Molecular diagnostics of lung cancer in the clinic
Introduction
Lung cancer is the number one cause of cancer deaths globally (1). Approximately 225,000 new cases of lung cancer are diagnosed annually in the United States (2). In current practice, the most common subtype—nearly half of all lung cancers—is adenocarcinoma (3). Adenocarcinoma is more common in smokers, however it is also the most common lung cancer subtype diagnosed in nonsmokers (4). Adenocarcinomas typically arise in the lung periphery and may be asymptomatic in their early stages. As a result, the majority of patients come to clinical attention with locally advanced or distantly metastatic disease; in the latter situation, fewer than 5% of patients live to 5 years despite therapy (1).
Key biomarkers in the clinic
The genetics of adenocarcinoma are diverse, but heavily influenced by the patient’s smoking history (5). The earliest recognized mutations in non-small cell lung cancers (NSCLC) were identified in KRAS and TP53. However the first clinically impactful genomic discovery was made in 2004, when mutations in the kinase domain of the epidermal growth factor receptor (EGFR) were described specifically in the tumors of lung cancer patients who responded to tyrosine kinase inhibitor (TKI) therapy directed at EGFR (6,7). These EGFR kinase domain mutations, >90% of which occur in exon 21 (L858R) and exon 19 (small insertions-deletions affecting the ELREA motif), lead to constitutive activation of downstream pro-growth, oncogenic signaling pathways. Fortuitously, these mutations also sensitize the tumor cells to EGFR TKIs and predict response to a broad spectrum of EGFR TKIs, such as first generation inhibitors erlotinib and gefitinib (8). EGFR mutations are identified almost exclusively in lung adenocarcinomas, occur more commonly in light or never smokers and are enriched in women and individuals of Asian ethnicity (9). EGFR is the second most commonly mutated driver oncogene in lung adenocarcinoma after KRAS in the United States—about 15% in Caucasians and African Americans—and is the most commonly mutated oncogene in lung adenocarcinoma in Asian populations (~60%) (10,11).
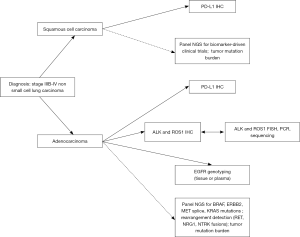
Rearrangements involving ALK and ROS1 were first described in lung adenocarcinoma in 2007 (12,13). Crizotinib, a commercially available inhibitor originally designed to target Met, proved effective against lung cancers harboring either ALK or ROS1 alterations (14,15) and has been approved for treatment of lung cancers with proven rearrangements. Both of these alterations are rare (<5% of lung cancers) but are enriched among light to never smokers and are seen almost exclusively in adenocarcinomas (16,17). Despite these clinicopathologic correlations that have been seen in EGFR, ALK and ROS1-altered lung tumors, it is clear that clinical features are neither highly sensitive nor specific for selecting patients for targeted inhibitors (18). Therefore, all patients with advanced lung adenocarcinoma should undergo testing for EGFR mutations and ALK and ROS1 rearrangements, irrespective of smoking status. In general, this testing is not indicated in patients with a diagnosis of squamous cell carcinoma or small cell carcinoma, however there are rare reports of, for example, de novo EGFR-mutated small cell carcinoma or ALK-rearranged squamous cell carcinoma in never smokers (19,20). Therefore, molecular testing is advised in patients with a histologic diagnosis that is out of keeping with their smoking history.
Relapse following targeted therapy is almost inevitable, and tends to occur after about a year of therapy on EGFR TKIs and after a median of 8 and 19 months, respectively, following first-line targeted therapy in the setting of ALK and ROS1 rearrangements (15,21). The mechanisms of resistance are relatively well defined. For EGFR, 50–60% of patients acquire the EGFR T790M mutation at the time of relapse (22). T790M reduces the efficacy of first generation EGFR inhibitors, but third generation inhibitors can overcome this resistance mutation, and one, osimertinib, has been FDA approved specifically for patients with a proven T790M mutation in the relapse setting (23). Other less common mechanisms of resistance include MET amplification, PIK3CA pathway activation, and small cell transformation (22). In ALK-rearranged patients, crizotinib resistance most commonly takes the form of a wide variety of secondary mutations occurring in the ALK kinase domain. Second and third generation ALK inhibitors can variably overcome these secondary mutations. While some authors have advocated for routine biopsy at relapse to define the mechanism of ALK inhibitor resistance (24), this practice is not widely employed, and alternative inhibitors are typically used empirically. Mechanisms of crizotinib resistance in the setting of ROS1 rearrangement are less-well defined, however mutations in ROS1 at codons 2032 and 2033 have been reported in individual cases (25,26).
Most recently, immune checkpoint blockade, or immunotherapy, has proven effective in a variety of tumor types, including lung cancers. Immunotherapeutics target some component of a regulatory network that keeps T cell response in check in inflammatory states; tumors can effectively hijack this network by, for example, upregulating surface PD-L1 expression to evade T cell-mediated anti-tumor responses. Approved immunotherapies in the lung include anti-programmed death-1 (PD-1) and anti programmed-death ligand-1 (PD-L1) antibodies (27). Biomarker analyses have shown that as a class the PD-1 and PD-L1 inhibitors appear to have greater efficacy in patients whose tumors express PD-L1, with a generally positive correlation between extent of expression and likelihood of response to treatment (28). Several drugs have been approved for NSCLC (both adenocarcinoma and squamous cell carcinoma) irrespective of the PD-L1 status (27), however the anti-PD-1 antibody, pembrolizumab, was approved for first line use only in patients with 50% or greater tumor cell expression of PD-L1 based on results of randomized controlled trials (29).
As a result, in the United States, there are four possible therapeutic options in the first line setting for patients with advanced (stage IIIb or IV) adenocarcinoma (Figure 1), and biomarker testing is required before deciding on the course of therapy. Retrospective analyses of PD-1 inhibitor trials have demonstrated that EGFR mutation and ALK rearrangement predict inferior response to immunotherapy (30); pembrolizumab is, therefore, approved only for patients with EGFR or ALK negative tumors. Data on immunotherapy efficacy in ROS1-rearranged tumors is limited at this time.
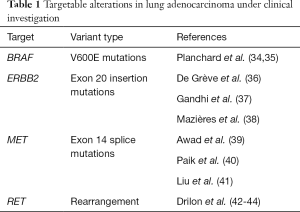
A number of other important oncogenic alterations are recognized in lung adenocarcinoma. These include uncommon but potentially targetable alterations in BRAF, ERBB2, MET and RET as well as common but difficult to target hotspot mutations in KRAS. In the vast majority of cancers, these alterations occur in a mutually exclusive fashion and represent independent oncogenic driver events (Figure 2) (31-33). A number of early clinical trials and case series support a role for routine testing of these targets (Table 1). In addition, there is increasing recognition of the prognostic importance of tumor suppressor genes, including adverse impact of TP53 mutations in patients with EGFR-mutated adenocarcinomas as well as dual RB1 and TP53 mutations as predictors of small cell transformation following EGFR TKI therapy (45,46). Other alterations, such as loss of SMARCA4, have been more broadly implicated as negative prognostic factors (47,48), whereas still others, such as loss function of STK11/LKB1, may predict specific therapeutic vulnerabilities among KRAS mutated tumors (49,50). Tumor mutational burden as defined by whole exome sequencing may predict response to immunotherapies (51), and some early evidence suggests that even smaller targeted panels (~300 genes) can generate informative mutation burden data (52).

Full table
The large number of clinically relevant genomic biomarkers in lung cancers presents a practical challenge to laboratories: sequential testing of up to eight different genes and ten or more targets within these genes is costly, time consuming, and demands significant amounts of tumor nucleic acid. In many cases the amount of available tumor tissue is less than needed to complete multiple molecular assays, as well as FISH and IHC (32). The current demands relating to biomarker testing demand a carefully coordinated effort between the oncologist, proceduralist, pathologist, and molecular laboratory to ensure proper specimen handling. For one, the indication for the biopsy should be clearly communicated down the line, so that the tissue is handled optimally for molecular testing, including minimized ischemic time, use of 10% buffered formalin as a standard fixative, and avoidance of fixatives that degrade nucleic acids (hydrochloric acid-based decalcification solution) or inhibit PCR (heavy metals such as B+). Secondly, the receiving pathologist should be informed if and when a diagnosis is already established in order to minimize diagnostic immunohistochemistry (18). In general, judicious use of immunohistochemistry is advised, with the core panel of diagnostic markers restricted to TTF-1 and p40, followed by other stains only if these are uninformative and the clinical picture is uncertain.
Next generation sequencing (NGS)
Advances in sequencing technology have yielded relatively cost-effective clinical testing platforms that leverage massively parallel, or NGS technologies; these platforms allow for multiplexing of gene targets over several orders of magnitude and can generate reliable results with anywhere from a few to several hundred nanograms of DNA (53). The choice of NGS platform will be informed by cost, anticipated test volume, breadth of genomic targets desired, and sensitivity desired. This latter variable dictates the depth of sequencing required. In general, the costs go up with number of targets (i.e., costs of whole genome > whole exome > targeted exome). Reagents, including chips for library preparation and flow cells for sequencing are expensive; these costs can be reduced by increasing the numbers of cases run at once (batching)—a step that is facilitated by tagging individual specimens with molecular barcodes to permit sample deconvolution during bioinformatic processing (53). Choice of platform also dictates the types of alterations that can be detected. In general, amplicon sequencing is preferred for targeted panels that are optimized for read depth and assay sensitivity, whereas hybrid capture sequencing is the approach of choice when breadth is desired (whole genome, whole exome, large targeted panels).
Both amplicon and hybrid capture assays can detect relative copy number alterations (i.e., amplification or gene deletion events), however hybrid capture sequencing is generally employed for this purpose, especially when more global copy change data is desired. In addition, the hybrid capture approach should be employed when translocation detection is desired, as DNA-based rearrangement detection requires that the assay capture large regions of intronic sequence, where breakpoints most commonly occur. Hybrid capture sequencing is sensitive and specific for translocation detection when the breakpoints and partner genes are well-defined for a given target (54). The sensitivity of this approach, however, is often limited for specimens with low tumor content relative to normal contaminating stromal cells. In addition, rearrangement detection is a thorny problem for bioinformatics, as short sequencing reads that capture two segments of the genome at once and support the presence of a rearrangement event may be discarded by some algorithms as “poorly mapped” (55). Algorithms must draw a distinction between poorly mapped reads that represent rearrangement, versus those that represent low quality sequencing, and their ability to do this defines their sensitivity and specificity and ultimately the reliability of the sequencing-based approach to rearrangement detection (56). Copy number calls, which are calculated based on a ratio of the read depth at a given locus in the sample relative to a known diploid normal sample, are also vulnerable to false negative and positive results. The former occurs in the context of low tumor content, the latter may occur when the DNA is heavily degraded. Given these limitations, parallel methods such as FISH should be available to confirm or refute unexpected or low quality findings, or for use in specimens with tumor content that is too low to generate a reliable translocation or copy number result by sequencing. IHC for protein overexpression, when available, may also help to confirm the significance of novel rearrangement breakpoints or partners. Labs may also consider use of RNA-based sequencing assays for translocation detection; significant technical advances, including the introduction of anchored multiplex PCR, now allow for fusion sequence detection from the short fragments of RNA present within formalin fixed tissues (57). Practically speaking, labs may then consider introducing a single large comprehensive hybrid-capture based panel, multiple smaller amplicon-sequencing based panels, or one smaller DNA based panel for mutation detection and another RNA-based panel for fusion detection, all supported by selected standalone assays for selected critical biomarkers (e.g., ALK, ROS1 FISH and/or IHC).
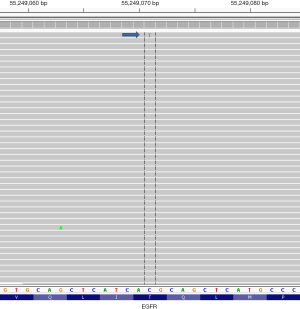
In contrast to traditional (Sanger) sequencing methods, where the readout appears as an averaging of the nucleotide content at any given position, in NGS each individual DNA molecule is separated in space and sequenced in a parallel fashion; bioinformatics tools then assemble the individual sequences, typically in relation to a reference genome (55). Using any of a variety of publically available genome viewing tools (Integrated Genome Viewer; Genome Viewer, both from the Broad Institute, Cambridge, MA, USA) the read “pile-ups” can be directly visualized and rare events easily observed. The ability to visualize the data in such a granular fashion can be hazardous, however, as it presents the risk of over-interpreting very low level mutational events as clinically significant when they may in fact represent technical artifact (Figure 3) (55). As a result, bioinformatics pipelines should be employed to generate automated calls; these are validated to optimize the assay sensitivity and specificity within the confines of the particular sequencing platform. Laboratorians should approach manual review of NGS data with caution and avoid enthusiastic endorsement of low level reads that fall below the limit of assay detection determined in the course of validation.
Liquid biopsy
So-called liquid biopsy refers to detection of tumor components in bodily fluids. These components may be viable cells (circulating tumor cells) or tumor DNA [circulating tumor DNA (ctDNA)]. Fluids that may contain these components include blood (plasma), urine, saliva, cerebrospinal fluid and liquid cytology specimens (58,59). This review will focus on the practice of ctDNA analysis within the cell free DNA (cfDNA) population in the plasma, as this is the area of most intense study and clinical assay development in lung cancer patients. Plasma testing is hugely attractive from a clinical perspective because tissue biopsies are often insufficient for molecular testing, thus necessitating repeat biopsy (32,54). Use of plasma testing may (I) allow a patient to forego additional invasive biopsy procedures; (II) reduce turnaround time by eliminating the need to schedule a procedure and to process the pathology specimen; (III) reduce sequencing artifact introduced by formalin fixation of tissue specimens; and (IV) be less biased by local tumor heterogeneity.
All individuals have some level of cfDNA detectable in the plasma—this is a product of normal cell apoptosis, with release of ~160 base pair, nucleosome-bound fragments of DNA into the circulation (58). Tumor cells also release their contents into the circulation, and the amount of detectable ctDNA is proportional to the overall burden of disease. Studies using droplet digital PCR (ddPCR) have shown that known tumor driver mutations are detectable in about 60% of lung cancer patients with a single metastatic site and 100% of those with four or more metastatic sites (60). Patients with early stage disease commonly do not have detectable levels of ctDNA using most currently available technologies (61,62). In general, ctDNA release from lung cancer is detectable at levels of 0.1% to 5% of total cfDNA (63). Therefore, highly sensitive techniques are needed in most patients to detect tumor specific alterations within the plasma.
Published approaches to ctDNA analysis in lung cancer have largely focused on EGFR mutations, including detection of activating hotspot mutations and the EGFR TKI resistance mutation T790M. Methods in routine use include real time PCR, ddPCR, and NGS. Indeed, the FDA has approved the cobas EGFR mutation test v2 (Roche Diagnostics, Indianapolis, IN, USA) using plasma as a substrate as a companion diagnostic for treatment with erlotinib in the first line setting and with osimertinib in the relapse setting. In acknowledgement of the limited sensitivity of plasma testing, the FDA approval advised that patients who are negative for these mutations in the plasma should undergo routine biopsy and repeat testing on the tumor tissue. Detection of EGFR mutations in the plasma predicts response to EGFR TKIs to the same degree as detecting mutations in tissue. Failure to clear the EGFR mutation in the blood after 8 weeks of combined platinum-based therapy plus erlotinib treatment is associated with an adverse prognosis (64). In the relapse setting, changes in the levels of T790M mutation in the plasma can be detected upon treatment with third generation inhibitors and in most cases these changes mirror the clinical status according to traditional radiographic staging (60,65). Scenarios in which the plasma levels of T790M decline but the patient experiences radiographic progression have been reported (65). This may reflect heterogeneity of resistance mechanisms, such as combined T790M mutation acquisition and small cell transformation.
Given the lower sensitivity of plasma-based testing relative to tissue, plasma genotyping assays should be designed to maximize the positive predictive value of the assay. This approach will minimize the risk of false positive results that might reduce confidence in a result and confound a clinician’s ability to decide on therapy (66). When a plasma assay is negative, however, follow-up plasma testing or tissue biopsy should be pursued to obtain tumor genotyping information.
Single gene assays can be powerful when employed from plasma specimens, as directed clinical questions in a population with a high pre-test probability (i.e., patients with relapsed EGFR-mutated lung cancer) can be answered rapidly and with confidence following a minimally-invasive blood draw. However, the use of single-gene assays limits the number of indications for which the technology can be used; most available assays are not amenable to large-scale multiplexing that might be necessary to provide more comprehensive information for a broader range of patients. Efforts to develop NGS-based approaches from plasma have led to a variety of promising academic and commercial assays. In principle, these are assays designed to maximize depth, rather than breadth of sequencing, so as to optimize assay sensitivity and clinical actionability. Focused bait design, modifications to the library preparation chemistry, sequencing to thousands-fold depth of coverage, and molecular barcoding to detect and suppress PCR errors have all been proposed as enhancements to NGS design for use in plasma specimens (62,67). Translocations and copy number alterations are also detectable in the plasma, given the appropriate design (68).
Conclusions
In routine practice, all patients with a diagnosis of advanced stage lung adenocarcinoma should undergo (I) EGFR mutational analysis that assesses, at a minimum, the L858 codon and the exon 19 deletion hotspot; (II) ALK rearrangement testing by immunohistochemistry, fluorescence in situ hybridization (FISH), RT-PCR or sequencing; (III) ROS1 rearrangement testing by FISH or molecular methods, with an option for prescreening by IHC; and (IV) IHC for PD-L1. When possible, use of a NGS assay that captures EGFR, ALK, and ROS1 as well as other less common but targetable (ERBB2, BRAF, MET, RET) and common but difficult to target (KRAS) oncogenes should be encouraged. Broader panels can detect a range of alterations in tumor suppressor genes such as TP53, STK11 and SMARCA4 that may have prognostic implications. Tumor mutation burden, whether derived from whole exome or large panel sequencing data (52), may predict response to immunotherapy (51). Use of plasma ctDNA in lieu of tumor tissue for molecular analysis allows a subset of patients to forego repeat biopsy due to insufficient tissue at diagnosis or at relapse. Plasma genotyping may also provide additional prognostic information as well as a mechanism to monitor solid tumor patients for response to therapy.
Acknowledgements
The authors would like to acknowledge the American Association for Cancer Research and it financial and material support in the development of the AACR Project GENIE registry, as well as members of the consortium for their commitment to data sharing. Interpretations are the responsibility of the author.
Footnote
Conflicts of Interest: The author has no conflicts of interest to declare.
References
- Torre LA, Siegel RL, Ward EM, et al. Global Cancer Incidence and Mortality Rates and Trends--An Update. Cancer Epidemiol Biomarkers Prev 2016;25:16-27. [Crossref] [PubMed]
- Cancer Stat Facts: Lung and Bronchus Cancer. Available online: https://seer.cancer.gov/statfacts/html/lungb.html
- Noone AM, Cronin KA, Altekruse SF, et al. Cancer incidence and survival trends by subtype using data from the Surveillance Epidemiology and End Results Program, 1992-2013. Cancer Epidemiol Biomarkers Prev 2017;26:632-41. [Crossref] [PubMed]
- Sun S, Schiller JH, Gazdar AF. Lung cancer in never smokers--a different disease. Nat Rev Cancer 2007;7:778-90. [Crossref] [PubMed]
- Imielinski M, Berger AH, Hammerman PS, et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell 2012;150:1107-20. [Crossref] [PubMed]
- Paez JG, Janne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 2004;304:1497-500. [Crossref] [PubMed]
- Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med 2004;350:2129-39. [Crossref] [PubMed]
- Ji H, Li D, Chen L, et al. The impact of human EGFR kinase domain mutations on lung tumorigenesis and in vivo sensitivity to EGFR-targeted therapies. Cancer Cell 2006;9:485-95. [Crossref] [PubMed]
- Sharma SV, Bell DW, Settleman J, et al. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer 2007;7:169-81. [Crossref] [PubMed]
- Campbell JD, Alexandrov A, Kim J, et al. Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. Nat Genet 2016;48:607-16. [Crossref] [PubMed]
- Li S, Choi YL, Gong Z, et al. Comprehensive Characterization of Oncogenic Drivers in Asian Lung Adenocarcinoma. J Thorac Oncol 2016;11:2129-40. [Crossref] [PubMed]
- Soda M, Choi YL, Enomoto M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature 2007;448:561-6. [Crossref] [PubMed]
- Rikova K, Guo A, Zeng Q, et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell 2007;131:1190-203. [Crossref] [PubMed]
- Kwak EL, Bang YJ, Camidge DR, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med 2010;363:1693-703. [Crossref] [PubMed]
- Shaw AT, Ou SH, Bang YJ, et al. Crizotinib in ROS1-Rearranged Non-Small-Cell Lung Cancer. N Engl J Med 2014;371:1963-71. [Crossref] [PubMed]
- Bergethon K, Shaw AT, Ignatius Ou SH, et al. ROS1 Rearrangements Define a Unique Molecular Class of Lung Cancers. J Clin Oncol 2012;30:863-70. [Crossref] [PubMed]
- Shaw AT, Yeap BY, Mino-Kenudson M, et al. Clinical features and outcome of patients with non-small-cell lung cancer who harbor EML4-ALK. J Clin Oncol 2009;27:4247-53. [Crossref] [PubMed]
- Lindeman NI, Cagle PT, Beasley MB, et al. Molecular testing guideline for selection of lung cancer patients for EGFR and ALK tyrosine kinase inhibitors: guideline from the College of American Pathologists, International Association for the Study of Lung Cancer, and Association for Molecular Pathology. J Mol Diagn 2013;15:415-53. [Crossref] [PubMed]
- Siegele BJ, Shilo K, Chao BH, et al. Epidermal growth factor receptor (EGFR) mutations in small cell lung cancers: Two cases and a review of the literature. Lung Cancer 2016;95:65-72. [Crossref] [PubMed]
- Shaozhang Z, Xiaomei L, Aiping Z, et al. Detection of EML4-ALK fusion genes in non-small cell lung cancer patients with clinical features associated with EGFR mutations. Genes Chromosomes Cancer 2012;51:925-32. [Crossref] [PubMed]
- Shaw AT, Kim DW, Nakagawa K, et al. Crizotinib versus chemotherapy in advanced ALK-positive lung cancer. N Engl J Med 2013;368:2385-94. [Crossref] [PubMed]
- Sequist LV, Waltman BA, Dias-Santagata D, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med 2011;3:75ra26. [Crossref] [PubMed]
- Jänne PA, Yang JC, Kim DW, et al. AZD9291 in EGFR inhibitor-resistant non-small-cell lung cancer. N Engl J Med 2015;372:1689-99. [Crossref] [PubMed]
- Gainor JF, Dardaei L, Yoda S, et al. Molecular Mechanisms of Resistance to First- and Second-Generation ALK Inhibitors in ALK-Rearranged Lung Cancer. Cancer Discov 2016;6:1118-33. [Crossref] [PubMed]
- Drilon A, Somwar R, Wagner JP, et al. A Novel Crizotinib-Resistant Solvent-Front Mutation Responsive to Cabozantinib Therapy in a Patient with ROS1-Rearranged Lung Cancer. Clin Cancer Res 2016;22:2351-8. [Crossref] [PubMed]
- Awad MM, Katayama R, McTigue M, et al. Acquired resistance to crizotinib from a mutation in CD74-ROS1. N Engl J Med 2013;368:2395-401. [Crossref] [PubMed]
- Postow MA, Callahan MK, Wolchok JD. Immune Checkpoint Blockade in Cancer Therapy. J Clin Oncol 2015;33:1974-82. [Crossref] [PubMed]
- Taube JM. Unleashing the immune system: PD-1 and PD-Ls in the pre-treatment tumor microenvironment and correlation with response to PD-1/PD-L1 blockade. Oncoimmunology 2014;3:e963413. [Crossref] [PubMed]
- Reck M, Rodriguez-Abreu D, Robinson AG, et al. Pembrolizumab versus Chemotherapy for PD-L1-Positive Non-Small-Cell Lung Cancer. N Engl J Med 2016;375:1823-33. [Crossref] [PubMed]
- Gainor JF, Shaw AT, Sequist LV, et al. EGFR Mutations and ALK Rearrangements Are Associated with Low Response Rates to PD-1 Pathway Blockade in Non-Small Cell Lung Cancer: A Retrospective Analysis. Clin Cancer Res 2016;22:4585-93. [Crossref] [PubMed]
- Gainor JF, Varghese AM, Ou SH, et al. ALK rearrangements are mutually exclusive with mutations in EGFR or KRAS: an analysis of 1,683 patients with non-small cell lung cancer. Clin Cancer Res 2013;19:4273-81. [Crossref] [PubMed]
- Sholl LM, Aisner DL, Varella-Garcia M, et al. Multi-institutional Oncogenic Driver Mutation Analysis in Lung Adenocarcinoma: The Lung Cancer Mutation Consortium Experience. J Thorac Oncol 2015;10:768-77. [Crossref] [PubMed]
- Sweeney SM, Cerami E, Baras A, et al. AACR project genie: Powering precision medicine through an international consortium. Cancer Discovery 2017;7:818-31. [Crossref] [PubMed]
- Planchard D, Besse B, Groen HJ, et al. Dabrafenib plus trametinib in patients with previously treated BRAF(V600E)-mutant metastatic non-small cell lung cancer: an open-label, multicentre phase 2 trial. Lancet Oncol 2016;17:984-93. [Crossref] [PubMed]
- Planchard D, Kim TM, Mazieres J, et al. Dabrafenib in patients with BRAF(V600E)-positive advanced non-small-cell lung cancer: a single-arm, multicentre, open-label, phase 2 trial. Lancet Oncol 2016;17:642-50. [Crossref] [PubMed]
- De Grève J, Teugels E, Geers C, et al. Clinical activity of afatinib (BIBW 2992) in patients with lung adenocarcinoma with mutations in the kinase domain of HER2/neu. Lung Cancer 2012;76:123-7. [Crossref] [PubMed]
- Gandhi L, Bahleda R, Tolaney SM, et al. Phase I study of neratinib in combination with temsirolimus in patients with human epidermal growth factor receptor 2-dependent and other solid tumors. J Clin Oncol 2014;32:68-75. [Crossref] [PubMed]
- Mazières J, Peters S, Lepage B, et al. Lung cancer that harbors an HER2 mutation: epidemiologic characteristics and therapeutic perspectives. J Clin Oncol 2013;31:1997-2003. [Crossref] [PubMed]
- Awad MM, Oxnard GR, Jackman DM, et al. MET Exon 14 Mutations in Non-Small-Cell Lung Cancer Are Associated With Advanced Age and Stage-Dependent MET Genomic Amplification and c-Met Overexpression. J Clin Oncol 2016;34:721-30. [Crossref] [PubMed]
- Paik PK, Drilon A, Fan PD, et al. Response to MET inhibitors in patients with stage IV lung adenocarcinomas harboring MET mutations causing exon 14 skipping. Cancer Discov 2015;5:842-9. [Crossref] [PubMed]
- Liu X, Jia Y, Stoopler MB, et al. Next-Generation Sequencing of Pulmonary Sarcomatoid Carcinoma Reveals High Frequency of Actionable MET Gene Mutations. J Clin Oncol 2016;34:794-802. [Crossref] [PubMed]
- Drilon A, Wang L, Hasanovic A, et al. Response to Cabozantinib in Patients with RET Fusion-Positive Lung Adenocarcinomas. Cancer Discov 2013;3:630-5. [Crossref] [PubMed]
- Drilon A, Rekhtman N, Arcila M, et al. Cabozantinib in patients with advanced RET-rearranged non-small-cell lung cancer: an open-label, single-centre, phase 2, single-arm trial. Lancet Oncol 2016;17:1653-60. [Crossref] [PubMed]
- Drilon A, Bergagnini I, Delasos L, et al. Clinical outcomes with pemetrexed-based systemic therapies in RET-rearranged lung cancers. Ann Oncol 2016;27:1286-91. [Crossref] [PubMed]
- Lee JK, Lee J, Kim S, et al. Clonal History and Genetic Predictors of Transformation Into Small-Cell Carcinomas From Lung Adenocarcinomas. J Clin Oncol 2017;35:3065-74. [Crossref] [PubMed]
- Clinical Lung Cancer Genome Project (CLCGP), Network Genomic Medicine (NGM). A genomics-based classification of human lung tumors. Sci Transl Med 2013;5:209ra153. [PubMed]
- Bell EH, Chakraborty AR, Mo X, et al. SMARCA4/BRG1 Is a Novel Prognostic Biomarker Predictive of Cisplatin-Based Chemotherapy Outcomes in Resected Non-Small Cell Lung Cancer. Clin Cancer Res 2016;22:2396-404. [Crossref] [PubMed]
- Herpel E, Rieker RJ, Dienemann H, et al. SMARCA4 and SMARCA2 deficiency in non-small cell lung cancer: immunohistochemical survey of 316 consecutive specimens. Ann Diagn Pathol 2017;26:47-51. [Crossref] [PubMed]
- Skoulidis F, Byers LA, Diao L, et al. Co-occurring genomic alterations define major subsets of KRAS-mutant lung adenocarcinoma with distinct biology, immune profiles, and therapeutic vulnerabilities. Cancer Discov 2015;5:860-77. [Crossref] [PubMed]
- Chen Z, Cheng K, Walton Z, et al. A murine lung cancer co-clinical trial identifies genetic modifiers of therapeutic response. Nature 2012;483:613-7. [Crossref] [PubMed]
- Rizvi NA, Hellmann MD, Snyder A, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 2015;348:124-8. [Crossref] [PubMed]
- Garofalo A, Sholl L, Reardon B, et al. The impact of tumor profiling approaches and genomic data strategies for cancer precision medicine. Genome Med 2016;8:79. [Crossref] [PubMed]
- Metzker ML. Sequencing technologies - the next generation. Nat Rev Genet 2010;11:31-46. [Crossref] [PubMed]
- Sholl LM, Do K, Shivdasani P, et al. Institutional implementation of clinical tumor profiling on an unselected cancer population. JCI Insight 2016;1:e87062. [Crossref] [PubMed]
- Rizzo JM, Buck MJ. Key principles and clinical applications of "next-generation" DNA sequencing. Cancer Prev Res (Phila) 2012;5:887-900. [Crossref] [PubMed]
- Abo RP, Ducar M, Garcia EP, et al. BreaKmer: detection of structural variation in targeted massively parallel sequencing data using kmers. Nucleic Acids Res 2015;43:e19. [Crossref] [PubMed]
- Zheng Z, Liebers M, Zhelyazkova B, et al. Anchored multiplex PCR for targeted next-generation sequencing. Nat Med 2014;20:1479-84. [Crossref] [PubMed]
- Sholl LM, Aisner DL, Allen TC, et al. Liquid Biopsy in Lung Cancer: A Perspective From Members of the Pulmonary Pathology Society. Arch Pathol Lab Med 2016;140:825-9. [Crossref] [PubMed]
- Asaka S, Yoshizawa A, Matsuda K, et al. A novel, rapid point-of-care test for lung cancer patients to detect epidermal growth factor receptor gene mutations by using real-time droplet-PCR and fresh liquid cytology specimens. Oncol Rep 2017;37:1020-6. [Crossref] [PubMed]
- Sacher AG, Paweletz C, Dahlberg SE, et al. Prospective Validation of Rapid Plasma Genotyping for the Detection of EGFR and KRAS Mutations in Advanced Lung Cancer. JAMA Oncol 2016;2:1014-22. [Crossref] [PubMed]
- Bettegowda C, Sausen M, Leary RJ, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med 2014;6:224ra24. [Crossref] [PubMed]
- Newman AM, Bratman SV, To J, et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med 2014;20:548-54. [Crossref] [PubMed]
- Bratman SV, Newman AM, Alizadeh AA, et al. Potential clinical utility of ultrasensitive circulating tumor DNA detection with CAPP-Seq. Expert Rev Mol Diagn 2015;15:715-9. [PubMed]
- Mok T, Wu YL, Lee JS, et al. Detection and Dynamic Changes of EGFR Mutations from Circulating Tumor DNA as a Predictor of Survival Outcomes in NSCLC Patients Treated with First-line Intercalated Erlotinib and Chemotherapy. Clin Cancer Res 2015;21:3196-203. [Crossref] [PubMed]
- Karlovich C, Goldman JW, Sun JM, et al. Assessment of EGFR Mutation Status in Matched Plasma and Tumor Tissue of NSCLC Patients from a Phase I Study of Rociletinib (CO-1686). Clin Cancer Res 2016;22:2386-95. [Crossref] [PubMed]
- Oxnard GR, Paweletz CP, Sholl LM. Genomic Analysis of Plasma Cell-Free DNA in Patients With Cancer. JAMA Oncol 2017;3:740-1. [Crossref] [PubMed]
- Newman AM, Lovejoy AF, Klass DM, et al. Integrated digital error suppression for improved detection of circulating tumor DNA. Nat Biotechnol 2016;34:547-55. [Crossref] [PubMed]
- Chabon JJ, Simmons AD, Lovejoy AF, et al. Circulating tumour DNA profiling reveals heterogeneity of EGFR inhibitor resistance mechanisms in lung cancer patients. Nat Commun 2016;7:11815. [Crossref] [PubMed]


