Beyond ALK and ROS1: RET, NTRK, EGFR and BRAF gene rearrangements in non-small cell lung cancer
Introduction
Over the past decade, molecular characterization of non-small cell lung carcinomas (NSCLCs) has uncovered molecularly defined subsets. Somatic molecular alterations in NSCLCs can lead to oncogene activation through multiple mechanisms, including point mutations, insertions, deletions and gene rearrangements. These alterations are generally mutually exclusive, with no more than one predominant driver in any given cancer (1,2). The hallmark of all of these alterations is oncogene addiction, in which cancers are driven primarily, or even exclusively, by aberrant oncogene signaling. Oncogenic gene rearrangements in NSCLC can lead to expression of oncogenic fusion proteins when a 5' partner forms an in-frame gene fusion with a 3' proto-oncogene (3-7). These fusions retain the kinase domain of the proto-oncogene, with expression and dimerization generally promoted by 5' partner. The result of this structure is expression of a constitutively activated fusion protein whose downstream signaling drives cell proliferation and survival in a ligand-independent manner (Figure 1) (8).
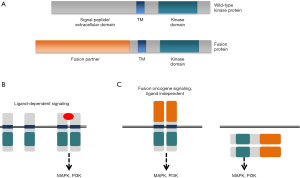
Targeted inhibition of oncogenic fusion proteins can result in potent cancer growth inhibition and regression of tumors in patients. To date, there are drugs approved by the U.S. Food and Drug Administration (FDA) and the European Medicines Agency (EMA) for NSCLCs containing gene fusions involving anaplastic lymphoma kinase (ALK) and ROS1 proto-oncogene receptor tyrosine kinase (ROS1) (9-12). However, other gene rearrangements involving other kinase-encoding genes, including RET, NTRK1, EGFR and BRAF have been described in NSCLC. Emerging data support the hypothesis that these fusions, like those involving ALK and ROS1, lead to oncogene dependency, and therefore may also offer an opportunity for targeted and effective anti-tumor therapy.
Rearranged during transfection (RET)
The RET proto-oncogene encodes a receptor tyrosine kinase that is expressed on tissues of neural crest origin and functions in development of the kidneys and enteric nervous system (13,14). Rearrangements involving RET have oncogenic potential, and occur in a subset of thyroid papillary carcinomas (15,16). Gene rearrangements involving RET have also been observed in approximately 1–2% of NSCLCs and are mutually exclusive with other oncogenic driver mutations such as ALK or ROS1 rearrangements or EGFR mutations (7,17-19). Multiple fusion partners with RET have been described. A recent global registry of patients with RET rearranged NSCLC reported that among 81 cases with identifiable fusion partners, 72% involved the kinesin family 5B gene (KIF5B), supporting previous observations that KIF5B is the most common RET fusion partner in NSCLC (7,17,19,20). The second most common fusion partner is CCDC6 (23%), though others including NCOA4 (2%), EPHA5 (1%) and PICALM (1%) have been reported (20,21).
The results from a global multicenter registry of 165 RET rearranged NSCLCs provide several valuable insights into the clinical and pathological features of this subtype of lung cancer (20). RET rearrangements were observed in males and females in equal proportions. Among patients in the global registry, 63% were never smokers, 24% were former smokers, and 10% were current smokers. 98% of cases were adenocarcinoma. The majority of patients (72%) had stage IV disease at the time of diagnosis, suggesting that RET rearranged NSCLCs may have high metastatic potential. Alternatively, it is possible that RET rearrangements are more readily identified in patients with metastatic disease because non-metastatic cases may less commonly undergo molecular testing.
Extrapolating from the paradigm of fusion oncogene dependence observed in ALK and ROS1-rearrangement NSCLCs, efforts have been made both preclinically and clinically to target RET in RET rearranged NSCLCs. In cell line models expressing exogenous KIF5B-RET, RET undergoes ligand independent tyrosine phosphorylation, and expression of the fusion construct has transforming activity (17). To date, no highly selective RET inhibitor has been tested clinically, and the majority of preclinical and clinical development has focused on use of multi-targeted kinase inhibitors. In preclinical cell line models expressing KIF5B-RET fusion transgenes, treatment with the multi-targeted kinase inhibitors vandetanib, sorafenib and sunitinib suppressed RET phosphorylation and inhibited cell viability in a dose-dependent manner (17,18).
Clinical activity of RET-directed therapy was first reported in 2013, when three patients with RET rearranged NSCLC were treated with cabozantinib (21). Two of these patients experienced partial responses by RECIST 1.1 criteria, and a third had prolonged stable disease. Extending from this work, a phase 2 trial was conducted to assess the activity of cabozantinib among patient with RET rearranged lung adenocarcinoma. Among 25 patients who were assessable for response, there were seven partial responses [overall response rate (ORR) 28%, 95% CI, 12–49]. The median duration of treatment was 4.7 months, and the median duration of response was 7.0 months. The median progression-free survival (PFS) was 5.5 months (22).
Similarly, vandetanib has shown single agent activity in RET rearranged NSCLC. A case reported in 2014 demonstrated activity of vandetanib in a patient with a CCDC6-RET rearrangement NSCLC (23). More recently, among 19 RET rearrangement NSCLC patients enrolled in a Japanese phase 2 single arm study, there was a response rate of 47% and a median PFS of 4.7 months (24).
Lenvatinib has also shown single agent activity, with an objective response rate of 16% among 25 RET-fusion positive adenocarcinoma patients, including 3 of 4 responses in patients who had received prior RET-directed therapy (25). In this study, a median PFS was 7.3 months (95% CI, 3.6–10.2) and the median duration of treatment was 16 weeks.
Other multi-target kinase inhibitors have also been used in RET-rearrangement NSCLC, including sunitinib (26), sorafenib (27), alectinib (28), nintedanib, ponatinib and regorafenib (20). Data for these agents are generally limited to case reports. There is no clear gold standard for treatment, and no direct comparison of RET inhibitors has been performed. Thus, it is difficult to identify the most active RET inhibitor based on the currently available clinical data.
One challenge facing clinical application of these RET inhibitors is toxicity. In the phase 2 trial of cabozantinib, dose reductions from the starting dose of 60 to 40 mg daily was required in 73% of patients, and to 20 mg daily in 15% of patients (22). Among RET rearrangement NSCLC patients, the most common reasons for dose reduction were palmar-plantar erythrodysesthesia, fatigue, and diarrhea (22). Such toxicities may be attributable to cabozantinib’s inhibitory activity of VEGFR2 and other kinases. Vandetanib is associated with diarrhea, rash, nausea, hypertension, fatigue, and electrocardiogram QT prolongation, as well as other effects (29).
It is unclear whether the multi-kinase activity of the RET inhibitors is an advantage, perhaps by affecting vasculature or other elements of the tumor micro-environment, or whether it is a liability, contributing to toxicity without improving activity, and potentially limiting clinical activity by preventing maximal RET inhibition. Development of highly-selective RET inhibitors, such as BLU-667 which is currently in clinical development (NCT03037385), may provide insights.
RXDX-105 is a small molecule RET inhibitor with potent inhibition of wild-type RET, RET fusions, and RET activating mutations in cell lines and in xenograft models (30). It also has activity against BRAF V600E (30-32). RXDX-105 spares VEGFR2 and VEGFR1, and therefore may also spare some of the toxicities seen with other multikinase inhibitors (30). In a patient with RET rearranged NSCLC with brain metastases, treatment with RXDX-105 led to a confirmed partial response and near complete resolution of previously noted subcentimeter brain metastases (30). A phase 1 trial of RXDX-105 is ongoing (NCT01877811).
Metastasis to the central nervous system (CNS) represents an important clinical challenge in RET rearrangement NSCLC. Several studies have specifically focused on this need. Vandetanib is thought to have limited blood-brain barrier penetration, but Minocha and colleagues hypothesized that this penetration may be improved by modulation pf P-gp/Abcb1- and Bcrp1/Abcg2-mediated efflux through the use of an mTOR inhibitor (33,34). Combining vandetanib and the mTOR inhibitor everolimus in RET rearranged NSCLC, the authors have observed promising systemic activity, with partial responses in 5 of 6 NSCLC patients with RET rearrangements identified by next generation sequencing (NGS) (33,35). Additionally and importantly, antitumor activity was also observed in the CNS, including in a patient with cabozantinib resistant disease (33,35). However, the combination of vandetanib and everolimus is associated with toxicity, including grade 3 diarrhea, rash and QTc prolongation (35). There is an ongoing phase 1 trial studying the combination in patients with advanced cancer (NCT01582191).
Alectinib is an orally available inhibitor of ALK, and also has RET inhibitory activity in cell-based and xenograft models of KIF5B-RET rearranged NSCLC (36). Alectinib also has a more favorable toxicity profile than many of the other multi-kinase RET inhibitors. A recent study reported that among four patients with metastatic RET rearranged NSCLC who were treated with alectinib, including three who had received prior RET TKIs, there were two objective radiographic responses (28). One patient with CNS metastases had improvement in both intracranial and extracranial disease on an escalated dose of 900 mg twice daily of alectinib (28).
Mechanisms of acquired resistance to RET inhibitors in patients are currently poorly understood. Repeat biopsies or analysis of circulating tumor DNA at the time of acquired resistance to RET inhibitors may be informative.
Neurotrophic tyrosine receptor kinase (NTRK)
The NTRK genes, NTRK1, NTRK2 and NTRK3, encode the tropomyosin receptor kinases (TRK), TRKA, TRKB and TRKC, respectively. The TRK proteins function during normal physiology as receptors for nerve growth factors (37-40). They signal through PI3 kinase, RAS/MAPK/ERK, and PLC-gamma pathways to mediate neuronal development (41-44).
A somatic chromosomal rearrangement resulting in expression of oncogenic fusion involving NTRK1 was first described in 1986 in a colorectal tumor sample (45). Since then, fusions involving NTRK1 have been described in a variety of other cancer types, including intrahepatic cholangiocarcinoma (46), papillary thyroid cancer (47), spitzoid neoplasms (48), glioblastoma (49) and sarcoma (50,51). Additionally, fusions involving NTRK2 and NTRK3 have been described in a variety of cancer types [reviewed in (44)]. Interestingly, the frequency of NTRK fusions is low (<5%) in common cancer types, but NTRK3 fusions can be frequent or nearly uniform in uncommon cancer types such as secretory breast carcinoma (52), mammary analog secretory carcinoma (53), and congenital fibrosarcomas (54,55).
Oncogenic fusions involving NTRK1 were first described in NSCLC in 2013. Using a targeted next-generation DNA sequencing (NGS) assay on a collection of 36 lung adenocarcinoma samples without other known oncogenic drivers, Vaishnavi and colleagues identified 2 cases with in-frame fusions involving NTRK1 (56). One fusion involved the 5' end of the myosin phosphatase-Rho-interacting protein gene (MPRIP) and the 3' end of NTRK1, including the tyrosine kinase domain. The second fusion involved the 5' end of CD74 and the 3' end of NTRK1, again with the tyrosine kinase domain intact. These fusions were tumorigenic in xenograft models. Additionally, Ba/F3 cells engineered to express a MPRIP-NTRK1 cDNA construct demonstrated auto-phosphorylation of TRKA and activation of downstream signaling pathways, while inhibition of TRK signaling with tyrosine kinase inhibitors blocked downstream signaling and reduced cell viability (56).
The frequency of NTRK fusions in NSCLC is not well established. Estimates range from 0.1% among all NSCLCs to 3% among patients without other identified driver mutations (8,56). Furthermore, the clinical and pathologic features of these patients are also not well characterized.
Several TRK inhibitors are now in clinical development. Larotrectinib (LOXO-101) is an oral tyrosine kinase inhibitor that has high selectivity and potency for TRKA, TRKB, and TRKC. Data from the phase 1 study of larotrectinib in solid-tumor patients was presented in late 2016 (57). The study had enrolled 59 patients. Of these, 8 patients had tumors harboring an NTRK gene fusion. The drug was well tolerated overall with the most common adverse effect (AE) being fatigue (37%). The most common grade 3/4 AE was anemia (8%). Among 7 NTRK fusion patients with evaluable disease, 6 had partial response and one had stable disease by RECIST assessment. The majority of NTRK fusion patients have remained on study for greater than 12 months (57). Larotrectinib is now being further investigated in an ongoing phase 2 study in subjects with NTRK fusion positive solid tumors (NCT02576431) and a pediatric phase 1/2 study (NCT02637687).
Entrectinib (RXDX-101) is an oral tyrosine kinase inhibitor with potent activity against ROS1, ALK, and TRKA, TRKB and TRKC (58). Among 119 patients treated on two phase 1 studies, entrectinib was very well tolerated (59). Among three patients with extra-cranial, TKI-treatment naive tumors harboring NTRK gene fusions, all had objective responses by RECIST criteria (59). One of these was a patient with NSCLC with a SQSTM1-NTRK1 fusion, who remained on treatment for over 18 months. Notably, in addition to having extensive extra-cranial disease, this patient had brain metastases at baseline and had a rapid and durable response to entrectinib in the brain (8,59). An additional patient with a glioneuronal tumor with a BCAN-NTRK1 fusion had a clinical and volumetric response to entrectinib (60). This promising activity within the CNS is highly relevant to patients with NSCLC, where CNS metastases are common. Entrectinib is now being further investigated in an ongoing basket phase 2 trial in subjects with solid tumors harboring NTRK1/2/3, ROS1 or ALK gene rearrangements (NCT02568267).
Several other TKIs have activity against TRKA/B/C. Others in clinical development include TSR-011 (NCT02048488), DS-6051b (NCT02279433), PLX7486 (NCT01804530). Notably, although crizotinib has some TRK inhibitory activity in preclinical models (56) its activity in patients with NTRK rearrangement tumors has not been well validated. While several multi-kinase inhibitors have some TRK inhibitory activity, this is likely more of a liability than a strength, given that highly potent and specific TRK inhibition is sufficient to induce dramatic clinical responses (57,59).
Although NTRK rearranged tumors have been highly sensitive to selective TRK inhibitors, several cases of acquired resistance to TRK inhibitors have been reported (61,62). In these cases, patients’ tumors initially responded well to the TRK inhibitor, but subsequently progressed, indicating outgrowth of a resistant subclone. Acquired resistance to entrectinib was described in a patient with colorectal cancer harboring an LMNA-NTRK1 rearrangement, and in a patient with a mammary analogue secretory tumor harboring an ETV6-NTRK3 fusion. In each case, resistant subclones had new mutations in the NTRK kinase domain, altering the protein structure to create steric hindrance to drug binding in the ATP binding pocket. These cases indicate that these tumors remain dependent on TRK signaling, and that resistance is mediated by preventing drug binding. Thus, strategies to overcome steric hindrance mutations may be effective in treating select cases of NTRK-rearrangement tumors with acquired resistance to first generation TKIs. It is not yet known how frequently NTRK rearranged tumors will develop resistance via alternative mechanisms, such as activation of a bypass signaling pathway or otherwise achieving TRK-independent resistance, as can be seen in ALK rearranged NSCLCs (63,64).
EGFR
The majority of clinically relevant EGFR mutations in lung cancer are exon 19 deletions or exon 21 missense mutations, which were first discovered in 2004 by sequencing EGFR in tumor biopsies from patients with response to the EGFR tyrosine kinase inhibitors gefitinib, or erlotinib (65). This discovery led to refined drug usage and development which over the past 13 years has improved outcomes for patients with lung cancer and activating/sensitizing EGFR mutations such that median overall survival is now measured in years (66). Next generation molecular pathology platforms have since detected EGFR gene fusions which involve a breakpoint somewhere in EGFR exon 23—intron 25 fused with either RAD51 or PURB, and the fusions are both activating and sensitizing. These gene fusions would not be discovered using conventional PCR-based methods for detecting classical EGFR mutations. A seminal paper in 2016 reported 5 cases of EGFR gene fusions, 4 of whom were treated with EGFR tyrosine kinase inhibitors with documented clinical and radiologic responses (67). Their technique utilized a next-generation sequencing assay that interrogated the entire coding region of EGFR (as well as introns 7, 15, 24, 25, and 26). The investigators surveyed over 10,000 lung cancers using this technique and found EGFR fusions in 5 cases (0.05%).
We at Massachusetts General Hospital Cancer Center are currently treating a 36-year-old man with virulent, chemorefractory lung cancer metastatic to brain, pleura and peritoneum who was found to have an EGFR exon 24-RAD51_exon 4 gene fusion using our next-generation sequencing platform (68). After discovery of the fusion, the patient demonstrated both clinical and radiologic improvement on erlotinib therapy and has remained on erlotinib for over 2 years with ongoing clinical and radiologic response.
In addition to EGFR fusions, an EGFR exon 18–25 kinase domain duplication has been reported in a 33-year-old male with lung cancer. This patient’s tumor responded to therapy with the EGFR inhibitor afatinib (69).
BRAF
BRAF is a RAF kinase which is downstream of RAS and signals via the MAPK pathway. The majority of activating BRAF mutations in cancer occurs within the kinase domain at amino acid V600, most commonly V600E. Genotyping studies of NSCLC have detected BRAF base substitutions in 2–5%, and about half of these mutations result in the BRAF amino acid substitution V600E. Most patients with BRAF mutation are former smokers, and non-V600E mutations are more common in heavy smokers. In the era before targeted therapy for BRAF, there was no difference in overall survival in patients based on the presence of BRAF mutation, or mutation subtypes (70-74).
In contrast to base substitutions, BRAF gene fusions are extremely rare and biologically distinct from BRAF V600E. In a study of comprehensive genomic profiling using the FoundationOne assay, BRAF fusions involving the intact in-frame BRAF kinase domain were discovered in 0.2% (8/4,013) of NSCLCs (75). In contrast, BRAF fusions are detected in 3% of melanomas; 2% of gliomas; 1% of thyroid cancers; and 0.3% of pancreatic carcinomas. All lung cancers with BRAF fusions were adenocarcinomas, or had adenocarcinoma features, and none were detected in squamous or small cell lung cancers (75).
BRAF fusions are similar to other kinase fusions in that they tend to be mutually exclusive of other activating mutations in the MAPK pathway (75). The 5' fusion partners identified to date include EPS15, NUP214, ARMC10, BTF3L4, AGK, GHR, ZC3HAV, and TRIM24 (75). Protein domain analysis shows that these fusions maintain the kinase domain of BRAF and the dimerization motif for BRAF at amino acids 506–509, however the RAS-binding domain is replaced by the various 5' partners (76). Early studies have determined that BRAF fusions are activating, and certain BRAF fusion variants can homodimerize with one another (77).
Distinguishing BRAF base substitutions from fusions may be particularly important with regards to selection of targeted drug therapy. Signaling through RAF/MEK/ERK is complex, dependent on RAF dimerization, and limited by feedback inhibition of RAS signal. BRAF V600E is unique in that it functions as an activated monomer independent of RAS signal and can be targeted with single-agent BRAF inhibitors. Prospective drug trials of single-agent BRAF inhibitors (vemurafenib or dabrafenib) in patients with metastatic NSCLC and BRAF V600E show response rates of 30–40% and median PFS of 5–7 months (78,79). Addition of the MEK inhibitor trametinib to dabrafenib in patients with BRAF V600E improves ORR to 63% and median PFS to 10 months (80). In contrast, all other activating BRAF mutants function as constitutive RAS-independent dimers not susceptible to single-agent RAF inhibitors (which do not inhibit dimers because their binding to one site in the dimer significantly reduces their affinity for the second site) (81). Thus, based on preclinical information, BRAF inhibitors may not be effective in BRAF fusion-driven malignancies, and in fact may actually promote cancer growth. Case reports of activity of single-agent MEK inhibitors in melanomas with BRAF fusion have been described (76).
Conclusions
RET, NTRK, EGFR and BRAF fusions in NSCLC represent recently discovered molecular subsets, and preclinical and early clinical data suggest that targeted inhibition of their activated protein products can cause tumor regressions. Further delineation of the most potent and selective kinase inhibitors, and the CNS activity of these drugs, is needed. However, these gene mutations are sufficiently rare that it may not be possible to conduct randomized trials to compare the activity of targeted therapies to traditional chemotherapy or immunotherapy for each subtype. There is no question that the overall survival of a patient with advanced lung cancer is improved when that patient has access to more than one effective therapy. There are now FDA approved drugs with different mechanisms of action for all lung cancer patients as first, second, and even third-line therapy (82). Discovery of actionable gene fusions opens doors for patients to try targeted therapies available by commercial supply, or to enroll them in an appropriate clinical trial. We anticipate that these expanded effective therapeutic options will lead to improved overall survival in these patients, as they have for patients with other molecularly targetable oncogenic alterations.
Taken individually, these genetic events are rare. However, taken together, and paired with universal comprehensive molecular profiling, there will be more and more patients who benefit from molecularly targeted therapy for their advanced lung cancer. Given the ever-increasing landscape of potentially actionable mutations in NSCLC, we advocate for multiplexed testing of all NSCLCs using next-generation gene sequencing platforms which identify both gene sequence alterations and gene fusions to increase discovery of targetable mutations in individual patients in the most efficient manner possible (68,83).
Acknowledgements
We thank our colleagues in thoracic oncology at the MGH Cancer Center.
Footnote
Conflicts of Interest: AF Farago serves as consulting or advisory role for Abbive, Pharmamar, Merrimack Pharmaceuticals, Takeda, Intervention Insights, and received honorarium from Foundation Medicine. CG Azzoli has no conflicts of interest to declare.
References
- Lin JJ, Ritterhouse LL, Ali SM, et al. ROS1 Fusions Rarely Overlap with Other Oncogenic Drivers in Non-Small Cell Lung Cancer. J Thorac Oncol 2017;12:872-7. [Crossref] [PubMed]
- Gainor JF, Varghese AM, Ou SH, et al. ALK rearrangements are mutually exclusive with mutations in EGFR or KRAS: an analysis of 1,683 patients with non-small cell lung cancer. Clin Cancer Res 2013;19:4273-81. [Crossref] [PubMed]
- Gainor JF, Shaw AT. Novel targets in non-small cell lung cancer: ROS1 and RET fusions. Oncologist 2013;18:865-75. [Crossref] [PubMed]
- Soda M, Choi YL, Enomoto M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature 2007;448:561-6. [Crossref] [PubMed]
- Rikova K, Guo A, Zeng Q, et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell 2007;131:1190-203. [Crossref] [PubMed]
- Takeuchi K, Choi YL, Soda M, et al. Multiplex reverse transcription-PCR screening for EML4-ALK fusion transcripts. Clin Cancer Res 2008;14:6618-24. [Crossref] [PubMed]
- Takeuchi K, Soda M, Togashi Y, et al. RET, ROS1 and ALK fusions in lung cancer. Nat Med 2012;18:378-81. [Crossref] [PubMed]
- Farago AF, Le LP, Zheng Z, et al. Durable Clinical Response to Entrectinib in NTRK1-Rearranged Non-Small Cell Lung Cancer. J Thorac Oncol 2015;10:1670-4. [Crossref] [PubMed]
- Shaw AT, Kim DW, Mehra R, et al. Ceritinib in ALK-rearranged non-small-cell lung cancer. N Engl J Med 2014;370:1189-97. [Crossref] [PubMed]
- Shaw AT, Ou SH, Bang YJ, et al. Crizotinib in ROS1-rearranged non-small-cell lung cancer. N Engl J Med 2014;371:1963-71. [Crossref] [PubMed]
- Shaw AT, Kim DW, Nakagawa K, et al. Crizotinib versus chemotherapy in advanced ALK-positive lung cancer. N Engl J Med 2013;368:2385-94. [Crossref] [PubMed]
- Shaw AT, Yeap BY, Solomon BJ, et al. Effect of crizotinib on overall survival in patients with advanced non-small-cell lung cancer harbouring ALK gene rearrangement: a retrospective analysis. Lancet Oncol 2011;12:1004-12. [Crossref] [PubMed]
- Nakamura T, Ishizaka Y, Nagao M, et al. Expression of the ret proto-oncogene product in human normal and neoplastic tissues of neural crest origin. J Pathol 1994;172:255-60. [Crossref] [PubMed]
- Schuchardt A, D’Agati V, Larsson-Blomberg L, et al. Defects in the kidney and enteric nervous system of mice lacking the tyrosine kinase receptor Ret. Nature 1994;367:380-3. [Crossref] [PubMed]
- Santoro M, Carlomagno F, Hay ID, et al. Ret oncogene activation in human thyroid neoplasms is restricted to the papillary cancer subtype. J Clin Invest 1992;89:1517-22. [Crossref] [PubMed]
- Takahashi M, Ritz J, Cooper GM. Activation of a novel human transforming gene, ret, by DNA rearrangement. Cell 1985;42:581-8. [Crossref] [PubMed]
- Kohno T, Ichikawa H, Totoki Y, et al. KIF5B-RET fusions in lung adenocarcinoma. Nat Med 2012;18:375-7. [Crossref] [PubMed]
- Lipson D, Capelletti M, Yelensky R, et al. Identification of new ALK and RET gene fusions from colorectal and lung cancer biopsies. Nat Med 2012;18:382-4. [Crossref] [PubMed]
- Wang R, Hu H, Pan Y, et al. RET fusions define a unique molecular and clinicopathologic subtype of non-small-cell lung cancer. J Clin Oncol 2012;30:4352-9. [Crossref] [PubMed]
- Gautschi O, Milia J, Filleron T, et al. Targeting RET in Patients With RET-Rearranged Lung Cancers: Results From the Global, Multicenter RET Registry. J Clin Oncol 2017;35:1403-10. [Crossref] [PubMed]
- Drilon A, Wang L, Hasanovic A, et al. Response to Cabozantinib in patients with RET fusion-positive lung adenocarcinomas. Cancer Discov 2013;3:630-5. [Crossref] [PubMed]
- Drilon A, Rekhtman N, Arcila M, et al. Cabozantinib in patients with advanced RET-rearranged non-small-cell lung cancer: an open-label, single-centre, phase 2, single-arm trial. Lancet Oncol 2016;17:1653-60. [Crossref] [PubMed]
- Falchook GS, Ordonez NG, Bastida CC, et al. Effect of the RET Inhibitor Vandetanib in a Patient With RET Fusion-Positive Metastatic Non-Small-Cell Lung Cancer. J Clin Oncol 2016;34:e141-4. [Crossref] [PubMed]
- Yoh K, Seto T, Satouchi M, et al. Vandetanib in patients with previously treated RET-rearranged advanced non-small-cell lung cancer (LURET): an open-label, multicentre phase 2 trial. Lancet Respir Med 2017;5:42-50. [Crossref] [PubMed]
- Velcheti V, Hida T, Reckamp KL, et al. Phase 2 study of lenvatinib (LN) in patients (Pts) with RET fusion-positive adenocarcinoma of the lung. Ann Oncol 2016;27:vi416-54. [Crossref]
- Wu H, Shih JY, Yang JC. Rapid Response to Sunitinib in a Patient with Lung Adenocarcinoma Harboring KIF5B-RET Fusion Gene. J Thorac Oncol 2015;10:e95-6. [Crossref] [PubMed]
- Horiike A, Takeuchi K, Uenami T, et al. Sorafenib treatment for patients with RET fusion-positive non-small cell lung cancer. Lung Cancer 2016;93:43-6. [Crossref] [PubMed]
- Lin JJ, Kennedy E, Sequist LV, et al. Clinical Activity of Alectinib in Advanced RET-Rearranged Non-Small Cell Lung Cancer. J Thorac Oncol 2016;11:2027-32. [Crossref] [PubMed]
- Wells SA Jr, Robinson BG, Gagel RF, et al. Vandetanib in patients with locally advanced or metastatic medullary thyroid cancer: a randomized, double-blind phase III trial. J Clin Oncol 2012;30:134-41. [Crossref] [PubMed]
- Li GG, Somwar R, Joseph J, et al. Antitumor Activity of RXDX-105 in Multiple Cancer Types with RET Rearrangements or Mutations. Clin Cancer Res 2017;23:2981-90. [Crossref] [PubMed]
- James J, Ruggeri B, Armstrong RC, et al. CEP-32496: a novel orally active BRAF(V600E) inhibitor with selective cellular and in vivo antitumor activity. Mol Cancer Ther 2012;11:930-41. [Crossref] [PubMed]
- Rowbottom MW, Faraoni R, Chao Q, et al. Identification of 1-(3-(6,7-dimethoxyquinazolin-4-yloxy)phenyl)-3-(5-(1,1,1-trifluoro-2-methylpropa n-2-yl)isoxazol-3-yl)urea hydrochloride (CEP-32496), a highly potent and orally efficacious inhibitor of V-RAF murine sarcoma viral oncogene homologue B1 (BRAF) V600E. J Med Chem 2012;55:1082-105. [Crossref] [PubMed]
- Subbiah V, Berry J, Roxas M, et al. Systemic and CNS activity of the RET inhibitor vandetanib combined with the mTOR inhibitor everolimus in KIF5B-RET re-arranged non-small cell lung cancer with brain metastases. Lung Cancer 2015;89:76-9. [Crossref] [PubMed]
- Minocha M, Khurana V, Qin B, et al. Co-administration strategy to enhance brain accumulation of vandetanib by modulating P-glycoprotein (P-gp/Abcb1) and breast cancer resistance protein (Bcrp1/Abcg2) mediated efflux with m-TOR inhibitors. Int J Pharm 2012;434:306-14. [Crossref] [PubMed]
- Cascone T, Subbiah V, Hess KR, et al. Significant systemic and CNS activity of RET inibitor vandetanib combined with mTOR inhibitor everolimus in patients with advanced NSCLC with RET fusion. J Clin Oncol 2016;34: abstr 9069.
- Kodama T, Tsukaguchi T, Satoh Y, et al. Alectinib shows potent antitumor activity against RET-rearranged non-small cell lung cancer. Mol Cancer Ther 2014;13:2910-8. [Crossref] [PubMed]
- Soppet D, Escandon E, Maragos J, et al. The neurotrophic factors brain-derived neurotrophic factor and neurotrophin-3 are ligands for the trkB tyrosine kinase receptor. Cell 1991;65:895-903. [Crossref] [PubMed]
- Kaplan DR, Martin-Zanca D, Parada LF. Tyrosine phosphorylation and tyrosine kinase activity of the trk proto-oncogene product induced by NGF. Nature 1991;350:158-60. [Crossref] [PubMed]
- Klein R, Nanduri V, Jing SA, et al. The trkB tyrosine protein kinase is a receptor for brain-derived neurotrophic factor and neurotrophin-3. Cell 1991;66:395-403. [Crossref] [PubMed]
- Klein R, Jing SQ, Nanduri V, et al. The trk proto-oncogene encodes a receptor for nerve growth factor. Cell 1991;65:189-97. [Crossref] [PubMed]
- Asakura K, Ueda A, Kawamura N, et al. Clioquinol inhibits NGF-induced Trk autophosphorylation and neurite outgrowth in PC12 cells. Brain Res 2009;1301:110-5. [Crossref] [PubMed]
- Kaplan DR, Stephens RM. Neurotrophin signal transduction by the Trk receptor. J Neurobiol 1994;25:1404-17. [Crossref] [PubMed]
- Stephens RM, Loeb DM, Copeland TD, et al. Trk receptors use redundant signal transduction pathways involving SHC and PLC-gamma 1 to mediate NGF responses. Neuron 1994;12:691-705. [Crossref] [PubMed]
- Vaishnavi A, Le AT, Doebele RC. TRKing down an old oncogene in a new era of targeted therapy. Cancer Discov 2015;5:25-34. [Crossref] [PubMed]
- Martin-Zanca D, Hughes SH, Barbacid M. A human oncogene formed by the fusion of truncated tropomyosin and protein tyrosine kinase sequences. Nature 1986;319:743-8. [Crossref] [PubMed]
- Ross JS, Wang K, Gay L, et al. New routes to targeted therapy of intrahepatic cholangiocarcinomas revealed by next-generation sequencing. Oncologist 2014;19:235-42. [Crossref] [PubMed]
- Greco A, Miranda C, Pierotti MA. Rearrangements of NTRK1 gene in papillary thyroid carcinoma. Mol Cell Endocrinol 2010;321:44-9. [Crossref] [PubMed]
- Wiesner T, He J, Yelensky R, et al. Kinase fusions are frequent in Spitz tumours and spitzoid melanomas. Nat Commun 2014;5:3116. [Crossref] [PubMed]
- Kim J, Lee Y, Cho HJ, et al. NTRK1 fusion in glioblastoma multiforme. PLoS One 2014;9:e91940. [Crossref] [PubMed]
- Doebele RC, Davis LE, Vaishnavi A, et al. An Oncogenic NTRK Fusion in a Patient with Soft-Tissue Sarcoma with Response to the Tropomyosin-Related Kinase Inhibitor LOXO-101. Cancer Discov 2015;5:1049-57. [Crossref] [PubMed]
- Stransky N, Cerami E, Schalm S, et al. The landscape of kinase fusions in cancer. Nat Commun 2014;5:4846. [Crossref] [PubMed]
- Tognon C, Knezevich SR, Huntsman D, et al. Expression of the ETV6-NTRK3 gene fusion as a primary event in human secretory breast carcinoma. Cancer Cell 2002;2:367-76. [Crossref] [PubMed]
- Bishop JA, Yonescu R, Batista D, et al. Utility of mammaglobin immunohistochemistry as a proxy marker for the ETV6-NTRK3 translocation in the diagnosis of salivary mammary analogue secretory carcinoma. Hum Pathol 2013;44:1982-8. [Crossref] [PubMed]
- Bourgeois JM, Knezevich SR, Mathers JA, et al. Molecular detection of the ETV6-NTRK3 gene fusion differentiates congenital fibrosarcoma from other childhood spindle cell tumors. Am J Surg Pathol 2000;24:937-46. [Crossref] [PubMed]
- Knezevich SR, McFadden DE, Tao W, et al. A novel ETV6-NTRK3 gene fusion in congenital fibrosarcoma. Nat Genet 1998;18:184-7. [Crossref] [PubMed]
- Vaishnavi A, Capelletti M, Le AT, et al. Oncogenic and drug-sensitive NTRK1 rearrangements in lung cancer. Nat Med 2013;19:1469-72. [Crossref] [PubMed]
- Hong DS, Dowlati A, Burris HA, et al. Clinical safety and activity from a phase 1 study of LOXO-101, a selective TRKA/B/C inhibitor, in solid-tumor patients with NTRK gene fusions. Ann Oncol 2016;27:ix46-51. [Crossref]
- Ardini E, Menichincheri M, Banfi P, et al. Entrectinib, a Pan-TRK, ROS1, and ALK Inhibitor with Activity in Multiple Molecularly Defined Cancer Indications. Mol Cancer Ther 2016;15:628-39. [Crossref] [PubMed]
- Drilon A, Siena S, Ou SI, et al. Safety and Antitumor Activity of the Multitargeted Pan-TRK, ROS1, and ALK Inhibitor Entrectinib: Combined Results from Two Phase I Trials (ALKA-372-001 and STARTRK-1). Cancer Discov 2017;7:400-9. [Crossref] [PubMed]
- Alvarez-Breckenridge C, Miller JJ, Nayyar N, et al. Clinical and radiographic response following targeting of BCAN-NTRK1 fusion in glioneuronal tumor. NPJ Precision Oncology 2017;1:5. [Crossref]
- Russo M, Misale S, Wei G, et al. Acquired Resistance to the TRK Inhibitor Entrectinib in Colorectal Cancer. Cancer Discov 2016;6:36-44. [Crossref] [PubMed]
- Drilon A, Li G, Dogan S, et al. What hides behind the MASC: clinical response and acquired resistance to entrectinib after ETV6-NTRK3 identification in a mammary analogue secretory carcinoma (MASC). Ann Oncol 2016;27:920-6. [Crossref] [PubMed]
- Gainor JF, Dardaei L, Yoda S, et al. Molecular Mechanisms of Resistance to First- and Second-Generation ALK Inhibitors in ALK-Rearranged Lung Cancer. Cancer Discov 2016;6:1118-33. [Crossref] [PubMed]
- Lin JJ, Riely GJ, Shaw AT. Targeting ALK: Precision Medicine Takes on Drug Resistance. Cancer Discov 2017;7:137-55. [Crossref] [PubMed]
- Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med 2004;350:2129-39. [Crossref] [PubMed]
- Paz-Ares L, Tan EH, O'Byrne K, et al. Afatinib versus gefitinib in patients with EGFR mutation-positive advanced non-small-cell lung cancer: overall survival data from the phase IIb LUX-Lung 7 trial. Ann Oncol 2017;28:270-7. [Crossref] [PubMed]
- Konduri K, Gallant JN, Chae YK, et al. EGFR Fusions as Novel Therapeutic Targets in Lung Cancer. Cancer Discov 2016;6:601-11. [Crossref] [PubMed]
- Zheng Z, Liebers M, Zhelyazkova B, et al. Anchored multiplex PCR for targeted next-generation sequencing. Nat Med 2014;20:1479-84. [Crossref] [PubMed]
- Gallant JN, Sheehan JH, Shaver TM, et al. EGFR Kinase Domain Duplication (EGFR-KDD) Is a Novel Oncogenic Driver in Lung Cancer That Is Clinically Responsive to Afatinib. Cancer Discov 2015;5:1155-63. [Crossref] [PubMed]
- Paik PK, Arcila ME, Fara M, et al. Clinical characteristics of patients with lung adenocarcinomas harboring BRAF mutations. J Clin Oncol 2011;29:2046-51. [Crossref] [PubMed]
- Marchetti A, Felicioni L, Malatesta S, et al. Clinical features and outcome of patients with non-small-cell lung cancer harboring BRAF mutations. J Clin Oncol 2011;29:3574-9. [Crossref] [PubMed]
- Cardarella S, Ogino A, Nishino M, et al. Clinical, pathologic, and biologic features associated with BRAF mutations in non-small cell lung cancer. Clin Cancer Res 2013;19:4532-40. [Crossref] [PubMed]
- Tissot C, Couraud S, Tanguy R, et al. Clinical characteristics and outcome of patients with lung cancer harboring BRAF mutations. Lung Cancer 2016;91:23-8. [Crossref] [PubMed]
- Villaruz LC, Socinski MA, Abberbock S, et al. Clinicopathologic features and outcomes of patients with lung adenocarcinomas harboring BRAF mutations in the Lung Cancer Mutation Consortium. Cancer 2015;121:448-56. [Crossref] [PubMed]
- Ross JS, Wang K, Chmielecki J, et al. The distribution of BRAF gene fusions in solid tumors and response to targeted therapy. Int J Cancer 2016;138:881-90. [Crossref] [PubMed]
- Hutchinson KE, Lipson D, Stephens PJ, et al. BRAF fusions define a distinct molecular subset of melanomas with potential sensitivity to MEK inhibition. Clin Cancer Res 2013;19:6696-702. [Crossref] [PubMed]
- Sievert AJ, Lang SS, Boucher KL, et al. Paradoxical activation and RAF inhibitor resistance of BRAF protein kinase fusions characterizing pediatric astrocytomas. Proc Natl Acad Sci U S A 2013;110:5957-62. [Crossref] [PubMed]
- Planchard D, Kim TM, Mazieres J, et al. Dabrafenib in patients with BRAF(V600E)-positive advanced non-small-cell lung cancer: a single-arm, multicentre, open-label, phase 2 trial. Lancet Oncol 2016;17:642-50. [Crossref] [PubMed]
- Hyman DM, Puzanov I, Subbiah V, et al. Vemurafenib in Multiple Nonmelanoma Cancers with BRAF V600 Mutations. N Engl J Med 2015;373:726-36. [Crossref] [PubMed]
- Planchard D, Besse B, Groen HJ, et al. Dabrafenib plus trametinib in patients with previously treated BRAF(V600E)-mutant metastatic non-small cell lung cancer: an open-label, multicentre phase 2 trial. Lancet Oncol 2016;17:984-93. [Crossref] [PubMed]
- Yao Z, Torres NM, Tao A, et al. BRAF Mutants Evade ERK-Dependent Feedback by Different Mechanisms that Determine Their Sensitivity to Pharmacologic Inhibition. Cancer Cell 2015;28:370-83. [Crossref] [PubMed]
- Masters GA, Temin S, Azzoli CG, et al. Systemic Therapy for Stage IV Non-Small-Cell Lung Cancer: American Society of Clinical Oncology Clinical Practice Guideline Update. J Clin Oncol 2015;33:3488-515. [Crossref] [PubMed]
- Azzoli CG, Engelman J, Fidias P, et al. Genotyping lung cancer is an investment in the future. J Clin Oncol 2014;32:3576-7. [Crossref] [PubMed]


