Tumor heterogeneity in small cell lung cancer defined and investigated in pre-clinical mouse models
Small cell lung carcinoma (SCLC) is a neuroendocrine subtype of lung cancer characterized by its fast growth and aggressive nature. SCLC development is strongly associated with heavy cigarette smoking. In a majority of cases, SCLC tumors have already metastasized to distant sites at the time of first diagnosis. Except in rare cases when tumors are diagnosed early, treatment options are limited and usually consist of several rounds of chemotherapy with cisplatin/carboplatin and etoposide. While this chemotherapy regimen often results in significant response and tumor shrinkage, relapse is usually quite rapid. A number of excellent reviews have recently discussed the key clinical features of SCLC and the current lack of efficient treatment despite a large number of clinical trials in the past three decades (1-4). New therapeutic approaches, including immunotherapies, are promising but at the time this review is written, there are still no approved targeted therapies for SCLC [reviewed in (5-8)].
Here we discuss emerging data in the field on the role of tumor heterogeneity in the growth of SCLC and the response of these tumors to therapy, and how mouse models have been used to identify such tumor heterogeneity and investigate its role. We use the term “intertumoral” heterogeneity to describe how SCLC tumors in patients or mice evolve differently at the genetic and epigenetic level during tumor progression and metastasis. We use the term “intratumoral” heterogeneity to describe the different subpopulations of cells within tumors at any given time; in this review, we mostly focus on different subpopulations of cancer cells and we only briefly discuss non-cancer cells such as immune cells in the tumor microenvironment.
Different approaches to model SCLC in mice
We consider two types of pre-clinical mouse models, genetically engineered mouse models (GEMMs) in which mutant mice develop autochthonous tumors upon targeted alterations in cancer genes, and mouse-derived allografts and patient-derived xenografts (PDXs) that grow in mice upon transplantation, either from tumors or from circulating tumor cells (CTCs) (Figure 1).
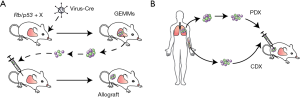
Autochthonous GEMMs of SCLC
GEMMs for SCLC and their relevance to the human disease have been reviewed in detail elsewhere (9-11). Briefly, these models are based on the near-ubiquitous inactivation of the RB and p53 tumor suppressors seen in human SCLC (12). Meuwissen, Berns, and colleagues first showed that deletion of the corresponding mouse genes in the lungs of adult mice leads to the development of tumors that are strikingly similar to human SCLC in their histology and their metastatic ability; the basic strategy has been to use adenoviral delivery of the Cre recombinase (Ad-Cre) in Rbflox/flox; p53flox/flox conditional knockout mice (13). Beyond loss of RB and p53, which is required for SCLC development, a number of other genes and signaling pathways are altered in SCLC tumors (12) and the roles of these genes/pathways can be tested in GEMMs. Ectopic expression of oncogenes such as Myc, Nfib or oncogenic forms of the Hedgehog pathway receptor Smo (14-17), or deletion of tumor suppressors such as the Rb family members p130 or Pten (18-20) in the Rb/p53 double knockout (DKO) model has led to new models with faster tumor development. With recent advances in genome engineering, including CRISPR/Cas9, it is likely that additional models will be soon available to the community. The addition of fluorescent reporters whose expression is turned on by the Cre recombinase in these mutant mice helps in tracking and purifying cancer cells as they grow from small lesions to fully metastatic tumors (21). Similarly, Cre-inducible expression of luciferase can be incorporated to monitor and quantify tumor development in situ (14,22).
Allograft and xenograft models
In autochthonous GEMMs, tumors initiate in the lungs of mice, grow in the lung microenvironment in animals with an intact immune system, and can spread to distant organs and tissues using mechanisms that are similar to SCLC patients. One disadvantage of GEMMs, however, is often the rather slow development of tumors (6–12 months in SCLC models). This issue can be in part solved by the use of allograft models: mouse tumors (primary tumors or metastases developing in GEMMs) that can be dissected, transplanted, and expanded in recipient mice. The use of syngeneic genetic background allows for studies in immunocompetent mice (23,24), which can be important for example when new immunotherapies are being evaluated.
Murine SCLC can of course only serve as a model for the human disease. Thus, it is important to develop parallel approaches to investigate the mechanisms underlying SCLC development and response to therapy in additional models, including PDXs (25,26). Currently, SCLC PDX models have only been studied in immunocompromised mice, which limits their utility for studies of the interaction between cancer cells and immune cells; the development of humanized xenograft models may eventually help resolve this issue (27,28). Nevertheless, classical PDX models can be used to investigate some of the roles of the innate immune system (23) and can prove extremely useful to investigate the response of human SCLC to chemotherapy or targeted therapies (29,30). It is also noteworthy, however, that it has been difficult to grow SCLC allografts or xenografts orthotopically [e.g., (23)] whereas tumors rapidly expanded outside the lungs, thus PDX studies are performed in a different microenvironment than the lungs. Tail vein injection of SCLC cells leads to the growth of tumors in the liver, which can provide a model for SCLC metastasis to the liver (21). A truly exciting development in the field has been the observation that SCLC patients often have more CTCs than what is found in other cancer types. This allows for the generation of CTC-derived explant (CDX) models (xenografts derived from CTCs), which are powerful tools in longitudinal studies, and the generation of large numbers of xenograft models from many patients (31,32).
GEMMs, allografts, and PDX/CDX models provide complementary systems to investigate the biology of SCLC, including tumor heterogeneity.
Intratumoral heterogeneity in SCLC
Evidence of intratumoral heterogeneity in SCLC was demonstrated more than 30 years ago by the heterogeneity of cell surface expression of a variety of antigens detectable with monoclonal antibodies (33). However, this heterogeneity has not been investigated at the functional level until more recently, including in mouse models. Emerging evidence indicates that neuroendocrine cancer cells can give rise to a number of non-neuroendocrine cancer cells that help promote tumor growth, including in response to chemotherapy (Figure 2).
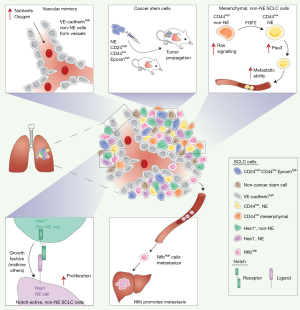
SCLC long-term propagating cells are neuroendocrine
Cancer stem cells (CSCs) are often viewed as the counterpart of normal adult stem cells in tumors. CSCs [also sometimes referred to as tumor re-initiating cells (TRICs), or tumor propagating cells (TPCs)] are thought to support long-term tumorigenic growth and determine the cellular heterogeneity of a primary tumor [see recent reviews (34,35)]. The establishment of cell lines in culture and even PDX models can result in the selection of specific subpopulations of cancer cells [e.g., (36,37)], thereby introducing bias in the identification of subpopulations that may exist in tumors in vivo. The analysis of human primary SCLC tumors is rendered difficult by rare surgeries and small biopsies. In contrast, in GEMMs, a large number of primary tumors at multiple stages of tumor development can be studied in vivo.
A number of cell surface molecules have been identified as possible markers for CSCs in SCLC cell lines in culture (38-43). However, when we examined cells for the ability to serially transplant and generate various subpopulations of tumor cells in a GEMM model of SCLC [Rb/p130/p53, triple knockout (TKO) (19)], we identified different markers, and CD24high CD44low EpCAMhigh SCLC cells were found to strongly enrich for CSCs (44). Importantly, these CD24high CD44low EpCAMhigh SCLC cells represent ~50% of live tumor cells, indicating that CSCs are not a rare subpopulation of cancer cells. The analysis of PDX/CDX models showed that the same markers identify tumor-propagating cells in human tumors, with similar frequencies. Notably, SCLC CSCs express high levels of markers of neuroendocrine differentiation, including Ascl1 (also known as Mash1), which is an oncogenic transcription factor in human SCLC cell lines (38,45).
These experiments do not exclude that subpopulations of cells exist within the neuroendocrine CD24high CD44low EpCAMhigh CSC pool, cells that may have even greater ability to self-renew and propagate tumors. Identifying such subpopulations using a combination of functional assays and single-cell analysis approaches may help refine our understanding of the cells that drive the long-term growth of SCLC.
The clinical relevance of these observations is best exemplified by recent studies showing that a subset of SCLC CSCs express Dll3, an atypical Notch ligand and Ascl1 transcriptional target, at their surface (46,47): targeting these cells with an antibody-drug conjugate may represent a potent way to eliminate SCLC CSCs and block the expansion of tumors in patients (30).
SCLC cells with a mesenchymal non-neuroendocrine phenotype
If, in a first approximation, ~50% of SCLC cells with strong neuroendocrine features have the capacity to act as stem cells for this neuroendocrine cancer type, this raises the question of the identity and the possible function of other subpopulations of cells in this tumor type. The Berns lab identified a population of non-neuroendocrine SCLC cells characterized by increased expression of mesenchymal markers (e.g., vimentin) and high levels of CD44 at the cell surface in Rb/p53 mutant mice (48,49). These CD44high SCLC cells can be generated by activation of signaling pathways such as Ras signaling. Their frequency in mouse tumors may only be a few percent, but these CD44high cells can significantly enhance the metastatic ability of neuroendocrine SCLC cells, in part using a paracrine mechanism involving secretion of FGF2 by CD44high cells and activation of the Pea3 transcription factor in neuroendocrine SCLC cells. The signals that induce neuroendocrine SCLC cells to become more mesenchymal include Ras signaling but are not fully understood. In addition, the frequency and the possible role of these CD44high SCLC cells in human SCLC remain to be determined. Finally, CD44high SCLC cells from the mutant mice can form tumors when transplanted in recipient mice (48) but it is unclear if similar cells have the same tumorigenic potential in humans.
Activation of Notch signaling within SCLC tumors and the generation of non-neuroendocrine SCLC cells
The genomic analysis of human SCLC tumors has identified recurrent mutations in the genes coding for Notch receptors; these mutations have not been functionally validated but are predicted to be most often loss-of-function events based on their location in the genes and their effects on the coding sequence (12). Accordingly, ectopic activation of Notch signaling can acutely inhibit the expansion of SCLC cells in culture and in mice (12,50-52). However, our recent work has shown that tumors in GEMMs of SCLC harbor a population of cells with activation of the Notch signaling pathway, including expression of the target gene Hes1 (52). These cells are cancer cells that have lost their neuroendocrine features, they do not express mesenchymal markers and are distinct from the CD44high SCLC cells (52). These non-neuroendocrine Hes1high cells are generated from neuroendocrine cancer cells after activation of Notch signaling within tumors. A mechanism underlying the switch from neuroendocrine to non-neuroendocrine is likely to be degradation or inhibition of Ascl1 expression (53,54) but also involves induction of Rest/NRSF, which acts as a repressor of neuronal and neuroendocrine programs in SCLC cells (52). Importantly, these Hes1high SCLC cells functionally interact with neuroendocrine tumor cells and promote their growth in ex vivo assays; these cells may also influence the growth of tumors under stress conditions such as chemotherapy. These studies provide a second example of intra-tumoral heterogeneity in which one subpopulation of cancer cells (non-neuroendocrine Hes1+ SCLC cells) can enhance the growth of neuroendocrine tumors. It is interesting to note that Notch signaling can act intrinsically as a tumor-suppressive mechanism in neuroendocrine SCLC cells but also as a pro-tumorigenic mechanism in a tumor-intrinsic, non-cell autonomous way in these tumors. Thus, strategies to activate or inhibit Notch therapeutically may both be beneficial to patients depending on the cellular make-up of their tumor.
Vascular mimicry in SCLC
The Dive lab recently described a third level of intratumoral heterogeneity in primary tumors and CDX models. A fraction of SCLC cells in tumors or in the circulation have markers of vascular mimicry, including expression of VE-Cadherin. Higher levels of vascular mimicry were associated with decreased overall survival in SCLC patients. Importantly, expression of VE-Cadherin is critical for vascular mimicry and reduced levels of VE-Cadherin result in slower tumor growth as well as increased efficiency of chemotherapy (55). It is possible that vascular mimicry helps supply nutrient and oxygen required for the expansion of neuroendocrine SCLC cells. Whether VE-Cadherin+ cells exist in SCLC GEMMs is not known. The signals that can induce the differentiation of SCLC cells into endothelial-like cells is also not yet known but may be linked to oxygen levels.
It is important to note that the CD44, Hes1, and VE-Cadherin markers do not take into account transition states between neuroendocrine and non-neuroendocrine states. The current studies also fail to examine thoroughly whether these states are reversible or even if some SCLC cells can switch from one non-neuroendocrine state to another. Lineage-tracing experiments in mice and single-cell studies will be critical to define all the subpopulations of cancer cells within SCLC tumors and their dynamics, including whether and how these currently known subpopulations overlap and interact with each other.
Non-cancer cells in SCLC tumors
The studies discussed above indicate that a number of subpopulations of cancer cells exist within SCLC tumors. Sections from mouse or human SCLC tumors often reveal few non-tumor cells such as immune cells or fibroblasts; indeed, quantification of tumor purity in genomic studies indicates that human primary SCLC tumors are more than 80% composed of cancer cells (12). Still, a number of human tumors contain immune cells and a higher number of T cells and macrophages correlates with favorable outcomes in patients (56). Long-term survivors of SCLC also maintained a high ratio of effector T cells to regulatory T cells (57). Our group has shown that inhibition of the “marker of self” CD47 [recently reviewed in (58)] can activate the innate immune system and induce the phagocytosis of SCLC in allograft and PDX models (23). Our goal here is not to provide an extensive discussion about the role of immune cells in SCLC development and as targets for therapy but to underscore that non-cancer cells can certainly play a key role in SCLC and probably interact with all the subpopulations of SCLC cells; the populations of immune cells and their interactions with other cells in tumors may also be affected by chemotherapy and other forms of treatment.
Inter-tumoral heterogeneity in SCLC
Intratumoral heterogeneity in SCLC tumors may be influenced by multiple factors, including tumor evolution over time. SCLC tumors have one of the highest mutation burdens among human tumors (12) and each tumor may have its own set of mutations. Still, emerging evidence shows that SCLC tumors may be separated into distinct subgroups based on their genetic profile and that this intertumoral heterogeneity may help design personalized therapeutic approaches.
Genomic studies of human SCLC and genetic diversity
A large number of studies over the past four decades have identified recurrent mutations in the genes coding for the Rb and p53 tumor suppressors in human SCLC. Genome sequencing efforts, including whole genome sequencing, have demonstrated that these mutations are nearly ubiquitous (12,59,60). These data and experiments showing that loss of both Rb and p53 is required for the development of tumors in mice (13) demonstrate that inactivation of these two genes is a critical requirement for SCLC initial development. However, SCLC tumors may then acquire different alterations as they evolve or as they progress to a metastatic state.
RNA-seq analysis indicates that a large fraction of human primary tumors (~75%) have high levels of neuroendocrine genes and low levels of Notch signaling; the remaining 25% have lower levels of some key neuroendocrine genes such as ASCL1 but it is unclear whether these gene expression differences truly represent distinct SCLC subtypes, especially since these differences are not correlated with specific genetic events (12). In addition, even though genes like EP300, CREBBP, TP73, or NOTCH1-4 are frequently mutated and genes like MYC family genes or IRS2 are recurrently amplified, there is also no clear pattern of overlapping or non-overlapping combinations between these events (12). This is possibly due to the still limited number of samples that have been analyzed in depth. Moreover, with the exception of Notch signaling (12), two Myc family members (c-Myc and L-Myc) (14), and Pten (18,20), very few of these recurrent alterations have been functionally tested in GEMMs or even PDX models. Thus, it is difficult to know if these genetic alterations represent distinct subtypes of SCLC that may have different growth properties or may respond to various therapies differently. However, mouse tumors acquire similar mutations as human tumors do during tumor progression, including loss of Pten, amplification of L-Myc, or loss-of-function mutations in Notch genes (12,18,59), providing further support of the relevance of these GEMMs.
c-Myc-driven “variant” SCLC
High levels of c-MYC are found in human tumors either as a result of DNA amplification or by unknown transcriptional mechanisms (12). Ectopic expression of c-Myc in preneoplastic, Rb/p53/p130 mutant neuroendocrine lung epithelial cells is sufficient to transform these cells (61). High levels of c-Myc (in its more stable form MycT58A) rapidly transform Rb/p53 mutant neuroendocrine lung epithelial cells (17). These Rb/p53/Myc mutant tumors express lower levels of neuroendocrine markers compared to classical SCLC GEMMs and express low levels of Ascl1 and high levels of NeuroD1 (17). Interestingly, these features are reminiscent of a so-called “variant” subtype of human SCLC (as opposed to a “classic” neuroendocrine phenotype), which was initially identified in SCLC cell lines (62-65). Importantly, this variant subtype of SCLC with high levels of c-Myc may be particularly sensitive to inhibitors of Aurora kinase (17,66), providing an elegant demonstration that genotype can determine therapeutic response in SCLC. MYC expression may also be a marker of sensitivity to CHK1 inhibitors (24).
Metastatic SCLC
Little is known about metastatic SCLC in large part because of limited tumor samples. This is changing with the development of CDX models from CTCs (which may be closely related to metastases) (31,32). However, GEMMs remain a key system in which to study the mechanisms of metastatic progression. Recently, three studies have highlighted a major role for the Nfib transcription factor in the metastatic progression of SCLC (14,15,21) (Figure 2). The Nfib gene is frequently amplified in mouse tumors as they become metastatic (21,67); in human tumors NFIB is more rarely amplified (12,59,60) but high expression is found in over 50% of metastases. We still do not know whether SCLC tumors with high levels of Nfib are more sensitive to specific targeted therapies but their faster growth may render them more sensitive to chemotherapy, at least initially (14).
Chemoresistant SCLC
While the majority of SCLC tumors in patients are initially responsive to chemotherapy, tumors usually relapse rapidly and when they do they are often resistant to the initial drugs as well as other chemotherapeutics. Chemoresistance may arise from the extensive mutational burden in SCLC tumors and the growth of resistant clones. In tumors that are intrinsically chemorefractory, recent evidence using CDX models indicates that specific copy-number aberrations are present (32). However, clonal analysis of SCLC tumors versus lung adenocarcinoma indicates that SCLC tumors have actually less subclonal diversity than these other tumors and the level of subclonal heterogeneity does not correlate with clinical stage (12). This clonal analysis suggests that genetic mechanisms such as point mutations may not be driving the evolution towards chemoresistance.
Notably, long-term tumor-propagating cells in a mouse model of SCLC are not more inherently chemoresistant than other cell subpopulations within tumors (44). These CSCs may even be more chemosensitive due to their fast proliferative rate. In contrast, slower-growing non-neuroendocrine SCLC cells in tumors, such as Notch-active SCLC cells may be inherently more resistant to chemotherapeutics as shown in a GEMM (discussed above) (52) and may help the neuroendocrine cells survive chemotherapy. SCLC cells with a mesenchymal phenotype may also be intrinsically more chemoresistant (68,69). Furthermore, interactions of SCLC cells with their microenvironment, including the extracellular matrix, may promote chemoresistance, although this has not been tested in animal models (70-74).
Recent data in PDX models by Poirier, Rudin, and colleagues suggest that epigenetic mechanisms may be at play during the acquisition of chemoresistance. Their data point to activation of the lysine methyltransferase EZH2 in chemoresistant PDXs and silencing of the DNA-damage repair SLFN11 as a major axis in the establishment of chemoresistance (29). SLFN11 expression may also mark tumors that are resistant to other drugs such as PARP inhibitors (75).
Conclusions
SCLC remains one of the most lethal forms of cancer, a disease for which therapeutic options are still extremely limited. Except for the very small number of SCLC patients diagnosed with early-stage cancer, survival rates are dismal. Recent work, in large part from animal models such as genetically-engineered mouse models and PDX/CDX models has shown new levels of cellular complexity in SCLC. The presence of various subpopulations of SCLC cells and their functional interactions may explain the outstanding plasticity of SCLC tumors and their striking metastatic potential. It will be essential in the future to relate this intratumoral heterogeneity with intertumoral heterogeneity, including how specific genetic or epigenetic events shape the cellular composition of tumors. For instance, tumors with mutations in NOTCH genes may harbor fewer non-neuroendocrine Hes1pos cells, but this has not been formally determined yet.
Combining these pre-clinical models with novel technologies such as single-cell analysis by RNA sequencing or mass cytometry (76) may prove useful to identify novel subsets of SCLC cells at different stages of SCLC evolution and after chemotherapy. Another key future step will be the development of better models to investigate how specific genetic events may lead to the development of distinct subtypes of SCLC tumors. One promising approach will be to use pre-neoplastic SCLC and test specific oncogenes or tumor suppressors (61); another approach may be to develop faster GEMMs using CRISPR/Cas9 approaches in vivo to manipulate specific genes and specific populations of cells. Faster modeling approaches may become critical as recent studies point to a number of new targets and possible biomarkers predictive of sensitivity or resistance to new therapeutics [e.g., AXL expression and resistance to Wee1 inhibition (77), p53 point mutations and chemotherapy response (78), or DNA methylation as a biomarker for the effects of LSD1 inhibitors (79)].
As an important note, it is striking that most studies with GEMMs study dozens of mice that often themselves carry dozens of clonally-distinct tumors while studies with allografts or xenografts nearly always involve at most a handful of independent models in each study. Mouse tumors do not accumulate many additional mutations beyond the initiating events and one way around this caveat might be to introduce mutations in genes whose loss will result in increased genetic instability (e.g., mismatch repair genes); this way, mouse tumors may resemble human tumors more. Conversely, the development of CDX models (directly from CTCs in the blood of patients) may allow the field to generate larger collections of human models, thereby limiting the caveats of working with a few samples that may not be representative of the disease in patients.
A better knowledge of intratumoral and intertumoral heterogeneity from animal models and from cell lines in culture is likely to be a requisite before molecular targeted therapies can be designed to fight SCLC.
Acknowledgements
The authors thank members of the Sage lab for helpful discussions.
Funding: This work was supported by the National Cancer Institute R01CA206540, R01CA201513 and U01CA213273 (to J Sage). YT Shue and JS Lim were supported by A*STAR Singapore. J Sage is the Harriet and Mary Zelencik Scientist in Children’s Cancer and Blood Diseases.
Footnote
Conflicts of Interest: YT Shue and JS Lim declare no financial interests. J Sage receives research funding from StemCentrx/Abbvie and is an inventor on patent application PCT/US2015/010650 “Targeted therapy for small cell lung cancer”.
References
- Kalemkerian GP. Small Cell Lung Cancer. Semin Respir Crit Care Med 2016;37:783-96. [Crossref] [PubMed]
- Pietanza MC, Byers LA, Minna JD, et al. Small cell lung cancer: will recent progress lead to improved outcomes? Clin Cancer Res 2015;21:2244-55. [Crossref] [PubMed]
- Carter BW, Glisson BS, Truong MT, et al. Small cell lung carcinoma: staging, imaging, and treatment considerations. Radiographics 2014;34:1707-21. [Crossref] [PubMed]
- Kalemkerian GP, Akerley W, Bogner P, et al. Small cell lung cancer. J Natl Compr Canc Netw 2013;11:78-98. [Crossref] [PubMed]
- Lehman JM, Gwin ME, Massion PP. Immunotherapy and Targeted Therapy for Small Cell Lung Cancer: There Is Hope. Curr Oncol Rep 2017;19:49. [Crossref] [PubMed]
- Sabari JK, Lok BH, Laird JH, et al. Unravelling the biology of SCLC: implications for therapy. Nat Rev Clin Oncol 2017;14:549-61. [Crossref] [PubMed]
- Patel SH, Rimner A, Cohen RB. Combining immunotherapy and radiation therapy for small cell lung cancer and thymic tumors. Transl Lung Cancer Res 2017;6:186-95. [Crossref] [PubMed]
- Bunn PA Jr, Minna JD, Augustyn A, et al. Small Cell Lung Cancer: Can Recent Advances in Biology and Molecular Biology Be Translated into Improved Outcomes? J Thorac Oncol 2016;11:453-74. [Crossref] [PubMed]
- Gazdar AF, Savage TK, Johnson JE, et al. The comparative pathology of genetically engineered mouse models for neuroendocrine carcinomas of the lung. J Thorac Oncol 2015;10:553-64. [Crossref] [PubMed]
- Semenova EA, Nagel R, Berns A. Origins, genetic landscape, and emerging therapies of small cell lung cancer. Genes Dev 2015;29:1447-62. [Crossref] [PubMed]
- Kwon MC, Berns A. Mouse models for lung cancer. Mol Oncol 2013;7:165-77. [Crossref] [PubMed]
- George J, Lim JS, Jang SJ, et al. Comprehensive genomic profiles of small cell lung cancer. Nature 2015;524:47-53. [Crossref] [PubMed]
- Meuwissen R, Linn SC, Linnoila RI, et al. Induction of small cell lung cancer by somatic inactivation of both Trp53 and Rb1 in a conditional mouse model. Cancer Cell 2003;4:181-9. [Crossref] [PubMed]
- Semenova EA, Kwon MC, Monkhorst K, et al. Transcription Factor NFIB Is a Driver of Small Cell Lung Cancer Progression in Mice and Marks Metastatic Disease in Patients. Cell Rep 2016;16:631-43. [Crossref] [PubMed]
- Wu N, Jia D, Ibrahim AH, et al. NFIB overexpression cooperates with Rb/p53 deletion to promote small cell lung cancer. Oncotarget 2016;7:57514-24. [PubMed]
- Park KS, Martelotto LG, Peifer M, et al. A crucial requirement for Hedgehog signaling in small cell lung cancer. Nat Med 2011;17:1504-8. [Crossref] [PubMed]
- Mollaoglu G, Guthrie MR, Bohm S, et al. MYC Drives Progression of Small Cell Lung Cancer to a Variant Neuroendocrine Subtype with Vulnerability to Aurora Kinase Inhibition. Cancer Cell 2017;31:270-85. [Crossref] [PubMed]
- McFadden DG, Papagiannakopoulos T, Taylor-Weiner A, et al. Genetic and clonal dissection of murine small cell lung carcinoma progression by genome sequencing. Cell 2014;156:1298-311. [Crossref] [PubMed]
- Schaffer BE, Park KS, Yiu G, et al. Loss of p130 accelerates tumor development in a mouse model for human small-cell lung carcinoma. Cancer Res 2010;70:3877-83. [Crossref] [PubMed]
- Cui M, Augert A, Rongione M, et al. PTEN is a Potent Suppressor of Small Cell Lung Cancer. Mol Cancer Res 2014;12:654-9. [Crossref] [PubMed]
- Denny SK, Yang D, Chuang CH, et al. Nfib Promotes Metastasis through a Widespread Increase in Chromatin Accessibility. Cell 2016;166:328-42. [Crossref] [PubMed]
- Jahchan NS, Dudley JT, Mazur PK, et al. A drug repositioning approach identifies tricyclic antidepressants as inhibitors of small cell lung cancer and other neuroendocrine tumors. Cancer Discov 2013;3:1364-77. [Crossref] [PubMed]
- Weiskopf K, Jahchan NS, Schnorr PJ, et al. CD47-blocking immunotherapies stimulate macrophage-mediated destruction of small-cell lung cancer. J Clin Invest 2016;126:2610-20. [Crossref] [PubMed]
- Sen T, Tong P, Stewart CA, et al. CHK1 inhibition in small cell lung cancer produces single-agent activity in biomarker-defined disease subsets and combination activity with cisplatin or olaparib. Cancer Res 2017;77:3870-84. [Crossref] [PubMed]
- Zitvogel L, Pitt JM, Daillere R, et al. Mouse models in oncoimmunology. Nat Rev Cancer 2016;16:759-73. [Crossref] [PubMed]
- Gould SE, Junttila MR, de Sauvage FJ. Translational value of mouse models in oncology drug development. Nat Med 2015;21:431-9. [Crossref] [PubMed]
- Byrne AT, Alferez DG, Amant F, et al. Interrogating open issues in cancer precision medicine with patient-derived xenografts. Nat Rev Cancer 2017;17:254-68. [Crossref] [PubMed]
- Morton JJ, Bird G, Refaeli Y, et al. Humanized Mouse Xenograft Models: Narrowing the Tumor-Microenvironment Gap. Cancer Res 2016;76:6153-8. [Crossref] [PubMed]
- Gardner EE, Lok BH, Schneeberger VE, et al. Chemosensitive Relapse in Small Cell Lung Cancer Proceeds through an EZH2-SLFN11 Axis. Cancer Cell 2017;31:286-99. [Crossref] [PubMed]
- Saunders LR, Bankovich AJ, Anderson WC, et al. A DLL3-targeted antibody-drug conjugate eradicates high-grade pulmonary neuroendocrine tumor-initiating cells in vivo. Sci Transl Med 2015;7:302ra136. [Crossref] [PubMed]
- Hodgkinson CL, Morrow CJ, Li Y, et al. Tumorigenicity and genetic profiling of circulating tumor cells in small-cell lung cancer. Nat Med 2014;20:897-903. [Crossref] [PubMed]
- Carter L, Rothwell DG, Mesquita B, et al. Molecular analysis of circulating tumor cells identifies distinct copy-number profiles in patients with chemosensitive and chemorefractory small-cell lung cancer. Nat Med 2017;23:114-9. [Crossref] [PubMed]
- Fargion S, Carney D, Mulshine J, et al. Heterogeneity of cell surface antigen expression of human small cell lung cancer detected by monoclonal antibodies. Cancer Res 1986;46:2633-8. [PubMed]
- O’Brien-Ball C, Biddle A. Reprogramming to developmental plasticity in cancer stem cells. Dev Biol 2017;430:266-74. [Crossref] [PubMed]
- Nassar D, Blanpain C. Cancer Stem Cells: Basic Concepts and Therapeutic Implications. Annu Rev Pathol 2016;11:47-76. [Crossref] [PubMed]
- Daniel VC, Marchionni L, Hierman JS, et al. A primary xenograft model of small-cell lung cancer reveals irreversible changes in gene expression imposed by culture in vitro. Cancer Res 2009;69:3364-73. [Crossref] [PubMed]
- Leong TL, Marini KD, Rossello FJ, et al. Genomic characterisation of small cell lung cancer patient-derived xenografts generated from endobronchial ultrasound-guided transbronchial needle aspiration specimens. PLoS One 2014;9:e106862. [Crossref] [PubMed]
- Jiang T, Collins BJ, Jin N, et al. Achaete-scute complex homologue 1 regulates tumor-initiating capacity in human small cell lung cancer. Cancer Res 2009;69:845-54. [Crossref] [PubMed]
- Sarvi S, Mackinnon AC, Avlonitis N, et al. CD133+ cancer stem-like cells in small cell lung cancer are highly tumorigenic and chemoresistant but sensitive to a novel neuropeptide antagonist. Cancer Res 2014;74:1554-65. [Crossref] [PubMed]
- Salcido CD, Larochelle A, Taylor BJ, et al. Molecular characterisation of side population cells with cancer stem cell-like characteristics in small-cell lung cancer. Br J Cancer 2010;102:1636-44. [Crossref] [PubMed]
- Wang P, Gao Q, Suo Z, et al. Identification and characterization of cells with cancer stem cell properties in human primary lung cancer cell lines. PLoS One 2013;8:e57020. [Crossref] [PubMed]
- Micke P, Basrai M, Faldum A, et al. Characterization of c-kit expression in small cell lung cancer: prognostic and therapeutic implications. Clin Cancer Res 2003;9:188-94. [PubMed]
- Rygaard K, Nakamura T, Spang-Thomsen M. Expression of the proto-oncogenes c-met and c-kit and their ligands, hepatocyte growth factor/scatter factor and stem cell factor, in SCLC cell lines and xenografts. Br J Cancer 1993;67:37-46. [Crossref] [PubMed]
- Jahchan NS, Lim JS, Bola B, et al. Identification and Targeting of Long-Term Tumor-Propagating Cells in Small Cell Lung Cancer. Cell Rep 2016;16:644-56. [Crossref] [PubMed]
- Augustyn A, Borromeo M, Wang T, et al. ASCL1 is a lineage oncogene providing therapeutic targets for high-grade neuroendocrine lung cancers. Proc Natl Acad Sci U S A 2014;111:14788-93. [Crossref] [PubMed]
- Henke RM, Meredith DM, Borromeo MD, et al. Ascl1 and Neurog2 form novel complexes and regulate Delta-like3 (Dll3) expression in the neural tube. Dev Biol 2009;328:529-40. [Crossref] [PubMed]
- Geffers I, Serth K, Chapman G, et al. Divergent functions and distinct localization of the Notch ligands DLL1 and DLL3 in vivo. J Cell Biol 2007;178:465-76. [Crossref] [PubMed]
- Calbo J, van Montfort E, Proost N, et al. A functional role for tumor cell heterogeneity in a mouse model of small cell lung cancer. Cancer Cell 2011;19:244-56. [Crossref] [PubMed]
- Kwon MC, Proost N, Song JY, et al. Paracrine signaling between tumor subclones of mouse SCLC: a critical role of ETS transcription factor Pea3 in facilitating metastasis. Genes Dev 2015;29:1587-92. [Crossref] [PubMed]
- Sriuranpong V, Borges MW, Ravi RK, et al. Notch signaling induces cell cycle arrest in small cell lung cancer cells. Cancer Res 2001;61:3200-5. [PubMed]
- Wael H, Yoshida R, Kudoh S, et al. Notch1 signaling controls cell proliferation, apoptosis and differentiation in lung carcinoma. Lung Cancer 2014;85:131-40. [Crossref] [PubMed]
- Lim JS, Ibaseta A, Fischer MM, et al. Intratumoural heterogeneity generated by Notch signalling promotes small-cell lung cancer. Nature 2017;545:360-4. [Crossref] [PubMed]
- Sriuranpong V, Borges MW, Strock CL, et al. Notch signaling induces rapid degradation of achaete-scute homolog 1. Mol Cell Biol 2002;22:3129-39. [Crossref] [PubMed]
- Chen H, Thiagalingam A, Chopra H, et al. Conservation of the Drosophila lateral inhibition pathway in human lung cancer: a hairy-related protein (HES-1) directly represses achaete-scute homolog-1 expression. Proc Natl Acad Sci U S A 1997;94:5355-60. [Crossref] [PubMed]
- Williamson SC, Metcalf RL, Trapani F, et al. Vasculogenic mimicry in small cell lung cancer. Nat Commun 2016;7:13322. [Crossref] [PubMed]
- Eerola AK, Soini Y, Paakko P. A high number of tumor-infiltrating lymphocytes are associated with a small tumor size, low tumor stage, and a favorable prognosis in operated small cell lung carcinoma. Clin Cancer Res 2000;6:1875-81. [PubMed]
- Koyama K, Kagamu H, Miura S, et al. Reciprocal CD4+ T-cell balance of effector CD62Llow CD4+ and CD62LhighCD25+ CD4+ regulatory T cells in small cell lung cancer reflects disease stage. Clin Cancer Res 2008;14:6770-9. [Crossref] [PubMed]
- Weiskopf K. Cancer immunotherapy targeting the CD47/SIRPalpha axis. Eur J Cancer 2017;76:100-9. [Crossref] [PubMed]
- Peifer M, Fernandez-Cuesta L, Sos ML, et al. Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat Genet 2012;44:1104-10. [Crossref] [PubMed]
- Rudin CM, Durinck S, Stawiski EW, et al. Comprehensive genomic analysis identifies SOX2 as a frequently amplified gene in small-cell lung cancer. Nat Genet 2012;44:1111-6. [Crossref] [PubMed]
- Kim DW, Wu N, Kim YC, et al. Genetic requirement for Mycl and efficacy of RNA Pol I inhibition in mouse models of small cell lung cancer. Genes Dev 2016;30:1289-99. [Crossref] [PubMed]
- Carney DN, Gazdar AF, Bepler G, et al. Establishment and identification of small cell lung cancer cell lines having classic and variant features. Cancer Res 1985;45:2913-23. [PubMed]
- de Leij L, Postmus PE, Buys CH, et al. Characterization of three new variant type cell lines derived from small cell carcinoma of the lung. Cancer Res 1985;45:6024-33. [PubMed]
- Gazdar AF, Carney DN, Nau MM, et al. Characterization of variant subclasses of cell lines derived from small cell lung cancer having distinctive biochemical, morphological, and growth properties. Cancer Res 1985;45:2924-30. [PubMed]
- Borromeo MD, Savage TK, Kollipara RK, et al. ASCL1 and NEUROD1 Reveal Heterogeneity in Pulmonary Neuroendocrine Tumors and Regulate Distinct Genetic Programs. Cell Rep 2016;16:1259-72. [Crossref] [PubMed]
- Sos ML, Dietlein F, Peifer M, et al. A framework for identification of actionable cancer genome dependencies in small cell lung cancer. Proc Natl Acad Sci U S A 2012;109:17034-9. [Crossref] [PubMed]
- Dooley AL, Winslow MM, Chiang DY, et al. Nuclear factor I/B is an oncogene in small cell lung cancer. Genes Dev 2011;25:1470-5. [Crossref] [PubMed]
- Horie M, Saito A, Ohshima M, et al. YAP and TAZ modulate cell phenotype in a subset of small cell lung cancer. Cancer Sci 2016;107:1755-66. [Crossref] [PubMed]
- Canadas I, Rojo F, Taus A, et al. Targeting epithelial-to-mesenchymal transition with Met inhibitors reverts chemoresistance in small cell lung cancer. Clin Cancer Res 2014;20:938-50. [Crossref] [PubMed]
- Sethi T, Rintoul RC, Moore SM, et al. Extracellular matrix proteins protect small cell lung cancer cells against apoptosis: a mechanism for small cell lung cancer growth and drug resistance in vivo. Nat Med 1999;5:662-8. [Crossref] [PubMed]
- Tripathi SC, Fahrmann JF, Celiktas M, et al. MCAM Mediates Chemoresistance in Small-Cell Lung Cancer via the PI3K/AKT/SOX2 Signaling Pathway. Cancer Res 2017;77:4414-25. [Crossref] [PubMed]
- Ye M, Wei T, Wang Q, et al. TSPAN12 promotes chemoresistance and proliferation of SCLC under the regulation of miR-495. Biochem Biophys Res Commun 2017;486:349-56. [Crossref] [PubMed]
- Kohmo S, Kijima T, Otani Y, et al. Cell surface tetraspanin CD9 mediates chemoresistance in small cell lung cancer. Cancer Res 2010;70:8025-35. [Crossref] [PubMed]
- Hartmann TN, Burger JA, Glodek A, et al. CXCR4 chemokine receptor and integrin signaling co-operate in mediating adhesion and chemoresistance in small cell lung cancer (SCLC) cells. Oncogene 2005;24:4462-71. [Crossref] [PubMed]
- Allison Stewart C, Tong P, Cardnell RJ, et al. Dynamic variations in epithelial-to-mesenchymal transition (EMT), ATM, and SLFN11 govern response to PARP inhibitors and cisplatin in small cell lung cancer. Oncotarget 2017;8:28575-87. [PubMed]
- Leelatian N, Doxie DB, Greenplate AR, et al. Single cell analysis of human tissues and solid tumors with mass cytometry. Cytometry B Clin Cytom 2017;92:68-78. [Crossref] [PubMed]
- Sen T, Tong P, Diao L, et al. Targeting AXL and mTOR Pathway Overcomes Primary and Acquired Resistance to WEE1 Inhibition in Small-Cell Lung Cancer. Clin Cancer Res 2017;23:6239-53. [Crossref] [PubMed]
- Akeno N, Reece AL, Callahan M, et al. Trp53 Mutants Drive Neuroendocrine Lung Cancer through Loss-of-Function Mechanisms with Gain-of-Function Effects on Chemotherapy Response. Mol Cancer Ther 2017;16:2913-26. [Crossref] [PubMed]
- Mohammad HP, Smitheman KN, Kamat CD, et al. A DNA Hypomethylation Signature Predicts Antitumor Activity of LSD1 Inhibitors in SCLC. Cancer Cell 2015;28:57-69. [Crossref] [PubMed]


