] [PubMed]
- Yamada R, Takahashi A, Torigoe T, et al. Preferential expression of cancer/testis genes in cancer stem-like cells: proposal of a novel sub-category, cancer/testis/stem gene. Tissue Antigens 2013;81:428-34. [Crossref] [PubMed]
- Yin B, Zeng Y, Liu G, et al. MAGE-A3 is highly expressed in a cancer stem cell-like side population of bladder cancer cells. Int J Clin Exp Pathol 2014;7:2934-41. [PubMed]
- Loriot A, Reister S, Parvizi GK, et al. DNA methylation-associated repression of cancer-germline genes in human embryonic and adult stem cells. Stem Cells 2009;27:822-4. [Crossref] [PubMed]
- Cartron PF, Blanquart C, Hervouet E, et al. HDAC1-mSin3a-NCOR1, Dnmt3b-HDAC1-Egr1 and Dnmt1-PCNA-UHRF1-G9a regulate the NY-ESO1 gene expression. Mol Oncol 2013;7:452-63. [Crossref] [PubMed]
- Hong JA, Kang Y, Abdullaev Z, et al. Reciprocal binding of CTCF and BORIS to the NY-ESO-1 promoter coincides with derepression of this cancer-testis gene in lung cancer cells. Cancer Res 2005;65:7763-74. [PubMed]
- Kang Y, Hong JA, Chen GA, et al. Dynamic transcriptional regulatory complexes including BORIS, CTCF and Sp1 modulate NY-ESO-1 expression in lung cancer cells. Oncogene 2007;26:4394-403. [Crossref] [PubMed]
- Cannuyer J, Van TA, Loriot A, et al. A gene expression signature identifying transient DNMT1 depletion as a causal factor of cancer-germline gene activation in melanoma. Clin Epigenetics 2015;7:114. [Crossref] [PubMed]
- Sen GL, Reuter JA, Webster DE, et al. DNMT1 maintains progenitor function in self-renewing somatic tissue. Nature 2010;463:563-7. [Crossref] [PubMed]
- Sigalotti L, Coral S, Altomonte M, et al. Cancer testis antigens expression in mesothelioma: role of DNA methylation and bioimmunotherapeutic implications. Br J Cancer 2002;86:979-82. [Crossref] [PubMed]
- Schrump DS. Targeting epigenetic mediators of gene expression in thoracic malignancies. Biochim Biophys Acta 2012;1819:836-45.
- Weiser TS, Guo ZS, Ohnmacht GA, et al. Sequential 5-Aza-2 deoxycytidine-depsipeptide FR901228 treatment induces apoptosis preferentially in cancer cells and facilitates their recognition by cytolytic T lymphocytes specific for NY-ESO-1. J Immunother 2001;24:151-61. [Crossref] [PubMed]
- Oliviero G, Brien GL, Waston A, et al. Dynamic Protein Interactions of the Polycomb Repressive Complex 2 during Differentiation of Pluripotent Cells. Mol Cell Proteomics 2016;15:3450-60. [Crossref] [PubMed]
- Pasini D, Di Croce L. Emerging roles for Polycomb proteins in cancer. Curr Opin Genet Dev 2016;36:50-8. [Crossref] [PubMed]
- Poynter ST, Kadoch C. Polycomb and trithorax opposition in development and disease. Wiley Interdiscip Rev Dev Biol 2016;5:659-88. [Crossref] [PubMed]
- Pasini D, Cloos PA, Walfridsson J, et al. JARID2 regulates binding of the Polycomb repressive complex 2 to target genes in ES cells. Nature 2010;464:306-10. [Crossref] [PubMed]
- Katoh M. Functional proteomics of the epigenetic regulators ASXL1, ASXL2 and ASXL3: a convergence of proteomics and epigenetics for translational medicine. Expert Rev Proteomics 2015;12:317-28. [Crossref] [PubMed]
- Hussain M, Rao M, Humphries AE, et al. Tobacco smoke induces polycomb-mediated repression of Dickkopf-1 in lung cancer cells. Cancer Res 2009;69:3570-8. [Crossref] [PubMed]
- Kondo Y, Shen L, Cheng AS, et al. Gene silencing in cancer by histone H3 lysine 27 trimethylation independent of promoter DNA methylation. Nat Genet 2008;40:741-50. [Crossref] [PubMed]
- Mantsoki A, Devailly G, Joshi A. CpG island erosion, polycomb occupancy and sequence motif enrichment at bivalent promoters in mammalian embryonic stem cells. Sci Rep 2015;5:16791. [Crossref] [PubMed]
- Harikumar A, Meshorer E. Chromatin remodeling and bivalent histone modifications in embryonic stem cells. EMBO Rep 2015;16:1609-19. [Crossref] [PubMed]
- Kemp CD, Rao M, Xi S, et al. Polycomb repressor complex-2 is a novel target for mesothelioma therapy. Clin Cancer Res 2012;18:77-90. [Crossref] [PubMed]
- Tan J, Yang X, Zhuang L, et al. Pharmacologic disruption of Polycomb-repressive complex 2-mediated gene repression selectively induces apoptosis in cancer cells. Genes Dev 2007;21:1050-63. [Crossref] [PubMed]
- LaFave LM, Beguelin W, Koche R, et al. Loss of BAP1 function leads to EZH2-dependent transformation. Nat Med 2015;21:1344-9. [Crossref] [PubMed]
- Bott M, Brevet M, Taylor BS, et al. The nuclear deubiquitinase BAP1 is commonly inactivated by somatic mutations and 3p21.1 losses in malignant pleural mesothelioma. Nat Genet 2011;43:668-72. [Crossref] [PubMed]
- Testa JR, Cheung M, Pei J, et al. Germline BAP1 mutations predispose to malignant mesothelioma. Nat Genet 2011;43:1022-5. [Crossref] [PubMed]
- Nasu M, Emi M, Pastorino S, et al. High Incidence of Somatic BAP1 alterations in sporadic malignant mesothelioma. J Thorac Oncol 2015;10:565-76. [Crossref] [PubMed]
- Kalakonda N, Fischle W, Boccuni P, et al. Histone H4 lysine 20 monomethylation promotes transcriptional repression by L3MBTL1. Oncogene 2008;27:4293-304. [Crossref] [PubMed]
- Trojer P, Cao AR, Gao Z, et al. L3MBTL2 protein acts in concert with PcG protein-mediated monoubiquitination of H2A to establish a repressive chromatin structure. Mol Cell 2011;42:438-50. [Crossref] [PubMed]
- Zauderer MG, Bott M, McMillan R, et al. Clinical characteristics of patients with malignant pleural mesothelioma harboring somatic BAP1 mutations. J Thorac Oncol 2013;8:1430-3. [Crossref] [PubMed]
- Baumann F, Flores E, Napolitano A, et al. Mesothelioma patients with germline BAP1 mutations have 7-fold improved long-term survival. Carcinogenesis 2015;36:76-81. [Crossref] [PubMed]
- Luchini C, Veronese N, Yachida S, et al. Different prognostic roles of tumor suppressor gene BAP1 in cancer: A systematic review with meta-analysis. Genes Chromosomes Cancer 2016;55:741-9. [Crossref] [PubMed]
- Li L, Davie JR. The role of Sp1 and Sp3 in normal and cancer cell biology. Ann Anat 2010;192:275-83. [Crossref] [PubMed]
- Rao M, Atay SM, Shukla V, et al. Mithramycin Depletes Specificity Protein 1 and Activates p53 to Mediate Senescence and Apoptosis of Malignant Pleural Mesothelioma Cells. Clin Cancer Res 2016;22:1197-210. [Crossref] [PubMed]
- Kadoch C, Copeland RA, Keilhack H. PRC2 and SWI/SNF Chromatin Remodeling Complexes in Health and Disease. Biochemistry 2016;55:1600-14. [Crossref] [PubMed]
- Pulice JL, Kadoch C. Composition and Function of Mammalian SWI/SNF Chromatin Remodeling Complexes in Human Disease. Cold Spring Harb Symp Quant Biol 2016;81:53-60. [Crossref] [PubMed]
- Yoshikawa Y, Sato A, Tsujimura T, et al. Biallelic germline and somatic mutations in malignant mesothelioma: Multiple mutations in transcription regulators including mSWI/SNF genes. Int J Cancer. 2015;136:560-71. [PubMed]
- Shukla S, Meeran SM. Epigenetics of cancer stem cells: Pathways and therapeutics. Biochim Biophys Acta 2014;1840:3494-502.
- Easwaran H, Johnstone SE, Van NL, et al. A DNA hypermethylation module for the stem/progenitor cell signature of cancer. Genome Res 2012;22:837-49. [Crossref] [PubMed]
- Jones PA. At the tipping point for epigenetic therapies in cancer. J Clin Invest 2014;124:14-6. [Crossref] [PubMed]
- Chiappinelli KB, Strissel PL, Desrichard A, et al. Inhibiting DNA Methylation Causes an Interferon Response in Cancer via dsRNA Including Endogenous Retroviruses. Cell 2015;162:974-86. [Crossref] [PubMed]
- Roulois D, Loo Yau H, Singhania R, et al. DNA-Demethylating Agents Target Colorectal Cancer Cells by Inducing Viral Mimicry by Endogenous Transcripts. Cell 2015;162:961-73. [Crossref] [PubMed]
- Wolff F, Leisch M, Greil R, et al. The double-edged sword of (re)expression of genes by hypomethylating agents: from viral mimicry to exploitation as priming agents for targeted immune checkpoint modulation. Cell Commun Signal 2017;15:13. [Crossref] [PubMed]
- Li H, Chiappinelli KB, Guzzetta AA, et al. Immune regulation by low doses of the DNA methyltransferase inhibitor 5-azacitidine in common human epithelial cancers. Oncotarget 2014;5:587-98. [Crossref] [PubMed]
- Kim K, Skora AD, Li Z, et al. Eradication of metastatic mouse cancers resistant to immune checkpoint blockade by suppression of myeloid-derived cells. Proc Natl Acad Sci U S A 2014;111:11774-9. [Crossref] [PubMed]
- Pathania R, Ramachandran S, Elangovan S, et al. DNMT1 is essential for mammary and cancer stem cell maintenance and tumorigenesis. Nat Commun 2015;6:6910. [Crossref] [PubMed]
- Yang J, Corsello TR, Ma Y. Stem cell gene SALL4 suppresses transcription through recruitment of DNA methyltransferases. J Biol Chem 2012;287:1996-2005. [Crossref] [PubMed]
- Yogelzang NJ, Herndon JE 2nd, Cirrincione C, et al. Dihydro-5-azacytidine in malignant mesothelioma. A phase II trial demonstrating activity accompanied by cardiac toxicity. Cancer and Leukemia Group B. Cancer 1997;79:2237-42. [Crossref] [PubMed]
- Schrump DS, Fischette MR, Nguyen DM, et al. Phase I study of decitabine-mediated gene expression in patients with cancers involving the lungs, esophagus, or pleura. Clin Cancer Res 2006;12:5777-85. [Crossref] [PubMed]
- Chabner BA, Johns DG, Coleman CN, et al. Purification and properties of cytidine deaminase from normal and leukemic granulocytes. J Clin Invest 1974;53:922-31. [Crossref] [PubMed]
- Lavelle D, Vaitkus K, Ling Y, et al. Effects of tetrahydrouridine on pharmacokinetics and pharmacodynamics of oral decitabine. Blood 2012;119:1240-7. [Crossref] [PubMed]
- Wentworth DF, Wolfenden R. On the interaction of 3,4,5,6-tetrahydrouridine with human liver cytidine deaminase. Biochemistry 1975;14:5099-105. [Crossref] [PubMed]
- Newman EM, Morgan RJ, Kummar S, et al. A phase I, pharmacokinetic, and pharmacodynamic evaluation of the DNA methyltransferase inhibitor 5-fluoro-2'-deoxycytidine, administered with tetrahydrouridine. Cancer Chemother Pharmacol 2015;75:537-46. [Crossref] [PubMed]
- Paik PK, Krug LM. Histone deacetylase inhibitors in malignant pleural mesothelioma: preclinical rationale and clinical trials. J Thorac Oncol 2010;5:275-9. [Crossref] [PubMed]
- Krug LM, Kindler HL, Calvert H, et al. Vorinostat in patients with advanced malignant pleural mesothelioma who have progressed on previous chemotherapy (VANTAGE-014): a phase 3, double-blind, randomised, placebo-controlled trial. Lancet Oncol 2015;16:447-56. [Crossref] [PubMed]
- Nguyen DM, Schrump WD, Chen GA, et al. Abrogation of p21 expression by flavopiridol enhances depsipeptide-mediated apoptosis in malignant pleural mesothelioma cells. Clin Cancer Res 2004;10:1813-25. [Crossref] [PubMed]
- Neuzil J, Swettenham E, Gellert N. Sensitization of mesothelioma to TRAIL apoptosis by inhibition of histone deacetylase: role of Bcl-xL down-regulation. Biochem Biophys Res Commun 2004;314:186-91. [Crossref] [PubMed]
- Ismail IH, Davidson R, Gagne JP, et al. Germline Mutations in BAP1 Impair Its Function in DNA Double-Strand Break Repair. Cancer Res 2014;74:4282-94. [Crossref] [PubMed]
- Fukuda T, Tsuruga T, Kuroda T, et al. Functional Link between BRCA1 and BAP1 through Histone H2A, Heterochromatin and DNA Damage Response. Curr Cancer Drug Targets 2016;16:101-9. [Crossref] [PubMed]
- Yu H, Pak H, Hammond-Martel I, et al. Tumor suppressor and deubiquitinase BAP1 promotes DNA double-strand break repair. Proc Natl Acad Sci U S A 2014;111:285-90. [Crossref] [PubMed]
- Hakiri S, Osada H, Ishiguro F, et al. Functional differences between wild-type and mutant-type BRCA1-associated protein 1 tumor suppressor against malignant mesothelioma cells. Cancer Sci 2015;106:990-9. [Crossref] [PubMed]
- Parrotta R, Okonska A, Ronner M, et al. A Novel BRCA1-Associated Protein-1 Isoform Affects Response of Mesothelioma Cells to Drugs Impairing BRCA1-Mediated DNA Repair. J Thorac Oncol 2017. [Epub ahead of print]. [Crossref] [PubMed]
- Guo ZS, Hong JA, Irvine KR, et al. De novo induction of a cancer/testis antigen by 5-aza-2'-deoxycytidine augments adoptive immunotherapy in a murine tumor model. Cancer Res 2006;66:1105-13. [Crossref] [PubMed]
- Covre A, Coral S, Nicolay H, et al. Antitumor activity of epigenetic immunomodulation combined with CTLA-4 blockade in syngeneic mouse models. Oncoimmunology 2015;4:e1019978. [Crossref] [PubMed]
- Kindler HL, Shulman KL. Metastatic colorectal cancer. Curr Treat Options Oncol 2001;2:459-71. [Crossref] [PubMed]
- Alley EW, Lopez J, Santoro A, et al. Clinical safety and activity of pembrolizumab in patients with malignant pleural mesothelioma (KEYNOTE-028): preliminary results from a non-randomised, open-label, phase 1b trial. Lancet Oncol 2017;18:623-30. [Crossref] [PubMed]
- Peng D, Kryczek I, Nagarsheth N, et al. Epigenetic silencing of TH1-type chemokines shapes tumour immunity and immunotherapy. Nature 2015;527:249-53. [Crossref] [PubMed]
- Thapa B, Salcedo A, Lin X, et al. The Immune Microenvironment, Genome-wide Copy Number Aberrations, and Survival in Mesothelioma. J Thorac Oncol 2017;12:850-9. [Crossref] [PubMed]


