The 180 splice variant of NCAM—containing exon 18—is specifically expressed in small cell lung cancer cells
Introduction
Small cell lung cancer (SCLC) represents 10% to 15% of clinical lung cancer cases. It is an aggressive malignancy strongly associated with smoking, annually killing an estimated 250,000 people worldwide. SCLC is decreasing in industrialized countries due to smoking cessation programs, but it is increasing in low- and middle-income countries (1).
In about two thirds of the cases SCLC is detected when metastatic lesions are already present (2). In those cases patients respond well to a first round of chemotherapy, but then invariably resistance to chemotherapy develops and the fast growth rate of resistant tumor cells will lead to death within a year from the initial diagnosis (1,3). Over the last decades survival rates for SCLC have not improved (4).
A limiting factor for the much needed improvement of therapies is the lack of biomarkers for SCLC. These markers, combined with serial ‘liquid biopsies’, would both allow early diagnosis and the monitoring of the effects of chemotherapy. Biomarkers could also be used to monitor the effect of new therapies.
In the past Neural Cell Adhesion Molecule (NCAM) has been investigated as a marker for SCLC and the wider group of neuroendocrine cancers to which SCLC belongs (5). NCAM, also known as CD56, is a member of the immunoglobulin superfamily (6). The protein is highly conserved between species where it is expressed by a variety of cells in the nervous system, in muscle cells (7) and on natural killer cells (8). In addition NCAM is expressed on a number of tumor cells, including SCLC (5). The presence of NCAM on SCLC is in line with the neuronal characteristics of these tumor cells (1,9).
Since NCAM is also present on normal cells it is not a ‘perfect’ tumor marker. Nevertheless, antibodies to NCAM conjugated to cytotoxic drugs have been studied in clinical trials as therapeutic agents. Patients with SCLC were treated with the anti-NCAM immunotoxin huN901-DM1, revealing acceptable toxicity and signs of clinical response (5). However, a later phase II study was discontinued due to poor results and a fatal outcome in one patient (http://investor.immunogen.com/releasedetail.cfm?ReleaseID=804315).
We took a closer look at the presence of the NCAM isoforms in SCLC and other cell types to investigate whether specific isoforms would be usable biomarkers. The human genome contains a single gene NCAM1 for NCAM, located in band q23 of chromosome 11 (10). This single gene encodes several isoforms via alternative splicing. The major isoforms are NCAM-120, NCAM-140 and NCAM-180, each named according to its apparent molecular mass. These three molecules share the same extracellular domain. NCAM-120 lacks the transmembrane domain, encoded by exon 16, present in NCAM-140 and NCAM-180 (6). The NCAM-180 has a cytoplasmic tail that is 272 amino acids longer than NCAM-140 in man (Figure 1). These amino acids are encoded by NCAM exon 18.

During the development of the brain, expression of NCAM-180 is restricted to neural cells (11,12). Results from NCAM knock-out mice demonstrate that NCAM is crucial for the normal development of the brain and neuronal plasticity in the adult brain (13). Searching the H. sapiens expressed sequence tag library of GenBank with the DNA sequence of human NCAM exon 18 yielded 7 hits with a query score of >50% (October 26, 2017), five from fetal brain, one from thalamus and one from teratocarcinoma.
In this paper we describe studies on the expression of NCAM-180 in a panel of cell lines, tumor tissues and controls. We found that NCAM exon 18 is expressed in SCLC cell lines and confirmed NCAM exon 18 expression in human SCLC tumor tissue biopsies. No expression was found in most other cancer cell lines and PBMC of healthy controls. Using a recombinant protein E18, induced and purified from E. coli, a monoclonal antibody specific for E18 was produced. This antibody was used to characterize the expression of NCAM-180 in SCLC cell lines. Finally, we studied the immunogenic potential of E18 in a mouse model.
Methods
Cell culture, RNA extraction and cDNA synthesis
A panel of 44 cell lines (Table 1; obtained from the American Type Culture Collection®, Manassas, USA) was used. Peripheral blood mononuclear cells (PBMC) of 10 healthy individuals were used as controls. Growing cells were harvested, cell pellets washed with phosphate buffered saline (PBS) and total RNA extracted using Trizol (Invitrogen, Paisley, UK) according to standard procedures. For each cell line 2 µg of RNA was reverse transcribed into cDNA using Omniscript reverse transcriptase (Qiagen). Briefly, the RNA was incubated for 60 min at 37 °C with reverse transcription reaction mixture containing 0.5 mM of each dNTP, 1 µM oligo-dT primer (Eurogentec SA, Seraing, Belgium), 10 units RNase inhibitor (Applied Biosystems, Foster City, CA, USA) and 4 units Omniscript reverse transcriptase (Roche, Applied science, Indianapolis, USA) in 20 µL total reaction volume. Human tumor biopsies: we used 10 sections (10 µm thickness) of the frozen biopsies for RNA extraction and subsequent cDNA synthesis. Frozen sections were homogenized using a Qiashredder homogenizer and total RNA was subsequently extracted using the RNeasy Micro kit (Qiagen). cDNA synthesis was performed using the First strand cDNA synthesis kit (Fermentas, St Leon-Rot, Germany). Quality control of the newly synthesized cDNA was done using a human β-actin-based PCR amplification reaction.

Full table
Differential expression PCR analysis of NCAM splice variants
PCR amplification was performed according to standard procedures. Briefly, 0.2 µL cDNA was added to an amplification mixture (20 µL) containing 2 µL 10× PCR gold buffer (Applied Biosystems Foster City, CA, USA), 1.6 µL MgCl2 (25 mM), 0.2 µL dNTP (10 mM, Promega, Madison USA), 0.1 µL forward primer (0.5 µM), 0.1 µL reverse primer (0.5 µM) and 0.2 µL Taq polymerase (AmpliTaq Gold, 250 Units, Applied Biosystems). Nucleotide sequences of forward and reverse primers for the two NCAM splice variants studied are shown in Figure 1. As a control primers for the β-actin mRNA (forward, 5'GCCAACCGCGAGAAGATGAC3'; reverse, 5'GATGGGCACAGTGTGGGTGAC3') were used. PCR reactions were performed in a T3 Thermocycler (Biometra, Westburg, Leusden, The Netherlands) and conditions were as follows: 30 s at 94 °C, 30 s at 60 °C, 1 min at 72 °C for 35 cycles, initiated by 12 min at 94 °C and terminated by 10 min at 72 °C. PCR products were loaded on 2% agarose gels. Detection was performed using Gelstar (Gelstar Nuclei acid gel stain, Cambrex, Rockland, ME, USA).
Production of recombinant E18 protein
His-tagged E18 protein was produced in E. coli BL21(DE3)pLysS using the pRSET based high-level recombinant protein expression system (Invitrogen Ltd, Paisley PA4 9RF, UK). For the isolation of the protein, bacterial pellets were lysed in 5 mL lysis buffer (50 mM NaH2PO4, 300 mM NaCl, pH 8.0) with lysozyme (1 mg/mL), DNase (5 µg/mL) and protease inhibitors (HALTTM Protease inhibitors EDTA-free, Thermo Fisher Scientific Inc., Waltham, MA, USA). The recombinant protein was purified on a Ni2+-NTA agarose column (Qiagen, Venlo, The Netherlands). The isolated protein was eluted by imidazole (300 mM) and renaturated by stepwise dialysis against PBS (4M, 2M, 1M, 0.5M urea, 1× PBS).
Production of E18-specific monoclonal antibodies
After medical ethical approval of the study protocol, Balb/c mice were immunized with purified His-tagged E18 protein from E. coli. We used subcutaneous immunization with 10 µg of protein in PBS. Mice were immunized 3 times with 3 weeks intervals. A final boost was given 3 days before the animals were sacrificed. Complete spleens were squeezed through sterile 70 µm nylon filters in culture medium and used to prepare hybridoma cells by standard procedures. Hybridoma clones were isolated and the secreted monoclonal antibodies were tested by ELISA. Positive clones were subcloned and then grown in a ‘miniPerm’ bioreactor according to standard procedures (14). The IgM monoclonal antibodies from the supernatant of the miniPerm culture were first isolated on a ConA affinity column (PBS, pH 7.4) and subsequently eluted using 0.5 M α-methyl mannoside in PBS of pH 7.4. The eluted fraction of the ConA column was subsequently purified on a Superose-6 gel filtration column and filtered through a 5-µm filter.
ELISA to determine E18-specific monoclonal antibody titers
To test the antigen specificity of the monoclonal antibodies ELISA plates were coated with 100 µL of a 10 µg/mL His-tagged E18 protein solution in 50 mM NaHCO3, pH 9.5. After overnight coating at 4 °C, plates were washed three times with washing buffer [PBS with 0.05% (v/v) Tween-20] and subsequently blocked for 2 h at 37 °C with blocking buffer (2% non-fat dry milk, Nutricia Protifar Plus) in washing buffer. The purified E18 monoclonal antibody was serially diluted in blocking buffer (0–130 ng/well) and incubated for 2 h at 37 °C. Plates were washed three times and bound antibodies detected using a polyclonal rabbit anti-mouse immunoglobulin conjugated with horseradish peroxidase (Dakocytomation, Denmark) (1:1,000 in blocking buffer). The conjugate was incubated for 1 h at 37 °C. Then the plates were washed and the substrate 3,3',5,5'-tetramethylbenzidine (Calbiochem) added for detection. The substrate was incubated for 15 minutes at 37 °C, the enzymatic reaction stopped with 0.5M H2SO4, and staining measured at 450 nm using a 96 multiwell plate based reading platform (Multiscan spectrum, Thermolab systems, Finland). To check for the presence of antibodies against the His-tag of the protein as used for the immunization, a control ELISA was performed with irrelevant His-tagged proteins produced and purified like the E18 protein.
Immunocytochemistry for E18 in SCLC cell lines
Antigen specificity of E18 antibodies was tested using immuno-cytochemistry. Therefore NCI-H69 and NCI-H82 cells were collected by centrifugation from the cell culture medium. Cell pellets were washed, resuspended in PBS and subsequently spotted on 18-wells, 4 mm diagnostic microscope slides (Cell-line, Brunschweig, Germany). The negative control cell line HCT116 was trypsinized, seeded in culture medium and grown overnight in a 12 well plate with a 18 mm glass slide on the bottom of the well. Before immunostaining, cells were permeabilized and fixed for 30 sec in methanol/acetone at −20 °C and incubated for 1h at room temperature with the primary antibodies against E18 or NCAM (RNL-1, 1 mg/mL, 1:100 diluted in PBS with 0.1% BSA; Nordic-Mubio, Susteren, The Netherlands). Then, cells were washed in PBS, and incubated for 1h at room temperature with the fluorescein isothiocyanate (FITC)-conjugated rabbit anti-mouse immunoglobulin (diluted 1:100 in PBS with 0.1% BSA). Stained cells were washed and stepwise de-hydrated in alcohol, cells were mounted in glycerol and viewed through a fluorescent microscope (Axioskop, Zeiss, Germany). Stained cells were also visualized with a Zeiss confocal imaging system [Stack scan mode, scaling: X: 0.1 µm, Y: 0.1 µm, stack size: X: 50.19 µm, Y: 32.43 µm, Scan zoom 2.8, Plan-apochromat objective (63×/1.40 Oil DIC M27, excitation at 488 nm].
Flow cytometry of NCI-H82 cells stained with the E18-specific monoclonal antibody
NCI-H82 cells, expressing NCAM-180, were stained in suspension with the purified E18 monoclonal antibody. The cells were harvested by centrifugation and washed twice with PBS. Half of the cells were permeabilized for 15 min with ice-cold methanol. Both, permeabilized and non-permeabilized cells were resuspended in buffer (PBS containing 0.1% NaN3 and 0.1% BSA). Staining was done for permeabilized and non-permeabilized cells (5×105 cells in 100 µL buffer) using the purified E18 monoclonal antibody (1.3 mg/mL, 200 µL, 1:400 diluted in PBS with 1% BSA). Permeabilized and non-permeabilized cells were stained for 1 h at 4 °C, washed twice with buffer, resuspended in 100 µL buffer and incubated for 1 h with 5 µL undiluted rabbit anti-mouse FITC (Dako, F0313). An incubation with secondary antibody alone was used as a control. Cells were washed twice and resuspended in 200 µL buffer. Fluorescence was measured using Cellquest software on a FACSCalibur (BD, FACSCalibur).
Immunisation and antigen specific cytotoxic T cells (CTL) measurement
Mice (Balb/c) were immunized with E18 protein and the E18-encoding DNA in the pCI mammalian expression vector (Promega). Immunization was subcutaneously with protein [12 µg protein in Abisco-100 (Isconova Sweden) or 10 µg protein nonformulated] and intramuscularly with DNA (2 times 50 µg DNA). Immunization was performed at the day 0 and day 21. Immunized animals were sacrificed at day 31. Mice spleens were isolated and squeezed to prepare a single cell suspension. Spleen cell suspensions were seeded for in vitro restimulation at a cell density of 1.5×106 cells/well (100 µL). In vitro restimulation was performed in four replicate samples for each spleen suspension. Restimulation was performed with a 9-mer peptides covering the E18 protein sequence (Pepscan, Lelystad, The Netherlands). A total of 67 peptides (9-mers, 5 amino acids overlap) were divided over 3 mixtures each containing 22 peptides (0.2 µg of each peptide/well). Control restimulation was performed with medium (negative control) and PMA/ionomycin (0.05 µg/mL/0.5 µg/mL) (positive control) and an irrelevant peptide mixture (overlapping peptides from the NCAM 7pi8 region, 17 peptides, 0.2 µg of each peptide/well). For all restimulations intracellular transport of cytokines was inhibited by adding 5 µg/mL Brefeldin A. Restimulation was performed for 4 hours at 37 °C in 5% CO2. Cell surface CD8 was stained using the FITC-labeled rat anti-mouse CD8 (BD Biosciences, Breda, The Netherlands). Cells were fixed and permeabilized according to standard procedures and subsequently stained for the intra-cellular cytokine IFN-γ using the APC-conjugated rat anti-mouse IFN-γ (BD, Biosciences). The proportion of IFN-γ secreting CD8+ positive cells as measured by FACS represents the number of antigen specific CD8+ T cells (CTL).
Results
Identification of coding regions in the NCAM1 gene and development of assays to detect NCAM isoforms
The NCAM protein sequence is present in GenBank under Accession number EAW67201. It consists of 1,119 amino acids and has, without posttranslational modifications, a calculated molecular weight of 119,447. The sequence of a complete transcript of the human NCAM1 gene, containing all 19 exons was not present in the databases. The sequences were reconstructed starting with the DNA sequence containing the NCAM region determined by Hattori et al. (Homo sapiens genomic DNA, chromosome 11q, clone:RP11-839D17, complete sequence, GenBank: AP000880.4). The region containing nucleotides 60,000–64,000 of the sequence was translated into protein. Exons 17, 18 and 19 were identified in the same reading frame (Figure 1). The contiguous sequence of the exon 17–19 translation products was identical to the corresponding part of EAW67201. The exon 18 encoded part of NCAM is 272 amino acids long and has a molecular weight of 26,134.
PCR primers were designed for differential expression PCR studies of exon 18. The positions of the primers are indicated in Figure 1. The primer sequences annealing within exon 18 result in a PCR amplicon of 604 bp upon expression of NCAM-180 and no PCR amplicon when the NCAM-140 kDa is expressed. A second PCR was done to study the expression of NCAM-140. The forward primer in exon 17 and a reverse primer in exon 19 result in a 180 bp PCR amplicon for the NCAM-140 splice variant. The expression of NCAM exon 18, was confirmed using the exon 17 forward primer with the exon 18 reverse primer, yielding a 715 bp amplicon for NCI-H82 cells. This PCR excludes that PCR amplicons as found in the PCR done with a forward and reverse primer within the exon 18 are due to a contamination of the cDNA sample with genomic DNA. When exon 18 is expressed, a 715 bp PCR product will be formed. Actin control PCRs were used throughout.
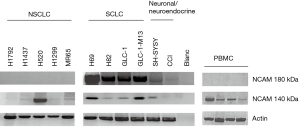
Expression of the exon 18 in SCLC cell lines
RT-PCR was performed to study expression of NCAM exon 18. Results are summarized in Table 1 and Figure 2. NCAM-180 expression was found in 1/9 NSCLC and 2/6 neuronal and neuroendocrine tumors. None of the leukemia cell lines, and among the non-lung cancer cell lines only the placenta choriocarcinoma (JAR) and the osteosarcoma (SJSA) expressed NCAM-180. Our data show a significant expression of exon 18 for all (5/5) SCLC cell lines studied (4 of these are shown in Figure 2). Exon 18 was also expressed in cell lines SH-SYSY and CCI (Figure 2). In none of the PBMC of healthy controls (0/10) exon 18 expression was found. We sequenced the expressed exon 18 in SCLC cell lines to check for potential sequence mutations compared to the sequence indicated in Figure 1. A completely identical nucleotide sequence (815 bp) was found for the SCLC cell lines.
NCAM-180 expression in SCLC tumors
To determine whether the results obtained in SCLC cell lines are a reflection of the in vivo situation SCLC tumor biopsies were investigated. RNA was extracted from 10 sections (10 µm) for each of the 10 frozen SCLC tumor biopsies. cDNA was synthesized and differential expression PCR performed as described before. We also amplified exon 18 both using the forward-reverse primer combination as well as the NCAM exon 17 forward primer combined with the exon 18 reverse primer. We used the cDNA of the NCI-H82 cell line as a positive control and RNA extracted from a squamous cell carcinoma biopsy as a negative control. Our PCR data show exon 18 expression in 8/10 (80%) of the SCLC tumors studied (Figure 3). The signals varied, from strong (S9) to medium (S1, S2, S3, and S10). For the samples S4-S8, a nested PCR was done. In this case, cDNA was preamplified using the NCAM exon 16 forward and exon 19 reverse primer, followed by further amplification with exon 17 forward and exon 18 reverse primers. This resulted in a signal in S4, S6 S7 but not in in S5 and S8. The differences in expression levels in the different SCLC tumor samples are difficult to interpret. No normalization was made for the size of the tumor nor the ratio of tumor versus normal tissue taken during biopsy sampling. Moreover, the amplification reactions were not quantitative. Nevertheless, our results clearly show the expression of NCAM exon 18 in human SCLC tumors. These data confirm the differential expression studies for in vitro cultured SCLC cell lines and indicate that E18 is a SCLC-specific tumor antigen.

Production and purification of recombinant E18 in E. coli
To facilitate further studies we produced recombinant E18 in E. coli. The E18 coding region was PCR-amplified from cDNA of NCI-H69 cells. The recombinant protein, including the His-tag is 314 amino acids, and the predicted molecular mass is 30.6 kDa (Figure S1). The purified protein was analyzed by SDS-PAGE, and the sequence was analyzed using liquid chromatography electrospray ionization tandem mass spectrometry. SDS-PAGE showed that the protein is partially present as a dimer. Sequence analysis however confirmed the E18 sequence of the protein. Why the protein formed dimers is not clear.

Monoclonal antibody production
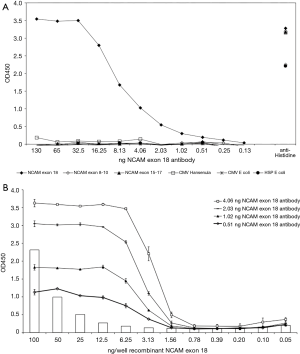
Balb/c mice were immunized with a truncated E18 protein (Figure S1). The hybridoma clone with the highest specificity was selected (MUMi-21B2. As shown in Supplementary Figure S2, panel A, the selected hybridoma clone produced antibodies binding to the E18 recombinant protein. No binding was found when ELISA plates were coated with His-tagged NCAM exon 8–10 or NCAM exon 15–17 proteins produced and purified from E. coli or other unrelated proteins. Thus the monoclonal antibody is specific for the E18 and is not cross reactive with the His-tag as expressed at the N-terminus of the purified proteins or with E. coli proteins potentially left as contaminants in the purified protein as used for immunization. Isotyping showed that the monoclonal antibody was of the IgM type. Sensitivity of the MUMi-21B2 monoclonal antibody was confirmed in a dose-response ELISA. Plates were coated with serially diluted recombinant protein. Serially diluted coating was confirmed using the anti-His polyclonal antibody detection (bars in Figure S1). As shown in the same Figure (bars in Figure S2, panel B), the purified monoclonal antibody specifically binds the serially diluted recombinant protein as coated to the plates. The data show that the optimal antibody concentration is about 4 ng antibody/well, and that the sensitivity is 10–20 ng/mL in the ELISA as used. Thus, the MUMi-21B2 monoclonal antibody is specific for the E18 protein with a sensitivity of 10–20 ng/mL. The monoclonal antibody recognizes an epitope in the recombinant protein produced in E. coli. In this host no posttranslational modification occurs. To determine whether the same epitope also is available in in vivo expressed eukaryotic proteins we performed immunocytochemistry to test the purified MUMi-21B2 monoclonal antibody for binding to E18 present in cultured SCLC cell lines.
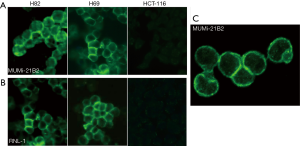
NCAM 180 staining in SCLC
The SCLC cell lines NCI-H69 and NCI-H82 showed clear expression of exon 18 in the RT-PCR. We performed immunocytochemistry on these cell lines with the MUMi-21B2 antibody. The cell lines were spotted on glass slides, fixed and permeabilized with ice-cold methanol and acetone and probed with the MUMi-21B2 monoclonal antibody. Staining with the RNL-1 monoclonal antibody, a commercially available NCAM-specific antibody, was used as a positive control. RNL-1 recognizes an extracellular epitope of the NCAM protein. The MUMi-21B2 antibody recognizes the cytoplasmic E18 part of the NCAM protein and only stains cells expressing the NCAM-180. Figure 4A, shows a strong reaction of the MUMi-21B2 monoclonal antibody, comparable to staining with antibody RNL-1 (Figure 4, panel B). No staining is observed in the HCT116 cell line, derived from colon carcinoma. The specific membrane bound localization of E18 is better shown in confocal images. Figure 4C shows NCI-H69 cells stained with the MUMi-21B2 monoclonal antibody. Going step-by-step through the depth of the stained cells, we observed a strong binding of the antibody at the cell-cell inter-phase of adherent cells.
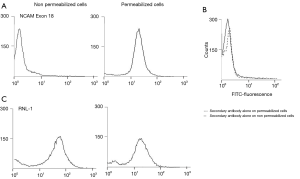
Cytoplasmic expression of E18
To confirm the intracellular location of E18 we used NCI-H82 cells. Binding of the monoclonal antibody MUMi-21B2 was evaluated for both permeabilized and non-permeabilized cells and fluorescence intensity was measured by flow cytometry. Our data clearly show that the MUMi-21B2 antibody binds to a cytoplasmic part of the NCAM protein. No binding of the antibody was found when non-permeabilized NCI-H82 cells were stained. As shown in Figure S3, panel A, staining of non-permeabilized NCI-H82 cells, results in a fluorescence intensity peak comparable to the fluorescence of staining with a secondary antibody alone (Figure S3, panel B). After permeabilization on the other hand, a shift of the peak to higher fluorescence intensity is found (Figure S3, panel A). This higher intensity is comparable to the intensity of the peak as measured for the RNL-1 antibody staining of NCI-H82 cells. For RNL-1 however, no difference in fluorescence is found for permeabilized versus non-permeabilized cells, because the RNL-1 monoclonal antibody binds to an extracellular NCAM epitope and the binding does not depend on the permeabilization of the NCI-H82 cells (Figure S3, panel C). These data demonstrate that MUMi-21B2 antibody specifically binds to a cytoplasmic part of the NCAM protein.
Antibody response in E18- immunized mice
Balb/c mice were immunized with E18 protein and plasmid DNA. SDS-PAGE analysis of the antigen revealed a pure protein, without dimers, with the expected molecular mass (32 kDa) (Figure 5A). The His-tagged recombinant protein antigen was inoculated either as suspension in PBS or formulated in Abisco-100. DNA immunization was performed by intramuscular injection of the mammalian expression vector pCI containing the exon 18 encoding DNA. Mice were immunized at day 0 and boosted on day 21. Read-out was performed on day 31 after the first immunization. Animals immunized with E18 protein both in PBS or formulated in Abisco-100 developed anti-E18 specific serum antibody titers as shown in Figure 5B group III and IV (dark bars). No binding of serum antibodies was shown when ELISA was performed with plates coated with His-tagged irrelevant protein that was also produced in E. coli [Figure 5B group III and IV (grey bars)]. The data show that no anti-histidine antibodies or no antibodies against E coli-linked protein were produced in the serum of the immunized animals. Immunization with plasmids encoding the exon 18 did not induce serum antibody titers, but when DNA-immunized mice were boosted with formulated E18 protein a clear induction of anti E18 antibody titers was found (Figure 5B group VI and VII). We conclude that immunization of mice with E18 protein induces a significant humoral immune response.
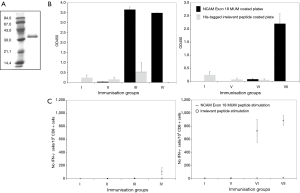
CTL response in E18-immunized mice
Balb/c mice were immunized as described. Mouse spleens were isolated and restimulated in vivo with overlapping nonamer peptides covering the amino acid sequence of E18 protein. A total of 67 overlapping peptides divided over 3 pools was used. We calculated the mean number of IFN-γ secreting cells per 105 CD8+ cells (Figure 5C group I-VII, 2 mice in each group). Our data show no or a low number of IFN-γ secreting CD8+ cells in the groups immunized with E18 (Figure 5C groups III and IV). For the DNA-immunized mice on the other hand intramuscular injection of the exon 18 encoding DNA clearly induced a significant CTL response (Figure 5C groups VI and VII). We found 727 IFN-γ secreting cells on a total of 105 CD8+ T cells (0.7%). There was a slight increase in the number of IFN-γ secreting CD8+ T cells for mice immunized with DNA and boosted with protein in Abisco-100 (887 IFN-γ secreting cells on 105 CD8+ cells, 0.9%). For all spleens as a negative control, an in vitro restimulation with an irrelevant peptide pool consisting of 17 overlapping peptides (9-mers, 5 amino acids overlap) covering a 76 amino acid sequence of the exon 7pi8 region of the NCAM protein was performed. For the recombinant protein as well as for the DNA-immunized animals no IFN-γ secretion of CD8+ T cells was found. Since T cells from immunized animals could not be restimulated with the irrelevant peptide pool, we conclude that immunization specifically induces E18-specific T cells, thereby confirming the specificity of the E18 immunization. We conclude that immunization with E18 and exon 18 encoding DNA induces an E18-specific T cell response. E18 protein immunization only resulted in a limited number of IFN-γ secreting T cells, whereas immunization with exon 18 encoding DNA induced a significant cellular E18-specific immune response.
Discussion
The results of this study show that the expression of NCAM exon 18 can be used as a tumor marker for SCLC. To determine the specificity of this marker more precisely, screening of larger panels of SCLC tissue compared to tissues of other tumor types have to be carried out. We have developed the tools to make these studies possible.
The precise biological consequences of expression of protein E18 are not clear. E18 could be a driver of cell proliferation an activity required during, but not after, embryonic development. Schor et al. (15) identified epigenetic modulation resulting in alternative splicing of NCAM and expression of E18 in neuroblastoma cells. A similar modulation could be the result of exposure to tobacco smoke. Ditlevsen et al. (16) reviewed the data on intracellular signaling by NCAM. They conclude that NCAM-180 can mediate neurite outgrowth. But other explanations are possible where expression of E18 is a result rather than the cause of carcinogenesis.
But even without understanding why E18 is expressed in SCLC and not in other tumor cells and normal cells, this finding can be used in the diagnosis and treatment of SCLC.
First the level of E18 circulating in the body can be used to follow the tumor load. This means that the effect of chemotherapy can be monitored. This could lead to a more refined chemotherapy that can be stopped when the required effect has been reached. After that regular monitoring should detect a relapse before clinical signs arise. At this point renewed or alternative treatment would be possible. There is a number of alternative treatments, but due to the rapid progression of the disease these are difficult to test in clinical trials (17-19). Real time monitoring of the tumor load in patients would enable personal medication. The recently developed possibilities to take frequent blood samples from patients and to analyze E18 or mRNA for E18 in these samples makes this approach possible (20,21).
Second, the presence of E18 in tumor cells offers a target for immunotherapy. Although E18 is intracellular, it will be processed by proteasomes and the resulting peptides will be presented on the cell surface. In this way CTLs can recognize the tumor cells and kill them. Our preliminary experiments in mice have shown that this approach has potential.
Acknowledgements
The authors like to thank Ms. Tamara Geerts, Ms. Annemie Valckx and Dr. Tim Perrera (Oncology Discovery Research and early development, Johnson and Johnson Pharmaceutical Research and Development, Beerse, Belgium), Ms. Sandra Claessen and Ms. Mieke Henfling (Department of Molecular Cell Biology, University Maastricht) for kindly providing the tumor cell line pellets. Dr. Marielle Thewissen, and Dr. Niels Hellings for the PBMC pellets of healthy controls (Hasselt University, Biomedical Research Institute and Transnationale Universiteit Limburg, School of Life Sciences, Diepenbeek, Belgium). Dr. Dendouga Najoua (MUbio Products B.V., Maastricht), Dr. Marcel Kuiper (Onyvax Limited, St George’s University of London, UK), Dr. Gereon Poschmann and Dr. Kai Stühler (Medical Proteome Centre, Bochum Germany) for peptide analysis of the recombinant E18protein. Drs. Steve Schoonooghe for technical help. Mr. Dirk Van Meensel (Carl Zeiss NV, Zaventem Belgium) for the help with the confocal imaging and imaging data processing.
Funding: This work was supported by a grant from the European Commission (COOP-CT-2004-512855). A.V.B. and M.D. were appointed by MUbio Products B.V. MUbio Products B.V. was the holder of the patent Cancer Vaccine (Application WO/2007/104511) that is presently owned by Catherinn B.V.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: SCLC tissue sections came from archival material at the Department of Genetics and Cell Biology. The animal experiments were carried out at CIRES and were approved by the Bezirksregierung Arnsberg under number 50.8735.1.
References
- Gazdar AF, Bunn PA, Minna JD. Small-cell lung cancer: what we know, what we need to know and the path forward. Nat Rev Cancer 2017;17:765. [Crossref] [PubMed]
- van Meerbeeck JP, Fennell DA, De Ruysscher DK. Small-cell lung cancer. Lancet 2011;378:1741-55. [Crossref] [PubMed]
- Rudin CM, Ismaila N, Hann CL, et al. Treatment of Small-Cell Lung Cancer: American Society of Clinical Oncology Endorsement of the American College of Chest Physicians Guideline. J Clin Oncol 2015;33:4106-11. [Crossref] [PubMed]
- Lally BE, Urbanic JJ, Blackstock AW, et al. Small cell lung cancer: have we made any progress over the last 25 years? Oncologist 2007;12:1096-104. [Crossref] [PubMed]
- Jensen M, Berthold F. Targeting the neural cell adhesion molecule in cancer. Cancer Lett 2007;258:9-21. [Crossref] [PubMed]
- Cunningham BA, Hemperly JJ, Murray BA, et al. Neural cell adhesion molecule: structure, immunoglobulin-like domains, cell surface modulation, and alternative RNA splicing. Science 1987;236:799-806. [Crossref] [PubMed]
- Gower HJ, Barton CH, Elsom VL, et al. Alternative splicing generates a secreted form of N-CAM in muscle and brain. Cell 1988;55:955-64. [Crossref] [PubMed]
- Farag SS, Caligiuri MA. Human natural killer cell development and biology. Blood Rev 2006;20:123-37. [Crossref] [PubMed]
- Onganer PU, Seckl MJ, Djamgoz MB. Neuronal characteristics of small-cell lung cancer. Br J Cancer 2005;93:1197-201. [Crossref] [PubMed]
- Nguyen C, Mattei MG, Mattei JF, et al. Localization of the human NCAM gene to band q23 of chromosome 11: the third gene coding for a cell interaction molecule mapped to the distal portion of the long arm of chromosome 11. J Cell Biol 1986;102:711-5. [Crossref] [PubMed]
- Pollerberg EG, Sadoul R, Goridis C, et al. Selective expression of the 180-kD component of the neural cell adhesion molecule N-CAM during development. J Cell Biol 1985;101:1921-9. [Crossref] [PubMed]
- Murray BA, Owens GC, Prediger EA, et al. Cell surface modulation of the neural cell adhesion molecule resulting from alternative mRNA splicing in a tissue-specific developmental sequence. J Cell Biol 1986;103:1431-9. [Crossref] [PubMed]
- Ronn LC, Hartz BP, Bock E. The neural cell adhesion molecule (NCAM) in development and plasticity of the nervous system. Exp Gerontol 1998;33:853-64. [Crossref] [PubMed]
- Falkenberg FW, Weichert H, Krane M, et al. In vitro production of monoclonal antibodies in high concentration in a new and easy to handle modular minifermenter. J Immunol Methods 1995;179:13-29. [Crossref] [PubMed]
- Schor IE, Fiszbein A, Petrillo E, et al. Intragenic epigenetic changes modulate NCAM alternative splicing in neuronal differentiation. EMBO J 2013;32:2264-74. [Crossref] [PubMed]
- Ditlevsen DK, Povlsen GK, Berezin V, et al. NCAM-induced intracellular signaling revisited. J Neurosci Res 2008;86:727-43. [Crossref] [PubMed]
- Mamdani H, Induru R, Jalal SI. Novel therapies in small cell lung cancer. Transl Lung Cancer Res 2015;4:533-44. [PubMed]
- Santarpia M, Daffina MG, Karachaliou N, et al. Targeted drugs in small-cell lung cancer. Transl Lung Cancer Res 2016;5:51-70. [PubMed]
- Lehman JM, Gwin ME, Massion PP. Immunotherapy and Targeted Therapy for Small Cell Lung Cancer: There Is Hope. Curr Oncol Rep 2017;19:49. [Crossref] [PubMed]
- Brinton LT, Sloane HS, Kester M, et al. Formation and role of exosomes in cancer. Cell Mol Life Sci 2015;72:659-71. [Crossref] [PubMed]
- Mamdani H, Ahmed S, Armstrong S, et al. Blood-based tumor biomarkers in lung cancer for detection and treatment. Transl Lung Cancer Res 2017;6:648-60. [Crossref] [PubMed]


