Small cell lung cancer tumors and preclinical models display heterogeneity of neuroendocrine phenotypes
Introduction
Traditionally, lung cancers are divided into small cell lung cancer (SCLC) and the other forms of lung cancer collectively referred to as non-SCLC (NSCLC). SCLC is a highly deadly form of lung cancer with early metastatic spread and a 5-year survival of about 7% (1). There have been no major clinical advances for about three decades. For these reasons the US Congress has designated it as a “Recalcitrant Cancer” (2). SCLC is a high grade neuroendocrine (NE) tumor (3), and most tumors and cell lines express the entire program of NE cell differentiation.
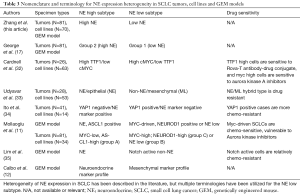
The World Health Organization (WHO) Classification recognizes SCLC as a relatively homogenous tumor, with only SCLC and combined SCLC subtypes (4). In the combined form, SCLC is admixed with one of the forms of NSCLC. Thus, other than minor variations, there is only one morphological form of SCLC officially recognized. However, about 30 years ago we described a “variant” form of SCLC cell line that differed from the typical or “classic” form of SCLC in being characterized by larger cells with distinct cell borders, a moderate amount of eosinophilic cytoplasm, and a single nucleus with a prominent central nucleolus and paranucleolar chromatin clearing (5,6). Classic lines often grew as non-adherent cell aggregates or as true spheroids. By contrast, variant cell lines had different growth characteristics, often growing as loosely aggregated or single cells, with varying degrees of substrate adherence, sometimes growing in single cell linear aggregates or “Indian file” pattern. Many variant lines were from post therapy tumors that had recurred, and Myc family amplification was more frequent than in the classic lines. The variant lines were more radioresistant than the classic lines with a prominent “shoulder” in the radioresponse curve, indicating the ability to repair and recover from lower radiation doses (7). Other investigators found similar features in SCLC cell lines that they had established (8,9). The Seneca Valley virus shows selective tropism for the variant subtype (10). In addition to tumors and cell lines, the variant subtype or its equivalent has been described in GEM models of SCLC (11,12). A summary of the published reports of the variant form and its properties are summarized in Table 1.

Full table
While the variant subtype represents a major morphological variation of SCLC, often with increased growth and cloning abilities, it is also associated with decreased or absent expression of NE cell features. Loss or decreased expression of NE features has also been described in tumors (17) and cell lines without any mention of altered morphological features. Loss of NE properties may be associated with alterations in the expression of the master transcription regulator ASCL1, a member of the basic helix-loop-helix (BHLH) family of transcription factors. The major purpose of this manuscript is to document the heterogeneity of SCLC, with respect to morphology, growth characteristics, other biological properties and the complex relationship of NE differentiation with respect to ASCL1, the Notch pathway and other inhibitors and promoters of NE differentiation. In order to do this quantitatively, we developed a scoring system for NE differentiation, which is outlined in this manuscript. We also document heterogeneity in expression of certain oncogenes associated with SCLC (Myc gene family, NFIB, MYB). We compare these features in tumors and cell lines and GEM models, as the latter are one of the major forms of in vitro models available for the study of a cancer such as SCLC when human tumor materials for study are scant (18).
Methods
Establishment of a NE score for lung cancers
Additional details about our methodology can be found elsewhere, where we used a similar approach to develop a gene expression identifier for the classification of NSCLC tumors (19). Multiple steps were performed to generate a NE score. First, expression array data (Affymetrix HuGene 1.0 ST) were compared between 8 matched normal adrenal medulla (NE) samples and 8 normal adrenal cortex (non-NE) samples. From a volcano plot, we identified 50 genes differentially expressed between the two groups (25 NE and 25 non-NE genes). We next applied this adrenal-based signature to a large panel of SCLC and NSCLC lines (profiled on Illumina WG6-V3) and identified 41 NE cell lines (31 SCLCs and 10 NE-NSCLCs) and 87 non-NE cell lines (NSCLCs only). These two groups of cell lines were then used to generate a different 50-gene lung cancer-specific NE signature from the most highly differentially expressed genes (25 genes overexpressed in the NE cell lines, 25 genes overexpressed in the non-NE cell lines). A quantitative NE score was generated from this signature using the formula: NE score = (correl NE – correl non-NE)/2 where correl NE (or non-NE) is the Pearson correlation between expression of the 50 genes in the test sample and expression of these genes in the NE (or non-NE) cell line group. This score has a range of −1 to +1 where a positive score predicts for NE while a negative score predicts for non-NE cell types. The higher the score in absolute value, the better the prediction.
Identification of SCLC cell lines
Many of the cell lines were initiated at the National Cancer Institute several years ago and the original pathology materials were not available for review. Our criteria for calling cell lines as SCLC were as follows: (I) pathological diagnosis of SCLC of the original diagnostic material; (II) cell culture with the growth properties characteristic of SCLC; (III) morphological features of classic or variant SCLC by examination of formalin fixed, paraffin embedded agarose cell blocks (20) or cell line derived xenografts (CCXs); and (IV) loss of Rb protein expression by Western blotting (21), or inactivating mutations of the RB1 gene by sequencing. As rare SCLC tumors lack inactivation of RB1 (17), in one case we identified a cell line with intact Rb protein expression as SCLC. The basis for SCLC designation in this case depended on the presence of other SCLC cell characteristics not discussed herein.
SCLC tumors and cell lines
RNASeq data were available for 81 resected SCLC tumors (17) and for 70 SCLC cell lines that we established and characterized in our laboratory (22,23). With the exception of one cell line, RB1 gene was inactivated in all cases as determined by whole exome sequencing for inactivating mutations or by Western blot for protein expression. Mutations of the TP53 gene were present in all cases. The provenance of all cell lines was confirmed by use of the GenePrint 10 kit for short tandem repeats (Promega), and confirmed free of mycoplasma contamination by the e-Myco plus kit (Boca Scientific).
Cell lines were examined in the living state by inverted microscopy to determine growth patterns. For determination of cytological morphology, cell lines were pelleted, fixed in formalin, paraffin embedded, sectioned and stained with hematoxylin and eosin. For some cases, histological slides of the original tumors from which cell lines were derived or CCXs were available for review.
We calculated the Pearson correlation coefficient between the NE score and genes or pathways known or suspected to be involved in the pathogenesis of SCLC. For SCLC tumors, the expression of highest expressed isoform was chosen and all the expression values were converted as log2 levels. P<0.05 is considered statistically significant.
GEM model
Mouse primary lung tumors from Rb1fl/flTrp53fl/flMycT58ALSL/LSL mice (RPM, Jackson Laboratory stock No. 029971) (11) were micro-dissected under sterile conditions. Individual tumors were processed to single cell suspension by mechanical and chemical separation with incubation in 0.25% Trypsin-EDTA (1X) Solution (Gibco, Waltham, MA, USA) for 20 min. Cell suspensions were filtered through a 100 µm cell strainer and re-suspended in RPMI media with 10% FBS, 1% Penicillin/Streptomycin and L-glutamine and grown in uncoated tissue culture flasks or coated 100 mm plates.
RNA isolation and RNA-Seq was performed as previously described (11) for primary mouse RPM tumors and cell lines. Mouse lung tumor RNA-Seq data are available at NCBI GEO: GSE89660. Eleven RPM tumors (histology of the tumors not available for review) and eight double negative cell lines were scored for NE and the Pearson correlation coefficient between the NE score and genes was calculated.
Results
Development and validation of a NE score for lung cancers
As described in Methodology, we developed a numeric score for evaluating the degree of NE differentiation in lung tumors and cell lines. The top 25 genes most strongly associated with NE differentiation and the top 25 genes not associated with NE differentiation were selected for inclusion into the NE signature score (Figure 1, Tables S1,S2). The score ranges from −1.0 to 1.0, with NE differentiation values ranging from 0 to 1.0, and non-NE scores (i.e., indicating absence of NE differentiation) ranging from 0 to –1.0. These genes clearly differentiated lung NE from non-NE cell lines (Figure 1).
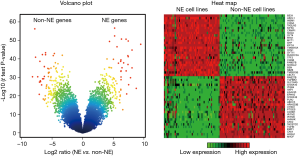
Full table
Full table
The top 25 genes for NE differentiation include a range of known and previously unknown NE and neural genes: neural (n=11), NE (n=2), both neural and NE (n=6), and other (n=6) (Table S3). Some of them, ASCL1, chromogranin A (CHGA) (24) and INSM1 (25) are in use as immunostains for the pathological diagnosis of SCLC and other NE tumors.
Full table
The top 25 genes negatively associated with NE differentiation (Table S4) had a wide range of functions including substrate attachment, cell migration, invasion and metastasis, calcium signaling, inflammation and immunity, fibrinolysis and blood clotting, HIPPO signaling, caveolae, G protein signaling, epithelial cell differentiation, CNS development, and the cell cycle. Note that, many of these genes have multiple functions.
Full table
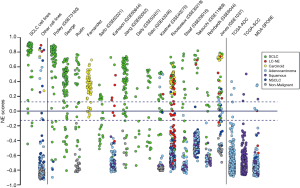
We evaluated the sensitivity and specificity of the score in public and private datasets of available NE and other lung tumors (Figure 2). Of the adenocarcinomas and squamous cell carcinomas in the well documented TCGA database, only 1% expressed positive NE scores. Of carcinoid tumors, 92% had high NE scores, while 8% had lower positive scores and only one had a negative score. The NSCLC cell lines demonstrated considerable heterogeneity. A subgroup of NSCLC lines, previously known to express NE features, including ASCL1 expression, had relatively high NE scores. We had referred to these lines as NSCLC-neuroendocrine or NSCLC-NE cell lines (28). On review of the original histology slides, CCXs and/or agar plugs, most, if not all, could be reclassified as lung NE tumors (other than SCLC), including atypical carcinoids and large cell NE carcinomas (data not shown). These cell lines will be described in detail elsewhere, and will not be discussed further in this report. Non-malignant tissues and immortalized respiratory epithelial cells (29) had negative scores. The signature is cross platform, with a very high concordance (98%) between RNASeq and microarray expression data and can also be applied to murine in vitro systems (GEM models and cell lines).
NE scores of SCLC tumors and cell lines
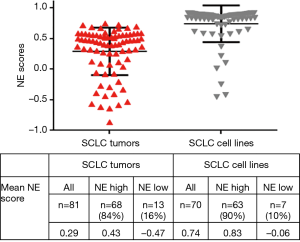
We applied the NE score to SCLC tumors and cell lines (Figure 3). We considered samples with values lower than 1 standard deviation (SD) as having low/negative values and the others as having high NE scores. By this criterion, 13/81 (16%) of tumors and 7/70 (10%) of cell lines had low NE scores.
Heterogeneity of SCLC
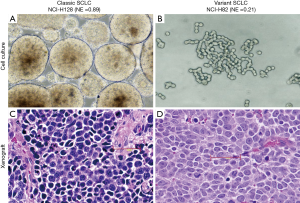
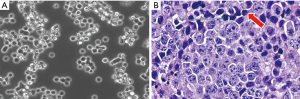
As images of tumors were not available, morphology and growth characteristics were limited to cell lines (Figure 4). Cell culture growth characteristics and pathological examination of cell line pellets or CCXs were performed on about 50 of the cell lines. Almost all cell lines in the high NE score group had in vitro growth characteristics of spheroids or irregular floating clusters, although some also had an adherent subpopulation. By contrast, all cell lines in the low NE score group grew in vitro as loosely attached cells or as mixtures of attached and loosely aggregated or single floating cells. Examination of FFPE cell line pellets indicated that the high NE cell lines had classic SCLC morphology with occasional giant cells, although a few had prominent nucleoli without perinucleolar chromatin clearing. The low NE score SCLC cell lines examined consisted of cells with variant features or mixtures of classic and variant cells.
The Myc driven GEM model we studied could also be divided into NE high (most tumor specimens) and low phenotypes (all cell lines after 14 or more days in culture). The cell lines were dual negative for Ascl1 and Neurod1, and grew as large, loosely aggregated or partially attached cells. When inoculated subcutaneously into mice, the tumors showed a minor classic morphology subtype, while most of the cells were larger and more closely resembled human large cell carcinomas rather than the variant form of SCLC (Figure 5).
Pathway deregulation in NE high and low subgroups
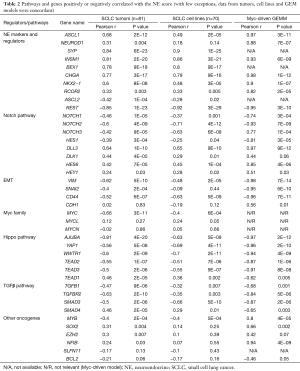
We performed Pearson correlations between the NE scores and the expression of genes/pathways related to NE differentiation and SCLC pathogenesis. Very similar or identical findings were present in human SCLC and GEM model tumors and cell lines (Table 2).
Full table
NE marker expression
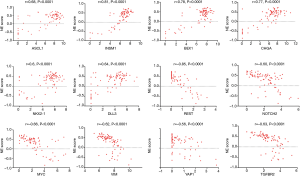
The NE signature is composed of the top genes overexpressed in a group of NE cell lines, so it is not surprising that the NE score has a strong positive correlation with several known NE genes, including ASCL1, NEUROD1, CHGA, INSM1, SYP and BEX1, as well as the related gene NKX2-1. Conversely the NE score was negatively correlated with REST and ASCL2 (Table 2).
Relationship between expression of ASCL1, NEUROD1, ASCL2 and the NE score
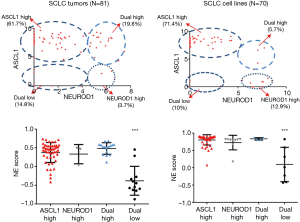
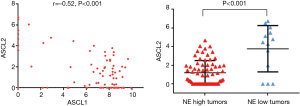
The basic loop-helix-loop transcription factors ASCL1 and NEUROD1, both of which play important roles in regulating expression of NE properties (16). We therefore correlated the expression of ASCL1 and NEUROD1 in tumors and cell lines (Figure 6, upper panel). The data suggested that the tumors and cell lines fell into four groups: (I) ASCL1 high; (II) NEUROD1 high; (III) dual high; or (IV) dual low. The largest group was the ASCL1 high group, although dual low groups were present in both tumors (14.8%) and cell lines (10%). Comparison with the NE scores (Figure 6, lower panel) indicated that there was a strong association between the low NE group and the dual low samples. As shown in Figure 7, the expression of ASCL2 was negatively correlated with ASCL1, and expressed in some of the NE low human SCLC tumors. However, ASCL2 was not expressed in the dual negative cell lines derived from the GEM model (data not shown).
Notch pathway
As seen in Table 2 and Figure 8, several Notch genes as well as HES1 were negatively correlated with the NE score, while others (DLL3, DLK1, HES6) were positively correlated.
Epithelial mesenchymal transition (EMT)
Expression of EMT associated genes VIM, SNAI2 and CD44 were negatively correlated with the NE score, while CDH1 showed no correlation.
Myc family
Of the three Myc family members, only MYC was negatively correlated with the NE score.
Hippo pathway
Expression of several members of the Hippo pathway were negatively correlated with the NE score, including AJUBA, YAP1, WWTR1, TEAD2, and TEAD3.
TGFβ pathway
Expression of the TGF beta pathway genes TGFB1, TGFBR2 and SMAD3 were negatively correlated with the NE score, while SMAD4 was positively correlated.
Other genes of interest
Other genes whose expressions were positively correlated with the NE score were SOX2, EZH2 and NFIB (borderline significance in tumors) while MYB was negatively correlated. Expression of BCL2 and SLFN11 were not correlated.
Discussion
According to the WHO classification, there is a single morphological form of SCLC, although it may be combined with NSCLC elements (4). Since the discovery by Bensch and coworkers that SCLC tumors (and bronchial carcinoids and pulmonary NE cells) had dense core granules (30), the ultrastructural hallmark of NE cells, SCLC has been regarded as a NE tumor. More than 30 years ago we described the variant form of SCLC cell lines, which had altered morphology and growth characteristics, and loss of some NE cell features (5,31). Since that time, the variant form has been described in human tumors and xenografts, and more recently in GEM models. A number of observers have described a modest number of changes associated with the variant subtype (Table 1).
While most SCLC tumors and cell lines morphologically resemble the WHO description of SCLC and express the entire spectrum of NE cell markers, our data indicate that an important subset have low or absent NE differentiation and may have altered (variant) morphology and altered growth characteristics. Other published works have noted heterogeneity in SCLC tumors and in vitro models, as summarized in Table 3, although a variety of terminologies have been applied to the identified or implied NE high and low phenotypes. In order to accurately correlate the effects of loss of NE features with expression of other genes and pathways, a quantitative measurement of NE cell properties was essential.
Full table
We have developed and validated a robust scoring system for NE differentiation in lung cancer tumors and cell lines. The 50 gene-based scoring system uses 25 genes whose expression was most highly correlated with NE differentiation, and 25 genes whose expression was most highly negatively correlated with NE differentiation. This system can also be applied to patient-derived xenografts (PDXs)(data not shown) and to GEM models (Table 2). In addition, the scoring system can cross platforms as it is applicable to various microarray and RNASeq derived data. The system has the advantage of including a positive score (0 to +1), indicating NE differentiation, and a negative score (0 to −1) indicating the degree of “non-NE” differentiation. The lower the negative score, the more confidence that NE differentiation is lacking. It should be noted that a negative score does not rule out the possibility that some NE markers are expressed. For instance, ASCL1, the reputed master regulator of NE differentiation, may be expressed in NE stem cells without expression of the NE cell program (36). As the scoring system was tailored for lung cancers, its application to other NE tumor and cell types remains to be determined.
The scoring system confirmed that almost all (99%) NSCLC tumors lack NE differentiation. While most SCLC tumors and cell lines morphologically resemble the WHO description of SCLC and express the entire spectrum of NE cell markers, our data indicate that an important subset of SCLC have low or absent NE differentiation and may have altered (variant) morphology and altered growth characteristics. We chose to call tumors and cell lines that had NE scores below 1 SD as samples with low scores, although some may be truly negative. For SCLC tumors, the images were not available for us to correlate NE differentiation (or its lack) with morphology. However, for cell lines, most of those with low scores had some or all the features of variant cells. We used the NE score to determine the relationship, positive or negative, between NE differentiation and several genes and pathways important in the pathogenesis of SCLC. Our studies confirmed some previously identified associations with NE differentiation or other pathways. It also identified several previously unknown gene associations. Of interest, in several pathways, individual genes were either positively or negatively correlated with NE differentiation, indicating complicated sets of checks and balances within specific pathways. It is important to point out that in this report we utilized gene expression data for our analyses, and that RNA and protein expression are not always concordant.
While our original observations indicated that variant cell lines often were derived from tumors recurring after initial response to therapy and that myc family overexpression was frequent, our current study indicates the relatively frequent presence of low or absent NE differentiation in (mostly) untreated series of resected SCLC tumors. Unfortunately, we did not have access to the morphology of these tumors, and were unable to assess the relationship of this subset with the variant morphology. As mentioned in Table 3, other citations have also noted an association between the low NE phenotype and responses to chemotherapy and targeted therapies.
Our current knowledge of what genes contribute to NE cell differentiation is currently limited to a modest number (perhaps 10–12). Our studies indicate that the actual gene number is far greater and includes many neural genes not previously regarded as having a NE cell association. A small number (6 of 25) of the top genes lack a previously known neural or NE association. The 25 genes most negatively associated with NE cell differentiation encompassed a wide range of functions/pathways.
ASCL1 is expressed in most SCLC tumors and cell lines and its expression is tightly linked to NE differentiation. Some tumors with equivocal or negative scores expressed ASCL1, suggesting the possibility that they represent NE or neural stem cells. NEUROD1, which has been suggested as an alternative regulator of NE differentiation in a subset of SCLC (10,16), was expressed in a large subset of tumors and cell lines, almost all of them with positive NE scores. However, SCLC cells expressing NEUROD1 usually also expressed ASCL1, making it difficult to assess the relationship of NEUROD1 with NE differentiation. Currently, it is not known whether “dual positive” SCLC cells express both transcription factors and whether they have heterogeneous expression of one or the other. Most tumors and all but one cell line lacking NE differentiation had low or absent expression of these transcription factors (“dual negative SCLC”), suggesting another alternative master regulator. We have identified expression of ASCL2 in some of the “dual negative” tumors and cell lines, with weak or modest expression in a minority of NE positive samples. ASCL2 is closely related to ASCL1, and belongs in the BHLH family of transcription factors, and is involved in the determination of the neuronal precursors in the peripheral nervous system and the central nervous system. It is also expressed in certain gastro-intestinal cells and tumors (37,38), and its expression is a prognostic factor in lung squamous cell and breast cancers (39,40), and is associated with metastases in osteosarcomas (41).
The Myc-driven GEM model we studied rapidly develops tumors that are initially Ascl1 associated, but soon undergo a change to a Neurod1 expressing low NE phenotype. Cell lines derived from microdissected tumors were initially non-adherent spheroids (similar to most human SCLC cell lines), but within 14 days became loosely aggregated and partially adherent like the human variant form. These cell lines were dual negative for expression of the master transcription factors Ascl1, Neurod1, and Ascl2. We studied the cell lines at this later, more advanced phenotype and found their properties to closely resemble the low NE phenotype in human tumors and cell lines. However, the low NE cell lines from the GEM model lacked expression of Ascl2, and the morphology of their xenografts more closely resembled that of human large cell carcinoma rather than the variant subtype. We presume such a progression may have occurred in vivo if the mice had survived for longer periods of time. In the original description of the model (11) the Neurod1 expression was associated with the low NE phenotype. In retrospect, including the data presented herein, we can postulate that Neurod1 expression (at least in this model) accounts for a third phenotype intermediate in expression of its NE properties between the high and low phenotypes.
Expressions of several other well-established NE cell markers were highly positively correlated with the NE scores. Chromogranin A (CHGA) is a member of the chromogranin/secretogranin family of NE secretory proteins and is located in secretory vesicles of neurons and endocrine cells. This gene product is a precursor to three biologically active peptides, vasostatin, pancreastatin, and parastatin, which are negative modulators of the NE system. It is one of the better known NE cell products, and is often used for the identification of NE cells by immunostain.
The zinc finger transcription factor insulin associated 1 (INSM1), along with NEUROD1 and FOXA2, maintain the mature gene expression program in beta-cells (42). It is also essential for the neuronal phenotype in some brain cells (43,44). INSM1 is a transcriptional repressor, blocking expression of the Notch signaling HES1 and REST (see below) by binding to its corepressors RCOR1 and RCOR2, thus maintaining ASCL1 expression (45,46). INSM1 binds to the HES1 promoter and represses HES1, which is a key negative regulator of NE differentiation. INSM1 is increasingly being used as an immunostaining marker for NE cells and tumors (25) and its expression is very tightly correlated with the NE score. The establishment of the NE scoring system identified BEX1 as the top NE associated gene. BEX1 (brain expressed X-linked 1) plays a role in neuronal and myoblast development and has been reported to be a marker for NE cells (47,48). However, it may play a wider role and be involved in cell cycle control and act as a tumor suppressor gene in some situations (49).
NKX2-1 (also known as TTF1) is a transcription factor with limited tissue expression (brain, thyroid and the lung). Its expression in the lung is mainly in club cells and alveolar type II cells where it is crucial for the development of the peripheral lung, as well as in bronchial NE cells (50). It is expressed in most adenocarcinomas of the lung, where it functions as a lineage specific oncogene but can also act as a tumor suppressor (51,52). High expression of NKX2-1 is characteristic of lung (and thyroid) NE cells and tumors (53), although its function in lung NE cells is not understood. Its expression in SCLC is tightly correlated with expression of ASCL1 and NE properties, and, thus, it may play a role in NE differentiation in the lung. NKX2-1 has also been tightly correlated with DLL3 expression (see below) in SCLC and this correlation was validated at the protein level further supporting our findings (32).
RE1 silencing transcription factor (REST) encodes a transcriptional repressor that represses neuronal and NE genes in non-neuronal and non-NE tissues and thus serves as a master negative regulator of neurogenesis (54,55), including SCLC cells (35). It is expressed in almost all non-neuronal and non-NE tissues, including NSCLC. Thus, as expected, it is strongly negatively correlated with the NE score.
Notch signaling is a key negative regulator of NE differentiation in SCLC (17,56), and may function as a tumor suppressor. Mutations in the Notch pathway genes are frequent in SCLC (17). Several Notch receptors are negatively correlated with the NE score, as is HES1, a negative regulator of NE differentiation. By contrast, HES6, an inhibitor of Notch signaling, is positively correlated. Non-NE Notch-active small-cell lung cancer cells are slow growing, consistent with a tumor-suppressive role for Notch, but these cells are also relatively chemoresistant and provide trophic support to NE tumor cells, consistent with a pro-tumorigenic role (35). The DLL3 gene codes for a Notch ligand are a transcriptional target of ASCL1 (57). Unlike the other Notch ligands, it does not stimulate Notch signaling, but functions as an inhibitor. DLL3 expression is tightly linked to expression of ASCL1 and NE differentiation. Rovalpituzumab tesirine (ROVA-T) is a DLL3-targeted antibody-drug conjugate, which has shown encouraging single agent anti-tumor activity in DLL3 expressing lung NE carcinomas (58). Our results would suggest that SCLC tumors that have lost NE (and DLL3) expression would be resistant to ROVA-T therapy. In addition to DLL3, expression of DLK1, another inhibitor of Notch signaling, was also associated with the high NE phenotype (17).
Epithelial to mesenchymal transition (EMT) is a fundamental developmental process that is reactivated in wound healing and in a variety of diseases including cancer where it promotes migration/invasion and metastasis, lack of cell to cell adhesion, resistance to treatment, immune evasion and generation and maintenance of cancer stem cells (59,60). Many of these properties are present in the low NE subtype. Reprogramming of the epigenome is associated with induction of EMT. Loss of the epithelial cell marker E-cadherin (CDH1) and gain of the intermediate filament vimentin (VIM) are among the hallmarks of EMT. Clonal cells having variant like morphology and with loss of NE features were isolated from cultures of the prototype GEM Model for SCLC (12). These clones showed EMT with expression of VIM. The NE score was strongly negatively associated with VIM, suggesting that the variant subtype undergoes EMT. In addition, SNAI2, commonly known as SLUG, a zinc finger transcriptional repressor, which downregulates expression of CDH1 in premigratory neural crest cells and has anti-apoptotic properties (60), was negatively correlated with NE differentiation, although the findings were only significant in SCLC tumors. These findings are consistent with earlier observations that variant SCLC cells have increased growth rates, decreased cell-to-cell adhesion and cloning efficiencies (see Table 1). EMT can be induced in different ways, most notably by a group of transcriptional repressor proteins including SNAI1 and SNAI2 which recognize specific DNA-binding sites within E-boxes, regulatory regions of their target genes. E-boxes are also bound by many BHLH developmental regulators leading to competition among these factors and EMT-inducing repressors. SNAI1 and SNAI2 compete with the stem cell factor ASCL2 for binding to an E-box, suggesting a possible link between SNAI2, ASCL2 and EMT in “dual negative” cells.
All three members of the myc family (MYC, MYCL and MYCN) are frequently amplified or over-expressed in SCLC tumors and cell lines (61-63). Over expression of family members is mutually exclusive. MYC expression (but not of the other family members) was strongly negatively correlated with the NE score, consistent with previous studies (11,32). MYC amplification in SCLC tumors is associated with shortened survival (64). Of major potential therapeutic importance, inhibition of AURKA and AURKB selectively inhibit growth of myc family over-expressing SCLC cell lines and causes polyploidy (11,65).
The Hippo signaling pathway is highly conserved and is a central regulator of organ size (66,67). Genetic perturbation of this pathway in mouse models results in massively enlarged organs accompanied by tumor formation. The pathway consists of a core kinase cassette consisting of a pair of serine/threonine kinases (MST1 and MST2) and related genes. The pathway limits tissue growth by phosphorylating the homologous oncogenes YAP1 and TAZ (also known as WWTR1) which results in their ubiquitin mediated proteolysis. YAP1 plays a critical role in the self-renewal of airway stem cells. Germline mutations of YAP1 predisposes for lung adenocarcinoma with high familial penetrance (68). YAP1 promotes tissue growth by regulating transcription factors including the TEADs and SMADs, and may be accompanied by chemoresistance (34). The main Hippo pathway effectors, YAP1 and TAZ are overexpressed in variant cell lines with loss of NE properties, accompanied by relatively high expression of two of the three TEAD genes (TEAD2 and TEAD3). In addition, AJUBA, an important negative regulator of HIPPO signaling, is negatively associated with NE properties, and may contribute to the high levels of YAP1 and TAZ in non-NE SCLC cell lines, as has been reported previously (69). Inhibition of YAP1 offers a potential therapeutic approach for SCLC cells that lack NE properties (66).
TGFβ is a cytokine that exerts multiple biological functions, and which has a bi-directional role in different cancers, serving either as an oncogene or a tumor suppressor gene. In SCLC it has been reported to function as a tumor suppressor and to suppress ASCL1, and the pathway is repressed by EZH2 (70). We found a negative correlation between the NE score and the TGFβ pathway, in particular with the genes TGFBR2 and SMAD3. These findings are consistent with ASCL1 (and the NE pathway) being down regulated in the low NE tumor group.
Other oncogenes also appeared to play important roles indistinguishing the high and low NE subgroups. The transcription factor MYB plays a key role as a regulator of stem cells in some tissues. It is over-expressed in several tumors, where it may act as an oncogene (71). MYB and NFIB fusion products are present in most adenoid cystic carcinomas (72). MYB over-expression was described in a small series of SCLC cell lines (73). Our findings confirm that over-expression of MYB occurs in a relatively high percentage of SCLC tumors and cell lines. MYB was negatively correlated with NE expression in tumors and cell lines, but had a positive correlation in the MYC driven GEM model.
Of interest, expression of some genes, important in the pathogenesis of SCLC, was not significantly correlated to NE differentiation. These included NFIB (74), SLFN11 (75,76), BCL2 (28), SOX2 and EZH2 (70,77).
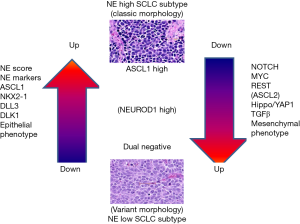
Figure 9 is a summary of the main differences between the NE high and low subtypes. As shown in a GEM model (11) or in a SCLC cell lines (33), this process is a dynamic one, with transition from NE high to NE low phenotype or toward a hybrid state. Whether changes in the opposite direction (NE low to NE high) can occur is currently unknown. The NE high subtype is associated with classic morphology, expression of the NE program of markers, epithelial cell phenotype, expression of NKX2-1 and the Notch pathway inhibitors DLL3 and DLK1. The NE low phenotype is associated with variant morphology, low or absent expression of the NE markers, activation of MYC, REST, Notch, Hippo and TGFβ pathways and profound EMT. In most cases, high NE expression is associated with expression of the master transcription factor ASCL1. In some cases, however, as exemplified by a GEM model and in human tumors and cell lines (11,16,17), the alternative neural transcription factor NEUROD1 is activated, either with or without expression of ASCL1. NEUROD1 expression is associated with varying levels of NE expression, but the low NE subtype usually has loss of both of these master transcription factors. In the low NE phenotype, in some cases, especially in human SCLC tumors, and some cell lines, the alternative master transcription factor ASCL2 is expressed.
Our findings indicate that the NE score can be utilized to separate NE high and low subsets of SCLC tumors and cell lines. Of interest, approximately similar frequencies of low NE samples were noted in resected tumors (16%, almost all from untreated patients) and from cell lines (10%, mostly derived from recurring tumors from previously treated patients). There was more than 90% concordance between the NE score and gene and pathway associations, indicating the high relevance of cell lines, most of which retained the typical appearances and high NE scores decades after their establishment.
Our findings are of high potential clinical importance. As we enter the era of targeted therapies, many of which are likely to target NE or related genes, understanding the heterogeneity of SCLC will be crucial for the identification of NE high and low subsets more likely to respond to specific therapies. Clearly, “tumor heterogeneity is a sort of written history of a particular cancer from which we can learn”, and “which could lead to more effective cancer therapies and prevention methods” (78).
Acknowledgements
Funding: This work was supported by grants from the National Cancer Institute, Bethesda, MD, USA: “Specialized Program in Research Excellence in Lung Cancer”, P50 CA70907 and the “Small Cell Lung Cancer Consortium Coordinating Center” U24CA213274, and 1R21CA216504-01A1 (TG Oliver).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Gazdar AF, Bunn PA, Minna JD. Small-cell lung cancer: what we know, what we need to know and the path forward. Nat Rev Cancer 2017;17:765. [Crossref] [PubMed]
- H.R. 733 — 112th Congress: Recalcitrant Cancer Research Act of 2012. . 2011. February 2, 2018. Available online: www.GovTrack.us
- Swarts DR, Ramaekers FC, Speel EJ. Molecular and cellular biology of neuroendocrine lung tumors: evidence for separate biological entities. Biochim Biophys Acta 2012;1826:255-71. [PubMed]
- Travis WD, Brambilla E. World Health Organization Classification of Lung and Pleural Tumors. 3rd ed. Berlin: Springer-Verlag, 2015.
- Gazdar AF, Carney DN, Nau MM, et al. Characterization of variant subclasses of cell lines derived from small cell lung cancer having distinctive biochemical, morphological, and growth properties. Cancer Res 1985;45:2924-30. [PubMed]
- Carney DN, Gazdar AF, Bepler G, et al. Establishment and identification of small cell lung cancer cell lines having classic and variant features. Cancer Res 1985;45:2913-23. [PubMed]
- Carney DN, Mitchell JB, Kinsella TJ. In vitro radiation and chemotherapy sensitivity of established cell lines of human small cell lung cancer and its large cell morphological variants. Cancer Res 1983;43:2806-11. [PubMed]
- de Leij L, Postmus PE, Buys CH, et al. Characterization of three new variant type cell lines derived from small cell carcinoma of the lung. Cancer Res 1985;45:6024-33. [PubMed]
- Berendsen HH, de Leij L, de Vries EG, et al. Characterization of three small cell lung cancer cell lines established from one patient during longitudinal follow-up. Cancer Res 1988;48:6891-9. [PubMed]
- Poirier JT, Dobromilskaya I, Moriarty WF, et al. Selective tropism of Seneca Valley virus for variant subtype small cell lung cancer. J Natl Cancer Inst 2013;105:1059-65. [Crossref] [PubMed]
- Mollaoglu G, Guthrie MR, Bohm S, et al. MYC Drives Progression of Small Cell Lung Cancer to a Variant Neuroendocrine Subtype with Vulnerability to Aurora Kinase Inhibition. Cancer Cell 2017;31:270-85. [Crossref] [PubMed]
- Calbo J, van Montfort E, Proost N, et al. A functional role for tumor cell heterogeneity in a mouse model of small cell lung cancer. Cancer Cell 2011;19:244-56. [Crossref] [PubMed]
- Mabry M, Nakagawa T, Nelkin BD, et al. v-Ha-ras oncogene insertion: a model for tumor progression of human small cell lung cancer. Proc Natl Acad Sci U S A 1988;85:6523-7. [Crossref] [PubMed]
- Scarpa S, Morstyn G, Carney DN, et al. Small cell lung cancer cell lines: pure and variant types can be distinguished by their extracellular matrix synthesis. Eur Respir J 1988;1:639-44. [PubMed]
- Doyle LA, Giangiulo D, Hussain A, et al. Differentiation of human variant small cell lung cancer cell lines to a classic morphology by retinoic acid. Cancer Res 1989;49:6745-51. [PubMed]
- Borromeo MD, Savage TK, Kollipara RK, et al. ASCL1 and NEUROD1 Reveal Heterogeneity in Pulmonary Neuroendocrine Tumors and Regulate Distinct Genetic Programs. Cell Rep 2016;16:1259-72. [Crossref] [PubMed]
- George J, Lim JS, Jang SJ, et al. Comprehensive genomic profiles of small cell lung cancer. Nature 2015;524:47-53. [Crossref] [PubMed]
- Gazdar AF, Hirsch FR, Minna JD. From Mice to Men and Back: An Assessment of Preclinical Model Systems for the Study of Lung Cancers. J Thorac Oncol 2016;11:287-99. [Crossref] [PubMed]
- Girard L, Rodriguez-Canales J, Behrens C, et al. An Expression Signature as an Aid to the Histologic Classification of Non-Small Cell Lung Cancer. Clin Cancer Res 2016;22:4880-9. [Crossref] [PubMed]
- Mansy SS, Abbas MA, Yehia HA, et al. Value of the innovated technique agarose cell block in improving the sensitivity of urine cytology in cases of bladder carcinoma. Ultrastruct Pathol 2006;30:379-85. [Crossref] [PubMed]
- Shimizu E, Coxon A, Otterson GA, et al. RB protein status and clinical correlation from 171 cell lines representing lung cancer, extrapulmonary small cell carcinoma, and mesothelioma. Oncogene 1994;9:2441-8. [PubMed]
- Phelps RM, Johnson BE, Ihde DC, et al. NCI-Navy Medical Oncology Branch cell line data base. J Cell Biochem Suppl 1996;24:32-91. [Crossref] [PubMed]
- Gazdar AF, Minna JD. NCI series of cell lines: An historical perspective. J Cell Biochem Suppl 1996;24:1-11. [Crossref] [PubMed]
- Travis WD. Pathology and diagnosis of neuroendocrine tumors: lung neuroendocrine. Thorac Surg Clin 2014;24:257-66. [Crossref] [PubMed]
- Rosenbaum JN, Guo Z, Baus RM, et al. INSM1: A Novel Immunohistochemical and Molecular Marker for Neuroendocrine and Neuroepithelial Neoplasms. Am J Clin Pathol 2015;144:579-91. [Crossref] [PubMed]
- Rudin CM, Durinck S, Stawiski EW, et al. Comprehensive genomic analysis identifies SOX2 as a frequently amplified gene in small-cell lung cancer. Nat Genet 2012;44:1111-6. [Crossref] [PubMed]
- Fernandez-Cuesta L, Peifer M, Lu X, et al. Frequent mutations in chromatin-remodelling genes in pulmonary carcinoids. Nat Commun 2014;5:3518. [Crossref] [PubMed]
- Augustyn A, Borromeo M, Wang T, et al. ASCL1 is a lineage oncogene providing therapeutic targets for high-grade neuroendocrine lung cancers. Proc Natl Acad Sci U S A 2014;111:14788-93. [Crossref] [PubMed]
- Sato M, Larsen JE, Lee W, et al. Human Lung Epithelial Cells Progressed to Malignancy through Specific Oncogenic Manipulations. Mol Cancer Res 2013;11:638-50. [Crossref] [PubMed]
- Bensch KG, Corrin B, Pariente R, et al. Oat-cell carcinoma of the lung. Its origin and relationship to bronchial carcinoid. Cancer 1968;22:1163-72. [Crossref] [PubMed]
- Carney DN, Gazdar AF, Nau M, et al. Biological heterogeneity of small cell lung cancer. Semin Oncol 1985;12:289-303. [PubMed]
- Cardnell RJ, Li L, Sen T, et al. Protein expression of TTF1 and cMYC define distinct molecular subgroups of small cell lung cancer with unique vulnerabilities to aurora kinase inhibition, DLL3 targeting, and other targeted therapies. Oncotarget 2017;8:73419-32. [Crossref] [PubMed]
- Udyavar AR, Wooten DJ, Hoeksema M, et al. Novel Hybrid Phenotype Revealed in Small Cell Lung Cancer by a Transcription Factor Network Model That Can Explain Tumor Heterogeneity. Cancer Res 2017;77:1063-74. [Crossref] [PubMed]
- Ito T, Matsubara D, Tanaka I, et al. Loss of YAP1 defines neuroendocrine differentiation of lung tumors. Cancer Sci 2016;107:1527-38. [Crossref] [PubMed]
- Lim JS, Ibaseta A, Fischer MM, et al. Intratumoural heterogeneity generated by Notch signalling promotes small-cell lung cancer. Nature 2017;545:360-4. [Crossref] [PubMed]
- Vasconcelos FF, Castro DS. Transcriptional control of vertebrate neurogenesis by the proneural factor Ascl1. Front Cell Neurosci 2014;8:412. [Crossref] [PubMed]
- Jubb AM, Hoeflich KP, Haverty PM, et al. Ascl2 and 11p15.5 amplification in colorectal cancer. Gut 2011;60:1606-7; author reply 7. [Crossref] [PubMed]
- Yan KS, Kuo CJ. Ascl2 reinforces intestinal stem cell identity. Cell Stem Cell 2015;16:105-6. [Crossref] [PubMed]
- Xu H, Zhao XL, Liu X, et al. Elevated ASCL2 expression in breast cancer is associated with the poor prognosis of patients. Am J Cancer Res 2017;7:955-61. [PubMed]
- Hu XG, Chen L, Wang QL, et al. Elevated expression of ASCL2 is an independent prognostic indicator in lung squamous cell carcinoma. J Clin Pathol 2016;69:313-8. [Crossref] [PubMed]
- Liu MH, Cui YH, Guo QN, et al. Elevated ASCL2 expression is associated with metastasis of osteosarcoma and predicts poor prognosis of the patients. Am J Cancer Res 2016;6:1431-40. [PubMed]
- Jia S, Ivanov A, Blasevic D, et al. Insm1 cooperates with Neurod1 and Foxa2 to maintain mature pancreatic beta-cell function. EMBO J 2015;34:1417-33. [Crossref] [PubMed]
- Jacob J, Storm R, Castro DS, et al. Insm1 (IA-1) is an essential component of the regulatory network that specifies monoaminergic neuronal phenotypes in the vertebrate hindbrain. Development 2009;136:2477-85. [Crossref] [PubMed]
- Rosenbaum JN, Duggan A, Garcia-Anoveros J. Insm1 promotes the transition of olfactory progenitors from apical and proliferative to basal, terminally dividing and neuronogenic. Neural Dev 2011;6:6. [Crossref] [PubMed]
- Jia S, Wildner H, Birchmeier C. Insm1 controls the differentiation of pulmonary neuroendocrine cells by repressing Hes1. Dev Biol 2015;408:90-8. [Crossref] [PubMed]
- Monaghan CE, Nechiporuk T, Jeng S, et al. REST corepressors RCOR1 and RCOR2 and the repressor INSM1 regulate the proliferation-differentiation balance in the developing brain. Proc Natl Acad Sci U S A 2017;114:E406-15. [Crossref] [PubMed]
- Khazaei MR, Halfter H, Karimzadeh F, et al. Bex1 is involved in the regeneration of axons after injury. J Neurochem 2010;115:910-20. [Crossref] [PubMed]
- Hofsli E, Wheeler TE, Langaas M, et al. Identification of novel neuroendocrine-specific tumour genes. Br J Cancer 2008;99:1330-9. [Crossref] [PubMed]
- Fernandez EM, Diaz-Ceso MD, Vilar M. Brain expressed and X-linked (Bex) proteins are intrinsically disordered proteins (IDPs) and form new signaling hubs. PLoS One 2015;10:e0117206. [Crossref] [PubMed]
- La Rosa S, Marando A, Gatti G, et al. Achaete-scute homolog 1 as a marker of poorly differentiated neuroendocrine carcinomas of different sites: a validation study using immunohistochemistry and quantitative real-time polymerase chain reaction on 335 cases. Hum Pathol 2013;44:1391-9. [Crossref] [PubMed]
- Snyder EL, Watanabe H, Magendantz M, et al. Nkx2-1 represses a latent gastric differentiation program in lung adenocarcinoma. Mol Cell 2013;50:185-99. [Crossref] [PubMed]
- Yamaguchi T, Hosono Y, Yanagisawa K, et al. NKX2-1/TTF-1: an enigmatic oncogene that functions as a double-edged sword for cancer cell survival and progression. Cancer Cell 2013;23:718-23. [Crossref] [PubMed]
- La Rosa S, Chiaravalli AM, Placidi C, et al. TTF1 expression in normal lung neuroendocrine cells and related tumors: immunohistochemical study comparing two different monoclonal antibodies. Virchows Arch 2010;457:497-507. [Crossref] [PubMed]
- Thiel G, Ekici M, Rossler OG. RE-1 silencing transcription factor (REST): a regulator of neuronal development and neuronal/endocrine function. Cell Tissue Res 2015;359:99-109. [Crossref] [PubMed]
- Gao Z, Ure K, Ding P, et al. The master negative regulator REST/NRSF controls adult neurogenesis by restraining the neurogenic program in quiescent stem cells. J Neurosci 2011;31:9772-86. [Crossref] [PubMed]
- Sriuranpong V, Borges MW, Ravi RK, et al. Notch signaling induces cell cycle arrest in small cell lung cancer cells. Cancer Res 2001;61:3200-5. [PubMed]
- Saunders LR, Bankovich AJ, Anderson WC, et al. A DLL3-targeted antibody-drug conjugate eradicates high-grade pulmonary neuroendocrine tumor-initiating cells in vivo. Sci Transl Med 2015;7:302ra136. [Crossref] [PubMed]
- Rudin CM, Pietanza MC, Bauer TM, et al. Rovalpituzumab tesirine, a DLL3-targeted antibody-drug conjugate, in recurrent small-cell lung cancer: a first-in-human, first-in-class, open-label, phase 1 study. Lancet Oncol 2017;18:42-51. [Crossref] [PubMed]
- Roche J, Gemmill RM, Drabkin HA. Epigenetic Regulation of the Epithelial to Mesenchymal Transition in Lung Cancer. Cancers (Basel) 2017;9:72. [Crossref] [PubMed]
- Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol 2014;15:178-96. [Crossref] [PubMed]
- Little CD, Nau MM, Carney DN, et al. Amplification and expression of the c-myc oncogene in human lung cancer cell lines. Nature 1983;306:194-6. [Crossref] [PubMed]
- Nau MM, Brooks BJ, Battey J, et al. L-myc, a new myc-related gene amplified and expressed in human small cell lung cancer. Nature 1985;318:69-73. [Crossref] [PubMed]
- Johnson BE, Brennan JF, Ihde DC, et al. myc family DNA amplification in tumors and tumor cell lines from patients with small-cell lung cancer. J Natl Cancer Inst Monogr 1992.39-43. [PubMed]
- Alves RC, Meurer RT, Roehe AV. MYC amplification is associated with poor survival in small cell lung cancer: a chromogenic in situ hybridization study. J Cancer Res Clin Oncol 2014;140:2021-5. [Crossref] [PubMed]
- Helfrich BA, Kim J, Gao D, et al. Barasertib (AZD1152), a Small Molecule Aurora B Inhibitor, Inhibits the Growth of SCLC Cell Lines In Vitro and In Vivo. Mol Cancer Ther 2016;15:2314-22. [Crossref] [PubMed]
- Pan D. YAPing Hippo Forecasts a New Target for Lung Cancer Prevention and Treatment. J Clin Oncol 2015;33:2311-3. [Crossref] [PubMed]
- Harvey KF, Zhang X, Thomas DM. The Hippo pathway and human cancer. Nat Rev Cancer 2013;13:246-57. [Crossref] [PubMed]
- Chen HY, Yu SL, Ho BC, et al. R331W Missense Mutation of Oncogene YAP1 Is a Germline Risk Allele for Lung Adenocarcinoma With Medical Actionability. J Clin Oncol 2015;33:2303-10. [Crossref] [PubMed]
- Horie M, Saito A, Ohshima M, et al. YAP and TAZ modulate cell phenotype in a subset of small cell lung cancer. Cancer Sci 2016;107:1755-66. [Crossref] [PubMed]
- Murai F, Koinuma D, Shinozaki-Ushiku A, et al. EZH2 promotes progression of small cell lung cancer by suppressing the TGF-beta-Smad-ASCL1 pathway. Cell Discov 2015;1:15026. [Crossref] [PubMed]
- Ramsay RG, Gonda TJ. MYB function in normal and cancer cells. Nat Rev Cancer 2008;8:523-34. [Crossref] [PubMed]
- Ferrarotto R, Heymach JV, Glisson BS. MYB-fusions and other potential actionable targets in adenoid cystic carcinoma. Curr Opin Oncol 2016;28:195-200. [Crossref] [PubMed]
- Griffin CA, Baylin SB. Expression of the c-myb oncogene in human small cell lung carcinoma. Cancer Res 1985;45:272-5. [PubMed]
- Denny SK, Yang D, Chuang CH, et al. Nfib promotes metastasis through a widespread increase in chromatin accessibility. Cell 2016;166:328-42. [Crossref] [PubMed]
- Allison Stewart C, Tong P, Cardnell RJ, et al. Dynamic variations in epithelial-to-mesenchymal transition (EMT), ATM, and SLFN11 govern response to PARP inhibitors and cisplatin in small cell lung cancer. Oncotarget 2017;8:28575-87. [PubMed]
- Gardner EE, Lok BH, Schneeberger VE, et al. Chemosensitive Relapse in Small Cell Lung Cancer Proceeds through an EZH2-SLFN11 Axis. Cancer Cell 2017;31:286-99. [Crossref] [PubMed]
- Hubaux R, Thu KL, Coe BP, et al. EZH2 promotes E2F-driven SCLC tumorigenesis through modulation of apoptosis and cell-cycle regulation. J Thorac Oncol 2013;8:1102-6. [Crossref] [PubMed]
- Campbell LL, Polyak K. Breast tumor heterogeneity: cancer stem cells or clonal evolution? Cell Cycle 2007;6:2332-8. [Crossref] [PubMed]


