Autochthonous murine models for the study of smoker and never-smoker associated lung cancers
Introduction
With the rise of smoking since World War I (1), lung cancer has the highest annual cancer incidence world-wide (2) and is the leading cause of cancer mortality in the USA. (3) and the world (2). Tobacco smoke contains >60 carcinogens including polycyclic aromatic hydrocarbons (PAHs), N-nitrosamines, aromatic amines, aldehydes, isoprenes, arsenic and cadmium (4).
Lung cancer among never-smokers, defined as those who have smoked <100 cigarettes in their lifetime (5), accounts for ~10–20% of cases (6-8), with more women than men (6). Risk factors include second hand smoke (9-12), radon (13,14), occupational exposures (asbestos, chromium, and arsenic) (15-17), indoor (18-20) and outdoor (21,22) air pollution, oncogenic viruses including human papilloma virus (HPV) (23-25), genetic factors (26-29), and a history of pulmonary disease (30,31). However, identification of the causative factors has remained elusive.
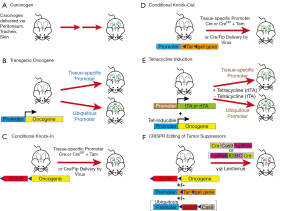
Murine cancer models are now a standard preclinical in vivo platform for the elucidation of cancer biology and the development of novel therapeutics. The models include xenograft or allograft (syngeneic) transplantation, carcinogen induced tumor models (CITMs), and genetically-engineered mouse models (GEMMs). In xenograft models, cancer cells are transplanted, subcutaneously or orthotopically, to establish tumors in vivo. In contrast, CITMs and GEMMs generate endogenous spontaneous cancers. Both models accurately reflect the histological progression of precancerous lesions to invasive cancers. CITMs may represent a more ‘realistic’ model of tobacco smoke-induced tumorigenesis than GEMMs. However, CITMs often require extended time periods for tumor development, particularly for invasive cancers. GEMMs, unlike CITMs, generate cancers through manipulation of specific oncoproteins or tumor suppressors (TSs). The precise and limited nature of genetic manipulation in GEMMs has yielded valuable biological insights on tumor initiation and progression. Several reviews of lung cancer GEMMs have been published recently (32-35). Here, we will focus primarily on CITMs and GEMMs of lung cancer and their relation to human never-smoker and smoker lung cancers. Methods of tumor induction in mouse models are presented in Figure 1.
Histology of lung cancers
Lung cancer is a heterogeneous group of epithelial malignancies with four major histological subtypes (36): small cell lung cancer (SCLC), adenocarcinoma, squamous cell carcinoma (SCC), and large cell carcinoma (LCC). The latter three are grouped together as non-small cell lung cancer (NSCLC). Adenocarcinoma occurs most frequently and accounts for ~45% of all lung cancers in the USA, followed by SCC (23%), SCLC (13%) and large cell carcinoma (1.7%) (37). Adenocarcinomas tend to be peripheral lesions whereas SCC and SCLC are more central. Most adenocarcinomas arise from type 2 pneumocytes in the alveoli (38,39). SCC most likely arises from CK5+ basal epithelial cells of bronchi (40). SCLC is thought to arise from neuroendocrine cells that often congregate into neuroendocrine body clusters located at junctions of bronchi (41). Both bronchi and neuroendocrine bodies are located more centrally.
Tobacco smoke is significantly associated with all four major histological subtypes although the association is strongest with SCLC and SCC (42-44). Adenocarcinoma is associated with both smokers and never-smokers (5) but is the most common histology among never-smokers (45-47). Tar in cigarette smoke is associated with SCC and SCLC (48,49) whereas nitrosamines such as 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) and PAHs are known to induce adenomas (50).
Human genomics
Lung SCC, SCLC, and adenocarcinoma, as smoking-associated cancers, rank among the top 5 cancers with the highest prevalence of somatic mutations (51) with higher frequencies of chromosomal abnormalities (52). Lung SCC, SCLC, and adenocarcinoma are notable for having a strong correlation with C>A transversion and C>T transition mutations (44,51). C>A transversions are primarily associated with smoking while C>T transitions are found in cancers of smokers and never-smokers (44).
Adenocarcinoma
Sequencing efforts (53,54) revealed that smoking-associated lung adenocarcinoma (LAD), when compared to those of never-smokers, had significantly higher mutation rates (10.5 vs. 0.6 mutations/Mb) (53), C>A transversions, and inversely correlated with C>T transitions (higher in never-smokers) (53,54). EGFR (53), PIK3CA and RB1 (54) mutations, ERRB2 insertions (54) and c-Ros oncogene 1 (ROS1) and anaplastic lymphoma kinase (ALK) fusions (53) were significantly correlated with never-smokers. Mutations in KRAS (53,54), BRAF (53), JAK2/3 (53), mismatch repair genes (53) and TS genes (e.g., TP53, STK11, SMARCA4, KEAP1) (54) were significantly correlated with smoking-associated LADs. Significant amplifications in NKX2-1, TERT, MDM2, KRAS, EGFR, MET, CCNE1, CCND1, TERC and MECOM and deletion in CDKN2A were noted (54), but not associated with smoking status.
Squamous cell lung cancer
Sequencing efforts from The Cancer Genome Atlas (TCGA) Research Network revealed significant rate of mutations in TP53, CDKN2A, PTEN, MLL2, RB1, PIK3CA, KEAP1, HLA-A, NFE2L2, and NOTCH1 (55). Copy number alterations of previously described SOX2, PDGFRA, KIT, EGFR, FGFR1, WHSCIL1, CCND1, and CDKN2A were identified. Amplifications of NFE2L2, MYC, CDK6, MDM2, BCL2L1, and EYS and deletions in FOXP1, PTEN, and NF1 were identified. Forty-seven percent of tumors had alterations in genes of the PI3K pathway, 24% in RAS pathway, and 28% in receptor tyrosine kinase (RTK) pathways. Many of the NOTCH1 mutations were truncating alterations suggesting a role for NOTCH1 loss-of-function (LOF). Currently, there are no clinically-available therapies that target these mutations in SCC although a number of targeted therapies are under clinical investigation.
SCLC
SCLC consists of two distinct histological subgroups that correlate with the expression of the basic helix-loop-helix (BHLH) transcription factors (TFs) ASCL1 and NEUROD1 (56): (I) classic SCLC constitute the majority of tumors and express high levels ASCL1 expression alone while (II) variant SCLC may express high levels of NEUROD1 +/− ASLC1 or low levels of NEUROD1/ASCL1 (57) although the precise definition of variant SCLC is in flux. ASCL1 is critical for neuronal and pulmonary neuroendocrine cell (PNEC) development and growth of SCLC (58). NEUROD1 is a neuronal differentiation TF (59), critical for the neural (60,61) and PNEC development. Classic ASCL1(+) SCLC present as small cells with high nuclear:cytoplasmic ratios, and fine granular chromatin (62). Variant NEUROD1+ SCLC are larger cells with prominent cytoplasm and nucleoli (62). ASCL1 targets include MYCL1, RET, SOX2, BCL2, NFIB, DLL3, 4 and JAG2. NEUROD1 primarily targets MYC (63).
All sequencing studies (64-67) identified high levels of C>A transversions consistent with smoking-associated cancers and nearly all tumor samples had inactivating alterations in TP53 and RB1. In contrast to adenocarcinoma (54) and SCC (55), few significant oncogenic driver mutations were identified (67). However, recurring amplifications in MYC family members (MYC, MYCN, and MYCL1), FGFR1, IRS2 were identified (67). SOX2 amplifications have also been reported (66). Other significantly mutated genes include CREBBP, COL22A1, RBL1, RBL2, TP73, FMN2, and NOTCH1 (67). No therapies that target these mutations are currently in clinical use although clinical trials of such agents are underway.
Mouse genomics
Adenocarcinoma
Whole exome sequencing (WES) revealed that carcinogen-induced adenomas had far more single nucleotide variations (SNVs) than KrasLA2 GEMM adenomas (68), with 728, 185, and 47 SNVs in MNU-, urethane-treated, and KrasLA2 mice, respectively (68). In addition to KrasG12D (MNU) and KrasQ61R (urethane) mutations, carcinogen-induced tumors frequently had mutations in genes commonly found in human adenocarcinomas including Arid1b, Atm, Crebbp, and Mll2. In contrast, KrasLA2 mice had frequent copy number alterations (CNAs) whereas CNAs and aneuploidies in CITMs were extremely infrequent. In most KrasLA2 adenomas, chromosome 6 (Chr6) gained an extra copy, where Kras is located. Other chromosomal changes include gains of chromosomes 2, 10, 12, 15 and 17 and losses of chromosomes 4, 9, 11, and 17. Frequently amplified genes included Kras, Ret, Met, Alk, Arid1b, and Akt1.
WES of adenomas from KrasG12D/+, KrasG12D/+;Trp53fl/fl, CCSP-rtTA;TetO-EGFRL858R, and CCSP-rtTA;TetO-MYC revealed a similarly low nonsynonymous mutation rate of 0.06, 0.07, 0.02, and 0.14 mutations/MB in comparison to 1.97 and 7.76 mutations/MB for human never-smoker and smoking-associated LAD, respectively (69). CNAs were common in all GEMMs with gains in chromosome 6, 2, 15, and 19 and losses of chromosome 9 and 14 for the Kras models, including amplification of the mutant KrasG12D gene. EGFR mutant GEMMs displayed a different pattern of CNAs with gain of Chr12 as the dominant feature for unclear reasons.
SCLC
Sequencing efforts of murine Trp53fl/fl;Rb1fl/fl SCLC identified chromosomal gains and losses as the dominant genetic aberrations (70). Murine SCLC had a point mutation rate of 0.91 per MB in contrast to 5.5–8.62 per MB of human SCLC (66,67). Murine SCLC most frequently exhibited hemizygous loss of Chr19 followed by gain of Chr14. Others changes include hemizygous loss of chromosomes 8, 12, 14 and gain of Chr16. Focal amplifications of Mycl1 and Nfib on Chr4 and deletions of Mir200a/b and Hes family members were noted. LOF mutations in Pten, located on Chr19, were frequently identified with hemizygous loss Chr19. Trp53fl/fl;Rb1fl/fl;Ptenfl/fl tumors harbored two copies of intact Chr19 whereas tumors from Trp53fl/fl;Rb1fl/fl;Ptenfl/+ mice contained hemizygous loss of Chr19 suggesting that Pten LOF mutations may drive the loss of Chr19 in murine Trp53fl/fl;Rb1fl/fl SCLC. In contrast, Rb1fl/fl;Trp53fl/fl;MycLSL/LSL mice that express a constitutively active MYC-T58A do not have any Mycl or Nfib amplifications (71), perhaps due to the sufficiency of MYC-T58A to drive tumor progression in the context of Trp53 and Rb1 loss.
Never-smoker lung cancer
Adenocarcinoma is the dominant histology of never-smoker lung cancers. Here, we will focus on prominent genetic alterations of adenocarcinomas and their corresponding mouse models (Table 1).

Full table
EGFR mutations
EGFR is a RTK that regulates multiple downstream pathways including RAS/MAP kinase pathway, PI3K pathway, and the JAK-STAT pathways (81). EGFR mutations occur primarily in never-smokers (53,54) and are the most common mutations in never-smoker LAD with ~15% of all LADs (54). Activating mutations occur in exons 18–21 of the tyrosine kinase domain (82-84) with L858 mutations and exon 19 deletions being most common (84). The tumors are highly susceptible to oral EGFR tyrosine kinase inhibitors (TKIs), such as erlotinib and gefitinib, and can lead to dramatic responses with durations of ~6–12 months. Secondary mutations in the EGFR tyrosine kinase domain account for 60% of acquired resistance to EGFR TKIs, of which, the T790M mutation accounts for 90% of resistant mutations (85). Osimertinib, a third generation, irreversible antagonist of the EGFR kinase domain, is FDA-approved for treatment-naïve LAD with activating-EGFR mutations and for second-line EGFR-T790M mutations after erlotinib or gefitinib therapy. An EGFR-C797S mutation confers resistance to osimertinib (86-88). Other mechanisms of resistance to TKIs include compensatory amplification of the MET gene leading to activation of the PI3K pathway (85), activating mutations in PIK3CA and BRAF, amplification of CRKL and ERK genes (85), and transformation to SCC (89) and SCLC (90,91).
EGFR mutant mouse models
In mutant EGFR GEMMs with tetracycline-inducible EGFR exon 19 deletion or EGFR-L858R mutants (72,73), the club cell secretory protein (CCSP or CC10) promoter regulated the expression of the reverse tetracycline transactivator (rtTA; Tet-On) (92,93). Treatment of both models with doxycycline resulted in LAD (72,73). Bi-transgenic L858R mutant mice generated adenocarcinoma in situ at 2 weeks with progression to multifocal adenocarcinoma with >4 weeks of doxycycline treatment. The tumors expressed surfactant protein C (SPC) but not CCSP consistent with an alveolar type 2 pneumocyte origin (72,73). Mice with exon 19 deletions developed tumors similarly to EGFR-L858 mice but at a slower rate. The tumors regressed when doxycycline treatment was withdrawn suggesting that the tumors are dependent on the mutant EGFR activity for growth. Furthermore, the tumors were sensitive to growth inhibition by erlotinib (72) or HKI-272 (73), two EGFR TKIs.
A GEMM with tetracycline-inducible expression of a dual mutant EGFR–L858R-T790M (EGFR-TL) under CCSP-rtTA control developed adenocarcinoma in situ with progression to both peripheral and bronchial adenocarcinoma (74). The T790M mutation induced resistance to erlotinib. The peripheral adenocarcinomas had bronchoalveolar features whereas the bronchial tumors were papillary adenocarcinomas. Peripheral tumors were strongly SPC(+) and CCSP(−) (74), suggesting a type II pneumocyte origin. In contrast, the bronchial tumors were CCSP(+) and SPC(−), suggesting a club cell origin. All tumors were resistant to erlotinib. Only peripheral tumors showed any responses with HKI-272, an irreversible EGFR TKI. Peripheral and bronchial tumors responded to treatment with rapamycin, an mTOR antagonist, and HKI-272 (74).
ALK fusions
ALK is an orphan RTK that was first identified in anaplastic large cell lymphomas and a subset of T-cell lymphomas (94). ALK fusions are found in ~3–7% of LADs, mostly among never-smokers (53). In LAD, echinoderm microtubule associated protein like 4 (EML4) is the primary fusion partner with ALK resulting in the constitutive activation of the ALK kinase (95). Smaller subsets of fusions with other proteins have also been identified (96). Crizotinib (97), an oral ATP-competitive TKI of ALK, MET, and ROS1 RTKs, is approved by the FDA for the treatment of advanced ALK-fusion LAD. Responses to crizotinib can be dramatic but only last ~6–12 months. Mutations in the ALK tyrosine kinase domain constitute ~30% of resistant cases with L1196M mutation being the most common (98,99). Other resistance mutations include C1156Y (100), G1202R (99), S1206Y (99), G1269A (98), and F1174L (101). Alternative resistance mechanisms include ALK amplification, activating KRAS mutations, EGFR activation, and c-KIT overexpression. Second generation ALK antagonists, ceritinib (102,103), brigatinib (104,105) and alectinib (106-108), are more potent ALK antagonists with activity against many of the crizotinib-resistance mutants including L1196M. The three antagonists have been approved by the FDA after progression on crizotinib. Alectinib has been approved by the FDA for initial therapy. Pemetrexed, an anti-metabolite chemotherapy, has activity against ALK-fusion LAD albeit with decreased response rates and shorter PFS than targeted agents (109-111).
ALK fusion mouse models
EML4-ALK expression, regulated by the SPC promoter (75) or CC10-rtTA with tetracycline (76), led to development of adenocarcinomas that were sensitive to ALK inhibition. No distant metastases were noted in either model. In SPCCreERT2; EML4-ALKLSL mice (Figure 1C), treatment with tamoxifen induced nuclear translocation of cre recombinase to initiate the expression of EML4-ALK in type II pneumocytes (77). Crizotinib-sensitive tumors developed within 1 week. Crizotinib-resistant EML4-ALK-L1196MLSL or -F1174LLSL mutant LAD, induced with adenovirus-expressing Cre recombinase (adeno-Cre), showed susceptibility to the combination of 17-DMAG, an HSP90 antagonist, and TAE684, a second-generation ALK antagonist (112).
Two groups independently used Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)/Cas9 technology to generate adenocarcinomas with endogenous EML4-ALK fusions. Adenoviruses or lentiviruses expressing Cas9 with single guide RNAs (sgRNAs) against EML4 intron 14 and ALK intron 19 were used to generate the fusion proteins (113,114). Tumors progress through papillary intrabronchial hyperplasia, atypical adenomatous hyperplasia (AAH), and adenocarcinoma within 8 weeks (113,114). Tumors were sensitive to crizotinib treatment (113).
ROS1 fusions
ROS1 is an RTK of the insulin receptor family with significant sequence and structural homology to ALK (115). ROS1 fusion proteins occur in ~1–2% of LADs (116-118) and are sensitive to crizotinib (116,119,120). CD74-ROS1 and SLC34A2-ROS1 are the most frequent alterations of the >20 ROS1 fusion proteins found in solid tumors (115,118,121).
ROS1 fusion mouse models
To date, three GEMMs of ROS1 fusion LAD have been developed: CD74-ROS1 (78), SDC4-ROS1 (78), and EZR-ROS1 (79) under the control of the SPC promoter. The mice develop papillary nodules within 3–4 weeks after birth and progress to adenocarcinoma, with decreased survival compared to wild type cohorts (78,79). Distant metastases were identified in <1% of CD74-ROS1 and SDC4-ROS1 mice (78).
PIK3CA mutations
PI3K is the upstream mediator of the PI3K-mTOR pathway and is one of the most commonly mutated pathways in human cancers (122). Mutations in PIK3CA occur in 7% of human LAD and are associated with the never-smoker group (54). The helical domain E545K and E542K mutations are the most common followed by the kinase domain H1047R mutations (54). These mutations occur in the p110 catalytic subunit of PI3K, leading to its activation and downstream signaling of the PI3K-mTOR pathway (123-125).
PIK3CA mouse models
Doxycycline treatment of CCSP-rtTA; TetO-PIK3CAH1047R mice induced expression of PIK3CAH1047R and developed LAD within 12 weeks of doxycycline treatment with regression of tumors upon doxycycline withdrawal (80). Treatment with the NVP-BEZ235, a dual PI3K-mTOR antagonist, decreased tumor growth and 18fluorodeoxyglucose (18FDG) avidity of the tumors on positron emission tomography (PET) scans (80).
Smoking-associated lung cancers
Tobacco smoke still causes the majority of LAD and is the primary cause of lung SCC and SCLC (42-44). Mouse models of smoking-associated lung cancer consist of three distinct groups: GEMMs, CITMs, and tobacco smoke-induced mouse models (TSMs). GEMMs are the most popular models due to the precise modulation of critical oncogenes and TSs with relatively short and consistent duration to tumor formation compared to TSMs and CITMs (Tables 2,3). TSMs have generally fallen out of favor due to time and labor-intensive procedures with long latency times and generation of primarily adenomas.
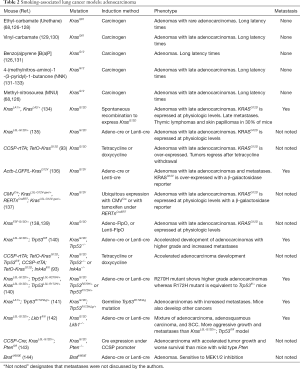
Full table
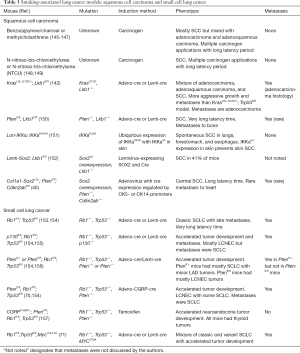
Full table
Adenocarcinoma
KRAS
KRAS mutations are the most common oncogenic driver alterations occurring in ~30% of LAD (54), are strongly associated with smoking (53,54), and confer a worse survival than tumors with wild-type KRAS (158,159). Activating KRAS mutations primarily occur in the glycine residues of codon 12 (G12, 91%) and 13 (G13, 6%), the glutamine of codon 61 (Q61, 2%) (160). Among the G12 mutations, substitution by cysteine (G12C, 44%) is most common, followed by valine (G12V, 23%) and aspartate (G12D, 17%) (160). There are no direct inhibitors of RAS proteins in clinical trials although small molecule irreversible antagonists against KRAS-G12C mutants have been reported (161-163).
Adenocarcinoma CITMs
The vast majority of CITMs are mutant Kras adenocarcinoma models induced by single cigarette smoke carcinogens. Commonly used carcinogens are benzo(a)pyrene [B(a)P] (126,131), ethyl carbamate (urethane) (126-128), vinyl carbamate (129,130), 4-(methylnitrosamino)-1-(3-pyridyl)-1butanone (NNK) and N’-nitrosonornicotine (NNN) (164). All of these compounds require cytochrome P450 enzymes found in lung epithelia for bioactivation (165), leading to formation of DNA adducts (132). The majority of induced lung nodules are adenomas with some adenocarcinoma formation within a year (68,126,133,164,166). B(a)P (126) and urethane induce C>A transversions and T>A transversions, respectively, in the Kras gene leading to G12 and Q61 mutations, respectively (68,126)—modeling the mutations seen in human LAD. NNK (133,166) and methylnitrosourea (MNU) (126) induce C>T transitions leading to KrasG12 mutations.
The frequency and latency of lung cancer incidence strongly depend on the inbred strain. A/J and SWR strains are the most susceptible, BALB/c and FVB have intermediate susceptibility and C57Bl/6, DBA, and AKR strains are relatively resistant (167,168).
Kras GEMMs
Collectively, mutant Kras GEMMs are currently one of the most commonly used autochthonous models. The first 5 models were reported in 2001: KrasLA1/+ (134), KrasLA2/+ (134), KrasLSL-G12D/+ (135), CCSP-rtTA; TetO-KrasG12D (93) and Actb-LGFPL-KrasG12V where LGFPL encodes for a loxp-GFP-polyA-loxp cassette (136). These models evolve from epithelial hyperplasia to lung adenomas and eventually adenocarcinomas. The KrasLA models spontaneously activate a latent KrasG12D allele (134); the LA1 mice activate the KrasG12D allele in half of the genetic recombinations whereas the LA2 mice activate the mutant Kras allele in all recombinations. Thus, the LA2 mice develop more tumors with shorter latency and survival times. The mice also develop thymic lymphomas and skin papillomas but not pancreatic or colon cancers despite the high frequency of Kras mutations in these cancers. The other models have greater lung specificity through use of CCSP-promoter or adeno-Cre or lentivirus-expressing Cre recombinase (lenti-Cre) administration via intranasal (135) or intratracheal routes. The CCSP-rtTA;TetO-KrasG12D (93) and Actb-LGFPL-KrasG12V (136) models express the mutant KRAS at supra-physiological levels that may account for their relative short latency of adenocarcinoma formation. The KrasLA (134) and KrasLSL-G12D/+ (135) mice express the mutant protein at physiologic levels as the mutations are positioned within the endogenous Kras locus. KrasLSL-G12V-geo/+ mice express KRAS-G12V at physiologic levels with a galactosidase reporter (137). The KrasLSL-G12D/+ is the most widely used model to generate mutant Kras LADs. Flp-FRT regulated KrasFSF-G12D/+ allele, where KrasG12D is induced by Flp recombinase, was developed to independently manipulate alleles controlled by other conditional systems in compound mutant mice (138,139).
Homology-directed repair (HDR) by CRISPR-Cas9 was used to induce tumors with twelve distinct KRAS-G12 and G13 mutations simultaneously in the same mouse (169) (Table 4). The adeno-associated viruses (AAVs) contained a sgRNA to target the KRAS region of interest, a Kras HDR template containing the mutation and a barcode, and Cre recombinase. Cell numbers of tumors derived from each Kras mutation were quantified using next generation sequencing (NGS) and Tuba-seq (170), a bioinformatics pipeline that tallies the barcode sequences as a surrogate for tumor size. KRAS-G12D, -G12R, and -G13R were the most potent oncogenes in the context of all three tested genotypes: Rosa26LSL-tdTomato/+;H11LSL-Cas9/+ (TC), TC;Trp53fl/fl, and TC;Lkb1fl/fl (169).

Full table
Mutant Kras strains have been the most common platforms to study TS loss. Conditional loss of p53 in KrasLSL-G12D/+;Trp53fl/f mice caused accelerated tumor development with higher grade (93,140) and increased metastases (140). Mice expressing the p53 contact mutant, Trp53LSL-R270H/fl, or the structural mutant, TrpLSL-R172H/fl, developed lung tumors with similar phenotypes to Trp53fl/fl (140). However, only the Trp53LSL-R270H/+ mutant, and not the TrpLSL-R172H/+ mutant, developed higher grade tumors than Trp53fll+, suggesting that the R270H mutant has greater oncogenic potential than the R172H mutant in KrasG12D LAD (140). KrasLA1/+;Trp53R270H∆g/+ mice have a germline TrpR270H mutation with loss of one intronic nucleotide (174). They develop LAD with metastases in 37% of mice but also develop mesotheliomas, sarcomas, carcinomas, and lymphomas (141). KrasLSL-G12D/+;Lkb1fl/fl mice with Lkb1 deletions (human STK11) developed a mixture of adenocarcinoma, adenosquamous carcinoma and SCC with greater tumor multiplicity and metastases than KrasLSL-G12D/+;Trp53fl/fl mice (142). Regional lymph node metastases occurred in 61% of mice but were all adenocarcinomas. CCSPCre;KrasLSL-G12D/+;Ptenfl/fl mice also showed increased tumor multiplicity and poorer survival compared to Pten+/+ mice (143).
BRAF
BRAF mutations occur in 7% of human LAD (54). BRAF acts downstream of KRAS as part of the MAPK signaling pathway (175). Conditional expression of mutant BRAFV600E after intranasal instillation of adeno-cre developed SPC(+), CCSP(−) adenomas suggestive of alveolar origin (144). In contrast to KrasLSL-G12D/+ mice, BrafLSL-V600E mice rarely progressed beyond adenomas. Treatment with PD0325901 (176), a MEK1/2 antagonist downstream of BRAF, inhibited adenoma formation.
CRISPR-mediated loss of TSs
Generation of GEMMs with loss of multiple TSs is time consuming and costly due to the requisite germline modifications and the number of crosses required to generate the desired genotype. To circumvent these problems, CRISPR/Cas9 technology has been utilized in GEMMs to generate lung cancers with TS loss (171,172), primarily in mutant Kras LAD GEMMs (Table 4). DNA constructs that encode for Cre recombinase to induce recombination of Loxp sites, a Cas9 nuclease and sgRNA to enable CRISPR-mediated gene editing have been packaged in lentiviruses for in vivo use. pSECC, the first generation construct, was utilized to induce loss of NKX2-1, APC, and PTEN proteins in LADs of KrasLSL-G12D/+ and KrasLSL-G12D/+;Trp53fl/fl mice (171). A second generation construct that improved lentiviral titers, LentiCRISPRv2Cre, was utilized to verify the TS properties of SETD2 in KrasLSL-G12D/+;Trp53fl/fl mice (172). Extending this concept further, lentiviral constructs encoding Cre recombinase, sgRNA against TSs of interest, a sgRNA-identifier, and a barcode unique to the construct were used to induce multiplexed TS loss simultaneously in the same mouse (170,173). Precise quantification of tumor cell number was performed using NGS and Tuba-seq bioinformatic analyses (170). Lentiviruses encoding sgRNAs against 11 putative TS genes were delivered simultaneously to KrasLSL-G12D/+;Rosa26LSL-tdTomato/+;H11LSL-Cas9/+ (KTC), KTC;Trp53fl/fl (KTCP), and KTC;Lkb1fl/fl (KTCL) mice. Lkb1, Setd2, and Rb1 were identified as the most potent TSs in KTC mice (170,173), Smada4, Arid1a, and Atm were TSs only in KTCP mice, and Rb1 and Apc were the only potent TSs in all three mouse genotypes (173). These results suggest that the tumor suppressive ability of genes is not universal but rather dependent on the context of other genetic alterations.
Squamous cell carcinoma
Lung SCC models have been the most difficult to establish among the major histological subtypes and consequently, the least studied.
Carcinogen-induced models
Repeated intratracheal instillation of high dose benzo(a)pyrene/charcoal (145,146) or 3-methylcolanthrene (147), up to 16 weeks, generated a mixture of adenomas, adenocarcinomas and SCC with SCC being the dominant histology. These experiments were technically difficult and not easily replicated. A third model used repeated cutaneous applications of N-nitroso-bis-chloroethylurea (148) or N-nitroso-tris-chloroethylurea (NTCU) given twice per week from 23 to 43 weeks (148,149). Mice developed early hyperplasia of bronchiole epithelia with transitions to metaplasia and SCC and adenosquamous carcinoma. The tumors expressed CCSP (148) and cytokeratin 5/6 [CK5/6, (149)], but not chromogranin (a neuroendocrine marker) nor surfactant apoprotein (an alveolar cell marker), consistent with SCC (148). RNA-seq transcriptome analysis of early preneoplastic airway epithelia after 24 weeks of NTCU treatment revealed upregulation of PI3K and Myc pathways (177).
GEMMs
The first GEMM of lung SCC was reported in KrasLSL-G12D/+;Lkb1fl/fl mice (142). However, two issues have prevented widespread use of this model for studying lung SCC. First, a mixture of adenocarcinoma, adenosquamous, and SCC were generated with 56% of mice developing SCC or mixed adenosquamous histology. Metastases were noted although they were adenocarcinomas rather than SCC. Second, KRAS mutations are rare in human lung SCC (55).
Ptenfl/fl;Lkb1fl/fl strain (150) was developed as LOF PTEN mutations are frequent in human SCC (55). All Ptenfl/fl;Lkb1fl/fl mice developed SCC within 40–50 weeks and strongly expressed the SCC markers p63, KRT5, and SOX2. A low frequency (4%) of axial skeletal metastasis was observed.
In a third model, ~20% of mice homozygous for the kinase dead IKKα-K44A mutant (IKKaKA/KA) developed spontaneous lung SCC but also developed significant skin papillomas and SCC (151). Expression of wt IKKα regulated by the skin-specific loricrin promoter (Lori-IKKα) rescued the skin phenotype of the IKKaKA/KA mice while lung SCC developed in all of the mice within 4–6 months of age. Interestingly, LKB1 expression was downregulated in the lung SCCs. The model also developed SCC in the forestomach and esophagus, and lacked both lymph nodes and metastases. The absence of lymph nodes may possibly account for lack of metastases.
SOX2 is often amplified in lung SCC (55,178) and several corresponding GEMMs have been developed. Lkbfl/fl mice treated with lentiviruses expressing SOX2 and Cre recombinase by intranasal inhalation developed multifocal SCC in 41% of mice within 6–10 months (152) whereas Trp53fl/fl mice developed adenocarcinoma. The tumors expressed the typical SCC markers and high levels of FGFR and pSTAT3. Another model used adenoviruses with promoter-specific expression of Cre recombinase in Sox2LSL;Ptenfl/fl;Cdkn2abfl/fl mice (40). Induced SOX2 expression in type 2 pneumocytes (SPC promoter), club cells (CC10 promoter), and tracheal-bronchial basal cells (CK5 or CK14 promoter) develop SCC in 100%, 100% and 73% of mice, respectively, within 7–8 months. Metastasis (to the heart) was noted in only one of six mice treated with adeno-CK5-Cre. The basal cell targeted adenoviruses generated central SCC similar to human lung SCC. Adenoviruses targeting club cells or type 2 pneumocytes generated peripheral lung SCC. As discussed previously, type 2 pneumocytes and a subset of club cells have been described as cells of origin for adenocarcinoma. These results suggest that SOX2 is a determinative oncogene for SCC in the context of Pten and Cdkn2ab loss regardless of the cell of origin. Further studies are needed to elucidate the mechanisms for SCC development when SOX2 is expressed in the context of Lkb1 or Pten/Cdkn2ab loss in contrast to adenocarcinoma development in the context of Trp53 loss.
SCLC
While transplant models of SCLC exist, GEMMs are the only available models of autochthonous SCLC. All current models are based on Rb1 and Trp53 loss analogous to human SCLC (66,67). Rb1fl/fl;Trp53fl/fl strain was the first reported SCLC GEMM by intratracheal administration of adeno-cre (153). Neuroendocrine hyperplasia and dysplasia developed between 4 and 6 months and SCLC developed between 7 and 12 months after infection. The tumors expressed high levels of ASCL1 (153) and L-MYC (179), consistent with human SCLC. Heterozygous loss of either Rb1 or Trp53 resulted in mixtures of neuroendocrine tumors and adenocarcinomas. Metastases to adrenal glands, bone, brain, liver and ovaries occurred in 42% of the mice. Additional loss of p130, a member of the RB family, to Rb1 and Trp53 led to accelerated development of neuroendocrine tumors within 3 months and liver metastases by 6 months (155). Triple knockout mice had more metastases than double knockout mice at 6 and 9 months (155).
Three models of SCLC in Ptenfl/fl;Rbfl/fl;Trp53fl/fl mice reported accelerated tumor formation: (I) induction via adeno-cre infection (156), (II) in PNECs via infection with adenovirus-expressing Cre under a CGRP-promoter specific (CGRP-Cre) (70) and (III) via tamoxifen administration to CGRPCreER/+;Ptenfl/fl;Rb1fl/fl;Trp53fl/fl (157).
Comparison of the various models revealed biases toward different histologies (154). Rb1fl/fl;Trp53fl/fl mice developed tumors that predominantly resemble human SCLC with a minor component of large cell neuroendocrine carcinoma (LCNEC). Rb1fl/fl;Trp53fl/fl;p130fl/fl mice developed mostly LCNEC early but then became dominated by SCLC at later time points. Metastases in this model were SCLC suggesting that SCLC may be a more aggressive tumor type despite a longer latency period. Ptenfl/fl;Rb1fl/fl;Trp53fl/fl and Ptenfl/+;Rb1fl/fl;Trp53fl/fl mice developed mixed SCLC and adenocarcinoma tumors with predominantly SCLC in Ptenfl/+ mice but not in Ptenfl/fl mice. Unlike the Rb1fl/fl;Trp53fl/fl or Ptenfl/+;Rb1fl/fl;Trp53fl/f models, metastases were not observed in the Ptenfl/fl;Rb1fl/fl;Trp53fl/fl after infection with adeno-cre (156). In Rb1fl/fl;Trp53fl/fl;Ptenfl/fl models using CGRP-cre adenovirus, LCNEC was the dominant histology with minor components of SCLC and adenocarcinoma but the metastases were SCLC (154).
Recently, a model of variant SCLC has been reported with the expression of constitutively active MYC-T58A combined with Rb1 and Trp53 loss (71). The mice developed tumors more quickly than those with Pten loss; they had shorter survival (average ~6 weeks) and a higher incidence of metastases. In contrast to previous models, tumors from Rb1fl/fl;Trp53fl/fl;MycLSL/LSL mice expressed high levels of NEUROD1, low levels of ASCL1 and had no Mycl amplifications, consistent with human variant SCLC.
Modeling the immune microenvironment in mouse models of lung cancer
Within the last 5 years, T cell immune checkpoint blockade (ICB) has transformed the therapeutic landscape of lung cancer. Immune checkpoints represent a myriad of molecules that regulate the activity of cytotoxic T cells (180). The two most intensely studied immune checkpoints are the inhibitory receptors programmed cell death 1 (PD-1), that binds to the programmed cell death 1 and 2 ligands (PD-L1; PD-L2), and cytotoxic T-lymphocyte antigen 4 (CTLA4), that binds to CD80 and CD86 ligands and competes with the activating receptor CD28 (180). Current immune checkpoint therapies are antibodies directed against PD-1 (nivolumab, pembrolizumab), PD-L1 (atezolizumab, durvalumab) or CTLA4 (ipilimumab, tremelimumab) that relieve the inhibitory activity of the checkpoints on cytotoxic T cell activation. Anti-CTLA4 antibodies also deplete regulatory T cells (181). Anti-PD-1/PD-L1 agents showed significantly improved progression-free survival (PFS) and overall survival (OS) rates in treatment-naïve (182,183) and previously-treated (184-187) patients with metastatic NSCLC and locally advanced NSCLC after chemoradiotherapy (188). Early phase1/2 trials with combination nivolumab (anti-PD-1) and ipilimumab (anti-CTLA-4) in treatment-naïve NSCLC (189) and pretreated-SCLC (190) patients showed promise although the toxicities were greater than nivolumab alone.
Despite these advances, identification of predictive biomarkers for ICB has been difficult. PD-L1 expression has not been a reliable biomarker for anti-PD-1/PDL-1 agents. Pembrolizumab has superior responses against NSCLC tumors with >50% PD-L1(+) cells by immunohistochemistry (IHC) (182,191). Nivolumab efficacy, however, had little correlation with PD-L1 staining (185,186). Although distinct PD-L1 tests were utilized for the two drugs, the assays are highly concordant (192). Thus, the cause for the divergent efficacies based on PD-L1 expression is still unknown.
Melanoma and NSCLC, two malignancies sensitive to ICB, have high total mutation burdens (TMBs) (193,194). Somatic mutations are thought to form “neoantigens” that may serve as epitopes that are recognized by T cells (195). NSCLC samples with high TMBs (> median value) had higher neoantigen levels and, when treated with pembrolizumab, correlated with PFS and durable clinical benefit (DCB), defined as partial or stable response >6 months (196). Consistent with these data, high TMB correlated with improved PFS in NSCLC patients treated with nivolumab compared to chemotherapy (197). No correlations between TMB and PD-L1 expression have been identified (198).
A striking distinction between human and GEMM lung cancers is the large discrepancy in the number of somatic mutations. Human never-smoker and smoking-associated lung cancers have 28- and 110-fold more nonsynonymous mutations, respectively, than GEMM lung cancers (69,70). The lack of mutations in GEMM lung cancers may present hurdles to study ICB in mice although some studies suggest that the transgenic sequences may be presented as antigens to T cells (199,200). Despite these limitations, mouse models have been valuable to elucidate the immune microenvironment with implications for human therapy. We will focus on GEMMs although syngeneic Lewis lung carcinoma transplant models have also been used. CITMs have not been used routinely for immune microenvironment studies although they may be attractive models given their greater number of somatic mutations than GEMMs (68).
Never-smoker models
In humans and GEMMs, mutant EGFR LAD exhibited reduced cytotoxic lymphocyte infiltration as compared to mutant Kras-driven tumors (201). In another study, EGFR pathway activation by EGFR Del19, L858R-T790M, and Del19-T790M mutants led to increased PD-L1 expression with corresponding decrease when treated with EGFR antagonists (202). Subsequent studies revealed similar correlations with human mutant EGFR, KRAS, EML4-ALK fusions and PD-L1 (203-206). The immune-triggering antigenic peptides in GEMMs still await identification.
The murine results are in contrast to human studies where EGFR-mutant tumors responded less favorably to ICB than wild type EGFR tumors (207,208). There may be several reasons for the differences. The murine tumors were treatment naïve whereas the human EGFR-mutant tumors had prior exposure to therapies such as EGFR TKIs or chemotherapy. Distinct mechanisms of resistance to those prior therapies may alter the microenvironment through changes in chemokine or cytokine expression in tumor cells (209,210). Lower mutational loads of EGFR-mutant tumors (69) may also account for the poor responses with ICB.
A DNA vaccine against cytoplasmic domain of ALK in EML4-ALK GEMMs delayed tumor growth through a cytotoxic T cell dependent mechanism and synergized with anti-PD-1 ICB (200). These studies led to the discovery of autoantibodies against ALK in patients with ALK positive tumors (211).
Smoking-associated models
Adenocarcinoma
Mutant Kras GEMM cancers showed poor tumor T cell infiltration and were resistant to ICB therapy (202,212-215). Intratumoral expression of antigenic luciferase (212) or treatment with immunogenic oxaliplatin/cyclophosphamide chemotherapy (213) in KrasLSL-G12D/+;Trp53fl/f mice induced tumor T cell infiltration and conferred sensitivity to ICB (213). In KrasG12D mice with IL-17 driven chronic inflammation, increased neutrophil infiltration led to ICB resistance (214). KrasG12D;Lkb1fl/fl adenosquamous tumors demonstrated a paucity of T cells, elevated pro-inflammatory cytokines and myeloid cells, reduced PD-L1 expression and were resistant to ICB (216) analogous to human cancers with KRAS and LKB1 mutations (216-218).
In KrasG12D/+ or KrasG12D;p53fl/fl mice, ICB in combination with immunogenic radiation (219) or chemotherapy (213) suppressed tumor growth. In the KrasLA2 model, mTOR inhibition was synergistic with ICB (215). Also, epigenetic modifiers HDAC6 inhibitor and JQ1 (bromodomain inhibitor) stimulated anti-tumor immunity in KrasG12D;p53 fl/fl adenocarcinoma (220).
Squamous cell lung cancer
Ptenfl/fl;Lkb1fl/fl SCCs have high levels of PD-L1 in the tumor propagating cells, increased tumor-associated neutrophils, and decreased tumor-associated macrophages. In the same study, increased density of neutrophils was also found in human SCCs as compared to adenocarcinomas (148). SCCs from Lori-IKKa;IKKaKA/KA mice had significantly increased CD4+ T cells and macrophages with mildly increased numbers of CD8+ T cells and neutrophils. Early depletion of macrophages inhibited SCC development (149).
SCLC
Few immuno-oncology studies in SCLC have been reported. Rb1fl/fl;Trp53fl/fl murine tumors had reduced T cell infiltration as compared to murine adenocarcinoma (201). CD47, a “don’t eat me signal” (221), is overexpressed in SCLC (222). Antibody-mediated CD47 blockade was therapeutically effective in transplant and GEMM SCLC models through activating macrophages (222).
Conclusions
GEMMs of never-smoker lung cancers may represent a fairly close approximation to human cancers. Never-smoker lung cancers have much lower rates of mutations and are less responsive to ICB than smoking-associated-lung cancers—analogous to the GEMMs. Furthermore, GEMMs of never-smokers have faithfully reproduced human sensitivity to TKIs or combination of TKIs (69) and provide a platform to test novel targeted therapies.
GEMMs of smoking-associated lung cancer have greater distinctions with their human counterparts. GEMM cancers are more resistant to ICB and have lower TMB than human cancers. Despite these discrepancies, the murine genetic aberrations lead to a phenotypic evolution from pre-cancerous lesions to carcinomas that are strikingly similar to human cancers. Examples, among many, include the gain of Mycl amplifications in Rb1fl/fl;Trp53fl/fl SCLC (70), heterogenous NEUROD1(+) and NEUROD1(−)/ASCL1(−) variant SCLC in Rb1fl/fl;Trp53fl/fl;MycLSL/LSL mice (71) and the T cell non-inflamed phenotype KrasG12D/+;Lkb1fl/fl tumors (216) mirror their analogous human cancers (67,216,217).
However, the need for better models is still present. SCC models are needed that represent the greater diversity of human genetic alterations. Models that more faithfully mimic the engagement of human cancers with the immune microenvironment are still needed. CITMs, with its higher TMB, may engage T cells more than GEMMs, although tumor histology is presently limited to adenomas. Also, models are needed that faithfully mimic the serious immune toxicities of ICB experienced by patients. Current murine models don’t exhibit such toxicities.
Despite these limitations, CITMs and GEMMs faithfully model human lung cancer pathogenesis and behavior from hyperplasia to metastases (for GEMMs). The development of de novo tumors in situ may also more closely model human lung cancers than transplant models of fully developed cancers placed into tumor-naïve microenvironments. Careful selection of CITMs and GEMMs provides a rich platform to elucidate the biology of lung cancers and to test novel therapeutic strategies.
Acknowledgements
Funding: The work is supported in part by a Career Enhancement Award through National Institutes of Health 5P50CA070907 (EA Akbay and J Kim), National Institutes of Health 1R01CA196851 (J Kim), Cancer Prevention and Research Institute of Texas Scholar Award RR160080 (EA Akbay), and American Cancer Society Research Scholar Award RSG-16-090-01-TBG (J Kim).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Spiro SG, Silvestri GA. One hundred years of lung cancer. Am J Respir Crit Care Med 2005;172:523-9. [Crossref] [PubMed]
- Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin 2015;65:87-108. [Crossref] [PubMed]
- Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin 2017;67:7-30. [Crossref] [PubMed]
- Chapter 5: Cancer. How Tobacco Smoke Causes Disease: The Biology and Behavioral Basis for Smoking-Attributable Disease: A Report of the Surgeon General. Publications and Reports of the Surgeon General. Atlanta (GA): US Department of Health and Human Services; 2010.
- Sun S, Schiller JH, Gazdar AF. Lung cancer in never smokers--a different disease. Nat Rev Cancer 2007;7:778-90. [Crossref] [PubMed]
- Parkin DM, Bray F, Ferlay J, et al. Global cancer statistics, 2002. CA Cancer J Clin 2005;55:74-108. [Crossref] [PubMed]
- Centers for Disease C. Prevention. Annual smoking-attributable mortality, years of potential life lost, and productivity losses--United States, 1997-2001. MMWR Morb Mortal Wkly Rep 2005;54:625-8. [PubMed]
- Thun MJ, Henley SJ, Burns D, et al. Lung cancer death rates in lifelong nonsmokers. J Natl Cancer Inst 2006;98:691-9. [Crossref] [PubMed]
- Nyberg F, Agudo A, Boffetta P, et al. A European validation study of smoking and environmental tobacco smoke exposure in nonsmoking lung cancer cases and controls. Cancer Causes Control 1998;9:173-82. [Crossref] [PubMed]
- Cohet C, Borel S, Nyberg F, et al. Exon 5 polymorphisms in the O6-alkylguanine DNA alkyltransferase gene and lung cancer risk in non-smokers exposed to second-hand smoke. Cancer Epidemiol Biomarkers Prev 2004;13:320-3. [Crossref] [PubMed]
- Gorlova OY, Zhang Y, Schabath MB, et al. Never smokers and lung cancer risk: a case-control study of epidemiological factors. Int J Cancer 2006;118:1798-804. [Crossref] [PubMed]
- Vineis P, Airoldi L, Veglia F, et al. Environmental tobacco smoke and risk of respiratory cancer and chronic obstructive pulmonary disease in former smokers and never smokers in the EPIC prospective study. BMJ 2005;330:277. [Crossref] [PubMed]
- Krewski D, Lubin JH, Zielinski JM, et al. A combined analysis of North American case-control studies of residential radon and lung cancer. J Toxicol Environ Health A 2006;69:533-97. [Crossref] [PubMed]
- Darby S, Hill D, Auvinen A, et al. Radon in homes and risk of lung cancer: collaborative analysis of individual data from 13 European case-control studies. BMJ 2005;330:223. [Crossref] [PubMed]
- Gottschall EB. Occupational and environmental thoracic malignancies. J Thorac Imaging 2002;17:189-97. [Crossref] [PubMed]
- Neuberger JS, Field RW. Occupation and lung cancer in nonsmokers. Rev Environ Health 2003;18:251-67. [Crossref] [PubMed]
- van Loon AJ, Kant IJ, Swaen GM, et al. Occupational exposure to carcinogens and risk of lung cancer: results from The Netherlands cohort study. Occup Environ Med 1997;54:817-24. [Crossref] [PubMed]
- Zhao Y, Wang S, Aunan K, et al. Air pollution and lung cancer risks in China--a meta-analysis. Sci Total Environ 2006;366:500-13. [Crossref] [PubMed]
- Lissowska J, Bardin-Mikolajczak A, Fletcher T, et al. Lung cancer and indoor pollution from heating and cooking with solid fuels: the IARC international multicentre case-control study in Eastern/Central Europe and the United Kingdom. Am J Epidemiol 2005;162:326-33. [Crossref] [PubMed]
- Ramanakumar AV, Parent ME, Siemiatycki J. Risk of lung cancer from residential heating and cooking fuels in Montreal, Canada. Am J Epidemiol 2007;165:634-42. [Crossref] [PubMed]
- Turner MC, Krewski D, Pope CA 3rd, et al. Long-term ambient fine particulate matter air pollution and lung cancer in a large cohort of never-smokers. Am J Respir Crit Care Med 2011;184:1374-81. [Crossref] [PubMed]
- Hamra GB, Guha N, Cohen A, et al. Outdoor particulate matter exposure and lung cancer: a systematic review and meta-analysis. Environ Health Perspect 2014;122:906-11. [PubMed]
- Cheng YW, Chiou HL, Sheu GT, et al. The association of human papillomavirus 16/18 infection with lung cancer among nonsmoking Taiwanese women. Cancer Res 2001;61:2799-803. [PubMed]
- Fei Y, Yang J, Hsieh WC, et al. Different human papillomavirus 16/18 infection in Chinese non-small cell lung cancer patients living in Wuhan, China. Jpn J Clin Oncol 2006;36:274-9. [Crossref] [PubMed]
- Ciotti M, Giuliani L, Ambrogi V, et al. Detection and expression of human papillomavirus oncogenes in non-small cell lung cancer. Oncol Rep 2006;16:183-9. [PubMed]
- Hung RJ, Boffetta P, Brockmoller J, et al. CYP1A1 and GSTM1 genetic polymorphisms and lung cancer risk in Caucasian non-smokers: a pooled analysis. Carcinogenesis 2003;24:875-82. [Crossref] [PubMed]
- Raimondi S, Boffetta P, Anttila S, et al. Metabolic gene polymorphisms and lung cancer risk in non-smokers. An update of the GSEC study. Mutat Res 2005;592:45-57. [Crossref] [PubMed]
- Zhou W, Liu G, Miller DP, et al. Polymorphisms in the DNA repair genes XRCC1 and ERCC2, smoking, and lung cancer risk. Cancer Epidemiol Biomarkers Prev 2003;12:359-65. [PubMed]
- Lo YL, Hsiao CF, Jou YS, et al. Polymorphisms of MLH1 and MSH2 genes and the risk of lung cancer among never smokers. Lung Cancer 2011;72:280-6. [Crossref] [PubMed]
- Brenner DR, Boffetta P, Duell EJ, et al. Previous lung diseases and lung cancer risk: a pooled analysis from the International Lung Cancer Consortium. Am J Epidemiol 2012;176:573-85. [Crossref] [PubMed]
- Denholm R, Schuz J, Straif K, et al. Is previous respiratory disease a risk factor for lung cancer? Am J Respir Crit Care Med 2014;190:549-59. [Crossref] [PubMed]
- Kwon MC, Berns A. Mouse models for lung cancer. Mol Oncol 2013;7:165-77. [Crossref] [PubMed]
- Inoue K, Fry E, Maglic D, et al. Genetically Engineered Mouse Models for Human Lung Cancer. In: Kayembe JM. editor. Oncogenesis, Inflammatory and Parasitic Tropical Diseases of the Lung. Rijeka, Croatia: InTech; 2013.
- Kellar A, Egan C, Morris D. Preclinical Murine Models for Lung Cancer: Clinical Trial Applications. Biomed Res Int 2015;2015. [Crossref] [PubMed]
- Gazdar AF, Hirsch FR, Minna JD. From Mice to Men and Back: An Assessment of Preclinical Model Systems for the Study of Lung Cancers. J Thorac Oncol 2016;11:287-99. [Crossref] [PubMed]
- Brambilla E, Travis WD, Colby TV, et al. The new World Health Organization classification of lung tumours. Eur Respir J 2001;18:1059-68. [Crossref] [PubMed]
- National Cancer Institute Surveillance E, and End Results Program. Cancer Stat Facts: Lung and Bronchus Cancer. Available online: https://seer.cancer.gov/statfacts/html/lungb.html
- Xu X, Rock JR, Lu Y, et al. Evidence for type II cells as cells of origin of K-Ras-induced distal lung adenocarcinoma. Proc Natl Acad Sci U S A 2012;109:4910-5. [Crossref] [PubMed]
- Sutherland KD, Song JY, Kwon MC, et al. Multiple cells-of-origin of mutant K-Ras-induced mouse lung adenocarcinoma. Proc Natl Acad Sci U S A 2014;111:4952-7. [Crossref] [PubMed]
- Ferone G, Song JY, Sutherland KD, et al. SOX2 Is the Determining Oncogenic Switch in Promoting Lung Squamous Cell Carcinoma from Different Cells of Origin. Cancer Cell 2016;30:519-32. [Crossref] [PubMed]
- Semenova EA, Nagel R, Berns A. Origins, genetic landscape, and emerging therapies of small cell lung cancer. Genes Dev 2015;29:1447-62. [Crossref] [PubMed]
- Khuder SA. Effect of cigarette smoking on major histological types of lung cancer: a meta-analysis. Lung Cancer 2001;31:139-48. [Crossref] [PubMed]
- Khuder SA, Mutgi AB. Effect of smoking cessation on major histologic types of lung cancer. Chest 2001;120:1577-83. [Crossref] [PubMed]
- Alexandrov LB, Ju YS, Haase K, et al. Mutational signatures associated with tobacco smoking in human cancer. Science 2016;354:618-22. [Crossref] [PubMed]
- Toh CK, Gao F, Lim WT, et al. Never-smokers with lung cancer: epidemiologic evidence of a distinct disease entity. J Clin Oncol 2006;24:2245-51. [Crossref] [PubMed]
- Wakelee HA, Chang ET, Gomez SL, et al. Lung cancer incidence in never smokers. J Clin Oncol 2007;25:472-8. [Crossref] [PubMed]
- Kawaguchi T, Takada M, Kubo A, et al. Gender, histology, and time of diagnosis are important factors for prognosis: analysis of 1499 never-smokers with advanced non-small cell lung cancer in Japan. J Thorac Oncol 2010;5:1011-7. [Crossref] [PubMed]
- Chapter 2. Cancer. The Health Consequences of Smoking: a Report of the Surgeon General. . Atlanta (GA): US Department of Health and Human Services; 2004.
- Hoffmann D, Djordjevic MV, Hoffmann I. The changing cigarette. Prev Med 1997;26:427-34. [Crossref] [PubMed]
- Hecht SS. Biochemistry, biology, and carcinogenicity of tobacco-specific N-nitrosamines. Chem Res Toxicol 1998;11:559-603. [Crossref] [PubMed]
- Alexandrov LB, Nik-Zainal S, Wedge DC, et al. Signatures of mutational processes in human cancer. Nature 2013;500:415-21. [Crossref] [PubMed]
- Sanchez-Cespedes M, Ahrendt SA, Piantadosi S, et al. Chromosomal alterations in lung adenocarcinoma from smokers and nonsmokers. Cancer Res 2001;61:1309-13. [PubMed]
- Govindan R, Ding L, Griffith M, et al. Genomic landscape of non-small cell lung cancer in smokers and never-smokers. Cell 2012;150:1121-34. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014;511:543-50. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature 2012;489:519-25. [Crossref] [PubMed]
- Gazdar AF, Bunn PA, Minna JD. Small-cell lung cancer: what we know, what we need to know and the path forward. Nat Rev Cancer 2017;17:725-37. [Crossref] [PubMed]
- Zhang W, Girard L, Zhang YA, et al. Small cell lung cancer tumors and preclinical models display heterogeneity of neuroendocrine phenotypes. Transl Lung Cancer Res 2018;7:32-49. [Crossref] [PubMed]
- Augustyn A, Borromeo M, Wang T, et al. ASCL1 is a lineage oncogene providing therapeutic targets for high-grade neuroendocrine lung cancers. Proc Natl Acad Sci U S A 2014;111:14788-93. [Crossref] [PubMed]
- Cho JH, Tsai MJ. The role of BETA2/NeuroD1 in the development of the nervous system. Mol Neurobiol 2004;30:35-47. [Crossref] [PubMed]
- Liu M, Pereira FA, Price SD, et al. Essential role of BETA2/NeuroD1 in development of the vestibular and auditory systems. Genes Dev 2000;14:2839-54. [Crossref] [PubMed]
- Miyata T, Maeda T, Lee JE. NeuroD is required for differentiation of the granule cells in the cerebellum and hippocampus. Genes Dev 1999;13:1647-52. [Crossref] [PubMed]
- Gazdar AF, Carney DN, Nau MM, et al. Characterization of variant subclasses of cell lines derived from small cell lung cancer having distinctive biochemical, morphological, and growth properties. Cancer Res 1985;45:2924-30. [PubMed]
- Borromeo MD, Savage TK, Kollipara RK, et al. ASCL1 and NEUROD1 Reveal Heterogeneity in Pulmonary Neuroendocrine Tumors and Regulate Distinct Genetic Programs. Cell Rep 2016;16:1259-72. [Crossref] [PubMed]
- Pleasance ED, Stephens PJ, O'Meara S, et al. A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature 2010;463:184-90. [Crossref] [PubMed]
- Peifer M, Fernandez-Cuesta L, Sos ML, et al. Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat Genet 2012;44:1104-10. [Crossref] [PubMed]
- Rudin CM, Durinck S, Stawiski EW, et al. Comprehensive genomic analysis identifies SOX2 as a frequently amplified gene in small-cell lung cancer. Nat Genet 2012;44:1111-6. [Crossref] [PubMed]
- George J, Lim JS, Jang SJ, et al. Comprehensive genomic profiles of small cell lung cancer. Nature 2015;524:47-53. [Crossref] [PubMed]
- Westcott PM, Halliwill KD, To MD, et al. The mutational landscapes of genetic and chemical models of Kras-driven lung cancer. Nature 2015;517:489-92. [Crossref] [PubMed]
- McFadden DG, Politi K, Bhutkar A, et al. Mutational landscape of EGFR-, MYC-, and Kras-driven genetically engineered mouse models of lung adenocarcinoma. Proc Natl Acad Sci U S A 2016;113:E6409-17. [Crossref] [PubMed]
- McFadden DG, Papagiannakopoulos T, Taylor-Weiner A, et al. Genetic and clonal dissection of murine small cell lung carcinoma progression by genome sequencing. Cell 2014;156:1298-311. [Crossref] [PubMed]
- Mollaoglu G, Guthrie MR, Bohm S, et al. MYC Drives Progression of Small Cell Lung Cancer to a Variant Neuroendocrine Subtype with Vulnerability to Aurora Kinase Inhibition. Cancer Cell 2017;31:270-85. [Crossref] [PubMed]
- Politi K, Zakowski MF, Fan PD, et al. Lung adenocarcinomas induced in mice by mutant EGF receptors found in human lung cancers respond to a tyrosine kinase inhibitor or to down-regulation of the receptors. Genes Dev 2006;20:1496-510. [Crossref] [PubMed]
- Ji H, Li D, Chen L, et al. The impact of human EGFR kinase domain mutations on lung tumorigenesis and in vivo sensitivity to EGFR-targeted therapies. Cancer Cell 2006;9:485-95. [Crossref] [PubMed]
- Li D, Shimamura T, Ji H, et al. Bronchial and peripheral murine lung carcinomas induced by T790M-L858R mutant EGFR respond to HKI-272 and rapamycin combination therapy. Cancer Cell 2007;12:81-93. [Crossref] [PubMed]
- Soda M, Takada S, Takeuchi K, et al. A mouse model for EML4-ALK-positive lung cancer. Proc Natl Acad Sci U S A 2008;105:19893-7. [Crossref] [PubMed]
- Chen Z, Sasaki T, Tan X, et al. Inhibition of ALK, PI3K/MEK, and HSP90 in murine lung adenocarcinoma induced by EML4-ALK fusion oncogene. Cancer Res 2010;70:9827-36. [Crossref] [PubMed]
- Pyo KH, Lim SM, Kim HR, et al. Establishment of a Conditional Transgenic Mouse Model Recapitulating EML4-ALK-Positive Human Non-Small Cell Lung Cancer. J Thorac Oncol 2017;12:491-500. [Crossref] [PubMed]
- Inoue M, Toki H, Matsui J, et al. Mouse models for ROS1-fusion-positive lung cancers and their application to the analysis of multikinase inhibitor efficiency. Carcinogenesis 2016;37:452-60. [Crossref] [PubMed]
- Arai Y, Totoki Y, Takahashi H, et al. Mouse model for ROS1-rearranged lung cancer. PLoS One 2013;8. [Crossref] [PubMed]
- Engelman JA, Chen L, Tan X, et al. Effective use of PI3K and MEK inhibitors to treat mutant Kras G12D and PIK3CA H1047R murine lung cancers. Nat Med 2008;14:1351-6. [Crossref] [PubMed]
- Normanno N, De Luca A, Bianco C, et al. Epidermal growth factor receptor (EGFR) signaling in cancer. Gene 2006;366:2-16. [Crossref] [PubMed]
- Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med 2004;350:2129-39. [Crossref] [PubMed]
- Paez JG, Janne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 2004;304:1497-500. [Crossref] [PubMed]
- Pao W, Miller V, Zakowski M, et al. EGF receptor gene mutations are common in lung cancers from "never smokers" and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci U S A 2004;101:13306-11. [Crossref] [PubMed]
- Ohashi K, Maruvka YE, Michor F, et al. Epidermal growth factor receptor tyrosine kinase inhibitor-resistant disease. J Clin Oncol 2013;31:1070-80. [Crossref] [PubMed]
- Song HN, Jung KS, Yoo KH, et al. Acquired C797S Mutation upon Treatment with a T790M-Specific Third-Generation EGFR Inhibitor (HM61713) in Non-Small Cell Lung Cancer. J Thorac Oncol 2016;11:e45-7. [Crossref] [PubMed]
- Ercan D, Choi HG, Yun CH, et al. EGFR Mutations and Resistance to Irreversible Pyrimidine-Based EGFR Inhibitors. Clin Cancer Res 2015;21:3913-23. [Crossref] [PubMed]
- Niederst MJ, Hu H, Mulvey HE, et al. The Allelic Context of the C797S Mutation Acquired upon Treatment with Third-Generation EGFR Inhibitors Impacts Sensitivity to Subsequent Treatment Strategies. Clin Cancer Res 2015;21:3924-33. [Crossref] [PubMed]
- Hou S, Han X, Ji H. Squamous Transition of Lung Adenocarcinoma and Drug Resistance. Trends Cancer 2016;2:463-6. [Crossref] [PubMed]
- Sequist LV, Waltman BA, Dias-Santagata D, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med 2011;3. [Crossref] [PubMed]
- Lee JK, Lee J, Kim S, et al. Clonal History and Genetic Predictors of Transformation Into Small-Cell Carcinomas From Lung Adenocarcinomas. J Clin Oncol 2017;35:3065-74. [Crossref] [PubMed]
- Tichelaar JW, Lu W, Whitsett JA. Conditional expression of fibroblast growth factor-7 in the developing and mature lung. J Biol Chem 2000;275:11858-64. [Crossref] [PubMed]
- Fisher GH, Wellen SL, Klimstra D, et al. Induction and apoptotic regression of lung adenocarcinomas by regulation of a K-Ras transgene in the presence and absence of tumor suppressor genes. Genes Dev 2001;15:3249-62. [Crossref] [PubMed]
- Morris SW, Kirstein MN, Valentine MB, et al. Fusion of a kinase gene, ALK, to a nucleolar protein gene, NPM, in non-Hodgkin's lymphoma. Science 1994;263:1281-4. [Crossref] [PubMed]
- Soda M, Choi YL, Enomoto M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature 2007;448:561-6. [Crossref] [PubMed]
- Hallberg B, Palmer RH. Mechanistic insight into ALK receptor tyrosine kinase in human cancer biology. Nat Rev Cancer 2013;13:685-700. [Crossref] [PubMed]
- Christensen JG, Zou HY, Arango ME, et al. Cytoreductive antitumor activity of PF-2341066, a novel inhibitor of anaplastic lymphoma kinase and c-Met, in experimental models of anaplastic large-cell lymphoma. Mol Cancer Ther 2007;6:3314-22. [Crossref] [PubMed]
- Doebele RC, Pilling AB, Aisner DL, et al. Mechanisms of resistance to crizotinib in patients with ALK gene rearranged non-small cell lung cancer. Clin Cancer Res 2012;18:1472-82. [Crossref] [PubMed]
- Katayama R, Shaw AT, Khan TM, et al. Mechanisms of acquired crizotinib resistance in ALK-rearranged lung Cancers. Sci Transl Med 2012;4. [Crossref] [PubMed]
- Choi YL, Soda M, Yamashita Y, et al. EML4-ALK mutations in lung cancer that confer resistance to ALK inhibitors. N Engl J Med 2010;363:1734-9. [Crossref] [PubMed]
- Sasaki T, Okuda K, Zheng W, et al. The neuroblastoma-associated F1174L ALK mutation causes resistance to an ALK kinase inhibitor in ALK-translocated cancers. Cancer Res 2010;70:10038-43. [Crossref] [PubMed]
- Marsilje TH, Pei W, Chen B, et al. Synthesis, structure-activity relationships, and in vivo efficacy of the novel potent and selective anaplastic lymphoma kinase (ALK) inhibitor 5-chloro-N2-(2-isopropoxy-5-methyl-4-(piperidin-4-yl)phenyl)-N4-(2-(isopropylsulf onyl)phenyl)pyrimidine-2,4-diamine (LDK378) currently in phase 1 and phase 2 clinical trials. J Med Chem 2013;56:5675-90. [Crossref] [PubMed]
- Friboulet L, Li N, Katayama R, et al. The ALK inhibitor ceritinib overcomes crizotinib resistance in non-small cell lung cancer. Cancer Discov 2014;4:662-73. [Crossref] [PubMed]
- Zhang S, Anjum R, Squillace R, et al. The Potent ALK Inhibitor Brigatinib (AP26113) Overcomes Mechanisms of Resistance to First- and Second-Generation ALK Inhibitors in Preclinical Models. Clin Cancer Res 2016;22:5527-38. [Crossref] [PubMed]
- Huang WS, Liu S, Zou D, et al. Discovery of Brigatinib (AP26113), a Phosphine Oxide-Containing, Potent, Orally Active Inhibitor of Anaplastic Lymphoma Kinase. J Med Chem 2016;59:4948-64. [Crossref] [PubMed]
- Sakamoto H, Tsukaguchi T, Hiroshima S, et al. CH5424802, a selective ALK inhibitor capable of blocking the resistant gatekeeper mutant. Cancer Cell 2011;19:679-90. [Crossref] [PubMed]
- Kinoshita K, Asoh K, Furuichi N, et al. Design and synthesis of a highly selective, orally active and potent anaplastic lymphoma kinase inhibitor (CH5424802). Bioorg Med Chem 2012;20:1271-80. [Crossref] [PubMed]
- Kodama T, Tsukaguchi T, Yoshida M, et al. Selective ALK inhibitor alectinib with potent antitumor activity in models of crizotinib resistance. Cancer Lett 2014;351:215-21. [Crossref] [PubMed]
- Shaw AT, Varghese AM, Solomon BJ, et al. Pemetrexed-based chemotherapy in patients with advanced, ALK-positive non-small cell lung cancer. Ann Oncol 2013;24:59-66. [Crossref] [PubMed]
- Lee HY, Ahn HK, Jeong JY, et al. Favorable clinical outcomes of pemetrexed treatment in anaplastic lymphoma kinase positive non-small-cell lung cancer. Lung Cancer 2013;79:40-5. [Crossref] [PubMed]
- Ma D, Hao X, Wang Y, et al. Clinical effect of pemetrexed as the first-line treatment in Chinese patients with advanced anaplastic lymphoma kinase-positive non-small cell lung cancer. Thorac Cancer 2016;7:452-8. [Crossref] [PubMed]
- Chen Z, Akbay E, Mikse O, et al. Co-clinical trials demonstrate superiority of crizotinib to chemotherapy in ALK-rearranged non-small cell lung cancer and predict strategies to overcome resistance. Clin Cancer Res 2014;20:1204-11. [Crossref] [PubMed]
- Maddalo D, Manchado E, Concepcion CP, et al. In vivo engineering of oncogenic chromosomal rearrangements with the CRISPR/Cas9 system. Nature 2014;516:423-7. [Crossref] [PubMed]
- Blasco RB, Karaca E, Ambrogio C, et al. Simple and rapid in vivo generation of chromosomal rearrangements using CRISPR/Cas9 technology. Cell Rep 2014;9:1219-27. [Crossref] [PubMed]
- Pal P, Khan Z. Ros1-1. J Clin Pathol 2017;70:1001-9. [Crossref] [PubMed]
- Bergethon K, Shaw AT, Ou SH, et al. ROS1 rearrangements define a unique molecular class of lung cancers. J Clin Oncol 2012;30:863-70. [Crossref] [PubMed]
- Davies KD, Le AT, Theodoro MF, et al. Identifying and targeting ROS1 gene fusions in non-small cell lung cancer. Clin Cancer Res 2012;18:4570-9. [Crossref] [PubMed]
- Takeuchi K, Soda M, Togashi Y, et al. RET, ROS1 and ALK fusions in lung cancer. Nat Med 2012;18:378-81. [Crossref] [PubMed]
- Shaw AT, Ou SH, Bang YJ, et al. Crizotinib in ROS1-rearranged non-small-cell lung cancer. N Engl J Med 2014;371:1963-71. [Crossref] [PubMed]
- Yasuda H, de Figueiredo-Pontes LL, Kobayashi S, et al. Preclinical rationale for use of the clinically available multitargeted tyrosine kinase inhibitor crizotinib in ROS1-translocated lung cancer. J Thorac Oncol 2012;7:1086-90. [Crossref] [PubMed]
- Rikova K, Guo A, Zeng Q, et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell 2007;131:1190-203. [Crossref] [PubMed]
- Yap TA, Bjerke L, Clarke PA, et al. Drugging PI3K in cancer: refining targets and therapeutic strategies. Curr Opin Pharmacol 2015;23:98-107. [Crossref] [PubMed]
- Miled N, Yan Y, Hon WC, et al. Mechanism of two classes of cancer mutations in the phosphoinositide 3-kinase catalytic subunit. Science 2007;317:239-42. [Crossref] [PubMed]
- Huang CH, Mandelker D, Schmidt-Kittler O, et al. The structure of a human p110alpha/p85alpha complex elucidates the effects of oncogenic PI3Kalpha mutations. Science 2007;318:1744-8. [Crossref] [PubMed]
- Zhao L, Vogt PK. Helical domain and kinase domain mutations in p110alpha of phosphatidylinositol 3-kinase induce gain of function by different mechanisms. Proc Natl Acad Sci U S A 2008;105:2652-7. [Crossref] [PubMed]
- You M, Candrian U, Maronpot RR, et al. Activation of the Ki-ras protooncogene in spontaneously occurring and chemically induced lung tumors of the strain A mouse. Proc Natl Acad Sci U S A 1989;86:3070-4. [Crossref] [PubMed]
- Nuzum EO, Malkinson AM, Beer DG. Specific Ki-ras codon 61 mutations may determine the development of urethan-induced mouse lung adenomas or adenocarcinomas. Mol Carcinog 1990;3:287-95. [Crossref] [PubMed]
- Kawano R, Nishisaka T, Takeshima Y, et al. Role of point mutation of the K-ras gene in tumorigenesis of B6C3F1 mouse lung lesions induced by urethane. Jpn J Cancer Res 1995;86:802-10. [Crossref] [PubMed]
- Foley JF, Anderson MW, Stoner GD, et al. Proliferative lesions of the mouse lung: progression studies in strain A mice. Exp Lung Res 1991;17:157-68. [Crossref] [PubMed]
- Massey TE, Devereux TR, Maronpot RR, et al. High frequency of K-ras mutations in spontaneous and vinyl carbamate-induced lung tumors of relatively resistant B6CF1 (C57BL/6J x BALB/cJ) mice. Carcinogenesis 1995;16:1065-9. [Crossref] [PubMed]
- Hecht SS, Isaacs S, Trushin N. Lung tumor induction in A/J mice by the tobacco smoke carcinogens 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone and benzo[a]pyrene: a potentially useful model for evaluation of chemopreventive agents. Carcinogenesis 1994;15:2721-5. [Crossref] [PubMed]
- Hecht SS. DNA adduct formation from tobacco-specific N-nitrosamines. Mutat Res 1999;424:127-42. [Crossref] [PubMed]
- Ronai ZA, Gradia S, Peterson LA, et al. G to A transitions and G to T transversions in codon 12 of the Ki-ras oncogene isolated from mouse lung tumors induced by 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) and related DNA methylating and pyridyloxobutylating agents. Carcinogenesis 1993;14:2419-22. [Crossref] [PubMed]
- Johnson L, Mercer K, Greenbaum D, et al. Somatic activation of the K-ras oncogene causes early onset lung cancer in mice. Nature 2001;410:1111-6. [Crossref] [PubMed]
- Jackson EL, Willis N, Mercer K, et al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev 2001;15:3243-8. [Crossref] [PubMed]
- Meuwissen R, Linn SC, van der Valk M, et al. Mouse model for lung tumorigenesis through Cre/lox controlled sporadic activation of the K-Ras oncogene. Oncogene 2001;20:6551-8. [Crossref] [PubMed]
- Guerra C, Mijimolle N, Dhawahir A, et al. Tumor induction by an endogenous K-ras oncogene is highly dependent on cellular context. Cancer Cell 2003;4:111-20. [Crossref] [PubMed]
- Young NP, Crowley D, Jacks T. Uncoupling cancer mutations reveals critical timing of p53 loss in sarcomagenesis. Cancer Res 2011;71:4040-7. [Crossref] [PubMed]
- Young NP, Jacks T. Tissue-specific p19Arf regulation dictates the response to oncogenic K-ras. Proc Natl Acad Sci U S A 2010;107:10184-9. [Crossref] [PubMed]
- Jackson EL, Olive KP, Tuveson DA, et al. The differential effects of mutant p53 alleles on advanced murine lung cancer. Cancer Res 2005;65:10280-8. [Crossref] [PubMed]
- Zheng S, El-Naggar AK, Kim ES, et al. A genetic mouse model for metastatic lung cancer with gender differences in survival. Oncogene 2007;26:6896-904. [Crossref] [PubMed]
- Ji H, Ramsey MR, Hayes DN, et al. LKB1 modulates lung cancer differentiation and metastasis. Nature 2007;448:807-10. [Crossref] [PubMed]
- Iwanaga K, Yang Y, Raso MG, et al. Pten inactivation accelerates oncogenic K-ras-initiated tumorigenesis in a mouse model of lung cancer. Cancer Res 2008;68:1119-27. [Crossref] [PubMed]
- Dankort D, Filenova E, Collado M, et al. A new mouse model to explore the initiation, progression, and therapy of BRAFV600E-induced lung tumors. Genes Dev 2007;21:379-84. [Crossref] [PubMed]
- Yoshimoto T, Hirao F, Sakatani M, et al. Induction of squamous cell carcinoma in the lung of C57BL/6 mice by intratracheal instillation of benzo[a]pyrene with charcoal powder. Gan 1977;68:343-52. [PubMed]
- Yoshimoto T, Inoue T, Iizuka H, et al. Differential induction of squamous cell carcinomas and adenocarcinomas in mouse lung by intratracheal instillation of benzo(a)pyrene and charcoal powder. Cancer Res 1980;40:4301-7. [PubMed]
- Henry CJ, Billups LH, Avery MD, et al. Lung cancer model system using 3-methylcholanthrene in inbred strains of mice. Cancer Res 1981;41:5027-32. [PubMed]
- Rehm S, Lijinsky W, Singh G, et al. Mouse bronchiolar cell carcinogenesis. Histologic characterization and expression of Clara cell antigen in lesions induced by N-nitrosobis-(2-chloroethyl) ureas. Am J Pathol 1991;139:413-22. [PubMed]
- Wang Y, Zhang Z, Yan Y, et al. A chemically induced model for squamous cell carcinoma of the lung in mice: histopathology and strain susceptibility. Cancer Res 2004;64:1647-54. [Crossref] [PubMed]
- Xu C, Fillmore CM, Koyama S, et al. Loss of Lkb1 and Pten leads to lung squamous cell carcinoma with elevated PD-L1 expression. Cancer Cell 2014;25:590-604. [Crossref] [PubMed]
- Xiao Z, Jiang Q, Willette-Brown J, et al. The pivotal role of IKKalpha in the development of spontaneous lung squamous cell carcinomas. Cancer Cell 2013;23:527-40. [Crossref] [PubMed]
- Mukhopadhyay A, Berrett KC, Kc U, et al. Sox2 cooperates with Lkb1 loss in a mouse model of squamous cell lung cancer. Cell Rep 2014;8:40-9. [Crossref] [PubMed]
- Meuwissen R, Linn SC, Linnoila RI, et al. Induction of small cell lung cancer by somatic inactivation of both Trp53 and Rb1 in a conditional mouse model. Cancer cell 2003;4:181-9. [Crossref] [PubMed]
- Gazdar AF, Savage TK, Johnson JE, et al. The comparative pathology of genetically engineered mouse models for neuroendocrine carcinomas of the lung. J Thorac Oncol 2015;10:553-64. [Crossref] [PubMed]
- Schaffer BE, Park KS, Yiu G, et al. Loss of p130 accelerates tumor development in a mouse model for human small-cell lung carcinoma. Cancer Res 2010;70:3877-83. [Crossref] [PubMed]
- Cui M, Augert A, Rongione M, et al. PTEN is a potent suppressor of small cell lung cancer. Mol Cancer Res 2014;12:654-9. [Crossref] [PubMed]
- Song H, Yao E, Lin C, et al. Functional characterization of pulmonary neuroendocrine cells in lung development, injury, and tumorigenesis. Proc Natl Acad Sci U S A 2012;109:17531-6. [Crossref] [PubMed]
- Svaton M, Fiala O, Pesek M, et al. The Prognostic Role of KRAS Mutation in Patients with Advanced NSCLC Treated with Second- or Third-line Chemotherapy. Anticancer Res 2016;36:1077-82. [PubMed]
- Mascaux C, Iannino N, Martin B, et al. The role of RAS oncogene in survival of patients with lung cancer: a systematic review of the literature with meta-analysis. Br J Cancer 2005;92:131-9. [Crossref] [PubMed]
- Cox AD, Fesik SW, Kimmelman AC, et al. Drugging the undruggable RAS: Mission possible? Nat Rev Drug Discov 2014;13:828-51. [Crossref] [PubMed]
- Ostrem JM, Peters U, Sos ML, et al. K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature 2013;503:548-51. [Crossref] [PubMed]
- Xiong Y, Lu J, Hunter J, et al. Covalent Guanosine Mimetic Inhibitors of G12C KRAS. ACS Med Chem Lett 2016;8:61-6. [Crossref] [PubMed]
- Janes MR, Zhang J, Li LS, et al. Targeting KRAS Mutant Cancers with a Covalent G12C-Specific Inhibitor. Cell 2018;172:578-89.e17. [Crossref] [PubMed]
- Vikis HG, Rymaszewski AL, Tichelaar JW. Mouse models of chemically-induced lung carcinogenesis. Front Biosci (Elite Ed) 2013;5:939-46. [Crossref] [PubMed]
- Anttila S, Raunio H, Hakkola J. Cytochrome P450-mediated pulmonary metabolism of carcinogens: regulation and cross-talk in lung carcinogenesis. Am J Respir Cell Mol Biol 2011;44:583-90. [Crossref] [PubMed]
- Kawano R, Takeshima Y, Inai K. Effects of K-ras gene mutations in the development of lung lesions induced by 4-(N-methyl-n-nitrosamino)-1-(3-pyridyl)-1-butanone in A/J mice. Jpn J Cancer Res 1996;87:44-50. [Crossref] [PubMed]
- Shimkin MB, Stoner GD. Lung tumors in mice: application to carcinogenesis bioassay. Adv Cancer Res 1975;21:1-58. [Crossref] [PubMed]
- Manenti G, Dragani TA. Pas1 haplotype-dependent genetic predisposition to lung tumorigenesis in rodents: a meta-analysis. Carcinogenesis 2005;26:875-82. [Crossref] [PubMed]
- Winters IP, Chiou SH, Paulk NK, et al. Multiplexed in vivo homology-directed repair and tumor barcoding enables parallel quantification of Kras variant oncogenicity. Nat Commun 2017;8:2053. [Crossref] [PubMed]
- Rogers ZN, McFarland CD, Winters IP, et al. A quantitative and multiplexed approach to uncover the fitness landscape of tumor suppression in vivo. Nat Methods 2017;14:737-42. [Crossref] [PubMed]
- Sánchez-Rivera FJ, Papagiannakopoulos T, Romero R, et al. Rapid modelling of cooperating genetic events in cancer through somatic genome editing. Nature 2014;516:428-31. [Crossref] [PubMed]
- Walter DM, Venancio OS, Buza EL, et al. Systematic In Vivo Inactivation of Chromatin-Regulating Enzymes Identifies Setd2 as a Potent Tumor Suppressor in Lung Adenocarcinoma. Cancer Res 2017;77:1719-29. [Crossref] [PubMed]
- Rogers ZN, McFarland CD, Winters IP, et al. Mapping the in vivo fitness landscape of lung adenocarcinoma tumor suppression in mice. Nat Genet 2018;50:483-6. [Crossref] [PubMed]
- Liu G, McDonnell TJ, Montes de Oca Luna R, et al. High metastatic potential in mice inheriting a targeted p53 missense mutation. Proc Natl Acad Sci U S A 2000;97:4174-9. [Crossref] [PubMed]
- Vakiani E, Solit DB. KRAS and BRAF: drug targets and predictive biomarkers. J Pathol 2011;223:219-29. [Crossref] [PubMed]
- Barrett SD, Bridges AJ, Dudley DT, et al. The discovery of the benzhydroxamate MEK inhibitors CI-1040 and PD 0325901. Bioorg Med Chem Lett 2008;18:6501-4. [Crossref] [PubMed]
- Xiong D, Pan J, Zhang Q, et al. Bronchial airway gene expression signatures in mouse lung squamous cell carcinoma and their modulation by cancer chemopreventive agents. Oncotarget 2017;8:18885-900. [PubMed]
- Bass AJ, Watanabe H, Mermel CH, et al. SOX2 is an amplified lineage-survival oncogene in lung and esophageal squamous cell carcinomas. Nat Genet 2009;41:1238-42. [Crossref] [PubMed]
- Calbo J, van Montfort E, Proost N, et al. A functional role for tumor cell heterogeneity in a mouse model of small cell lung cancer. Cancer cell 2011;19:244-56. [Crossref] [PubMed]
- Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer 2012;12:252-64. [Crossref] [PubMed]
- Simpson TR, Li F, Montalvo-Ortiz W, et al. Fc-dependent depletion of tumor-infiltrating regulatory T cells co-defines the efficacy of anti-CTLA-4 therapy against melanoma. J Exp Med 2013;210:1695-710. [Crossref] [PubMed]
- Reck M, Rodriguez-Abreu D, Robinson AG, et al. Pembrolizumab versus Chemotherapy for PD-L1-Positive Non-Small-Cell Lung Cancer. N Engl J Med 2016;375:1823-33. [Crossref] [PubMed]
- Langer CJ, Gadgeel SM, Borghaei H, et al. Carboplatin and pemetrexed with or without pembrolizumab for advanced, non-squamous non-small-cell lung cancer: a randomised, phase 2 cohort of the open-label KEYNOTE-021 study. Lancet Oncol 2016;17:1497-508. [Crossref] [PubMed]
- Herbst RS, Baas P, Kim DW, et al. Pembrolizumab versus docetaxel for previously treated, PD-L1-positive, advanced non-small-cell lung cancer (KEYNOTE-010): a randomised controlled trial. Lancet 2016;387:1540-50. [Crossref] [PubMed]
- Brahmer J, Reckamp KL, Baas P, et al. Nivolumab versus Docetaxel in Advanced Squamous-Cell Non-Small-Cell Lung Cancer. N Engl J Med 2015;373:123-35. [Crossref] [PubMed]
- Borghaei H, Paz-Ares L, Horn L, et al. Nivolumab versus Docetaxel in Advanced Nonsquamous Non-Small-Cell Lung Cancer. N Engl J Med 2015;373:1627-39. [Crossref] [PubMed]
- Rittmeyer A, Barlesi F, Waterkamp D, et al. Atezolizumab versus docetaxel in patients with previously treated non-small-cell lung cancer (OAK): a phase 3, open-label, multicentre randomised controlled trial. Lancet 2017;389:255-65. [Crossref] [PubMed]
- Antonia SJ, Villegas A, Daniel D, et al. Durvalumab after Chemoradiotherapy in Stage III Non-Small-Cell Lung Cancer. N Engl J Med 2017;377:1919-29. [Crossref] [PubMed]
- Hellmann MD, Rizvi NA, Goldman JW, et al. Nivolumab plus ipilimumab as first-line treatment for advanced non-small-cell lung cancer (CheckMate 012): results of an open-label, phase 1, multicohort study. Lancet Oncol 2017;18:31-41. [Crossref] [PubMed]
- Antonia SJ, Lopez-Martin JA, Bendell J, et al. Nivolumab alone and nivolumab plus ipilimumab in recurrent small-cell lung cancer (CheckMate 032): a multicentre, open-label, phase 1/2 trial. Lancet Oncol 2016;17:883-95. [Crossref] [PubMed]
- Garon EB, Rizvi NA, Hui R, et al. Pembrolizumab for the treatment of non-small-cell lung cancer. N Engl J Med 2015;372:2018-28. [Crossref] [PubMed]
- Hirsch FR, McElhinny A, Stanforth D, et al. PD-L1 Immunohistochemistry Assays for Lung Cancer: Results from Phase 1 of the Blueprint PD-L1 IHC Assay Comparison Project. J Thorac Oncol 2017;12:208-22. [Crossref] [PubMed]
- Topalian SL, Hodi FS, Brahmer JR, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med 2012;366:2443-54. [Crossref] [PubMed]
- Brahmer JR, Tykodi SS, Chow LQ, et al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N Engl J Med 2012;366:2455-65. [Crossref] [PubMed]
- Shukuya T, Carbone DP. Predictive Markers for the Efficacy of Anti-PD-1/PD-L1 Antibodies in Lung Cancer. J Thorac Oncol 2016;11:976-88. [Crossref] [PubMed]
- Rizvi NA, Hellmann MD, Snyder A, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 2015;348:124-8. [Crossref] [PubMed]
- Carbone DP, Reck M, Paz-Ares L, et al. First-Line Nivolumab in Stage IV or Recurrent Non-Small-Cell Lung Cancer. N Engl J Med 2017;376:2415-26. [Crossref] [PubMed]
- Rizvi H, Sanchez-Vega F, La K, et al. Molecular Determinants of Response to Anti-Programmed Cell Death (PD)-1 and Anti-Programmed Death-Ligand (PD-L)-Ligand 1 Blockade in Patients With Non-Small-Cell Lung Cancer Profiled With Targeted Next-Generation Sequencing. J Clin Oncol 2018;36:633-41. [Crossref] [PubMed]
- Ebben JD, Lubet RA, Gad E, et al. Epidermal growth factor receptor derived peptide vaccination to prevent lung adenocarcinoma formation: An in vivo study in a murine model of EGFR mutant lung cancer. Mol Carcinog 2016;55:1517-25. [Crossref] [PubMed]
- Voena C, Menotti M, Mastini C, et al. Efficacy of a Cancer Vaccine against ALK-Rearranged Lung Tumors. Cancer Immunol Res 2015;3:1333-43. [Crossref] [PubMed]
- Busch SE, Hanke ML, Kargl J, et al. Lung Cancer Subtypes Generate Unique Immune Responses. J Immunol 2016;197:4493-503. [Crossref] [PubMed]
- Akbay EA, Koyama S, Carretero J, et al. Activation of the PD-1 pathway contributes to immune escape in EGFR-driven lung tumors. Cancer Discov 2013;3:1355-63. [Crossref] [PubMed]
- Azuma K, Ota K, Kawahara A, et al. Association of PD-L1 overexpression with activating EGFR mutations in surgically resected nonsmall-cell lung cancer. Ann Oncol 2014;25:1935-40. [Crossref] [PubMed]
- Ota K, Azuma K, Kawahara A, et al. Induction of PD-L1 Expression by the EML4-ALK Oncoprotein and Downstream Signaling Pathways in Non-Small Cell Lung Cancer. Clin Cancer Res 2015;21:4014-21. [Crossref] [PubMed]
- Hong S, Chen N, Fang W, et al. Upregulation of PD-L1 by EML4-ALK fusion protein mediates the immune escape in ALK positive NSCLC: Implication for optional anti-PD-1/PD-L1 immune therapy for ALK-TKIs sensitive and resistant NSCLC patients. Oncoimmunology 2015;5. [Crossref] [PubMed]
- Coelho MA, de Carne Trecesson S, Rana S, et al. Oncogenic RAS Signaling Promotes Tumor Immunoresistance by Stabilizing PD-L1 mRNA. Immunity 2017;47:1083-99.e6. [Crossref] [PubMed]
- Sundar R, Cho BC, Brahmer JR, et al. Nivolumab in NSCLC: latest evidence and clinical potential. Ther Adv Med Oncol 2015;7:85-96. [Crossref] [PubMed]
- Lee CK, Man J, Lord S, et al. Checkpoint Inhibitors in Metastatic EGFR-Mutated Non-Small Cell Lung Cancer-A Meta-Analysis. J Thorac Oncol 2017;12:403-7. [Crossref] [PubMed]
- Yao Z, Fenoglio S, Gao DC, et al. TGF-beta IL-6 axis mediates selective and adaptive mechanisms of resistance to molecular targeted therapy in lung cancer. Proc Natl Acad Sci U S A 2010;107:15535-40. [Crossref] [PubMed]
- Lee HJ, Zhuang G, Cao Y, et al. Drug resistance via feedback activation of Stat3 in oncogene-addicted cancer cells. Cancer Cell 2014;26:207-21. [Crossref] [PubMed]
- Awad MM, Mastini C, Blasco RB, et al. Epitope mapping of spontaneous autoantibodies to anaplastic lymphoma kinase (ALK) in non-small cell lung cancer. Oncotarget 2017;8:92265-74. [Crossref] [PubMed]
- DuPage M, Cheung AF, Mazumdar C, et al. Endogenous T cell responses to antigens expressed in lung adenocarcinomas delay malignant tumor progression. Cancer Cell 2011;19:72-85. [Crossref] [PubMed]
- Pfirschke C, Engblom C, Rickelt S, et al. Immunogenic Chemotherapy Sensitizes Tumors to Checkpoint Blockade Therapy. Immunity 2016;44:343-54. [Crossref] [PubMed]
- Akbay EA, Koyama S, Liu Y, et al. Interleukin-17A Promotes Lung Tumor Progression Through Neutrophil Attraction to Tumor Sites and Mediating Resistance to PD-1 Blockade. J Thorac Oncol 2017;12:1268-79. [Crossref] [PubMed]
- Lastwika KJ, Wilson W 3rd, Li QK, et al. Control of PD-L1 Expression by Oncogenic Activation of the AKT-mTOR Pathway in Non-Small Cell Lung Cancer. Cancer Res 2016;76:227-38. [Crossref] [PubMed]
- Koyama S, Akbay EA, Li YY, et al. STK11/LKB1 Deficiency Promotes Neutrophil Recruitment and Proinflammatory Cytokine Production to Suppress T-cell Activity in the Lung Tumor Microenvironment. Cancer Res 2016;76:999-1008. [Crossref] [PubMed]
- Skoulidis F, Byers LA, Diao L, et al. Co-occurring genomic alterations define major subsets of KRAS-mutant lung adenocarcinoma with distinct biology, immune profiles, and therapeutic vulnerabilities. Cancer Discov 2015;5:860-77. [Crossref] [PubMed]
- Skoulidis F, Hellmann MD, Awad MM, et al. STK11/LKB1 co-mutations to predict for de novo resistance to PD-1/PD-L1 axis blockade in KRAS-mutant lung adenocarcinoma. J Clin Oncol 2017;35:9016.
- Herter-Sprie GS, Koyama S, Korideck H, et al. Synergy of radiotherapy and PD-1 blockade in Kras-mutant lung cancer. JCI Insight 2016;1. [Crossref] [PubMed]
- Adeegbe DO, Liu Y, Lizotte PH, et al. Synergistic Immunostimulatory Effects and Therapeutic Benefit of Combined Histone Deacetylase and Bromodomain Inhibition in Non-Small Cell Lung Cancer. Cancer Discov 2017;7:852-67. [Crossref] [PubMed]
- Matlung HL, Szilagyi K, Barclay NA, et al. The CD47-SIRPalpha signaling axis as an innate immune checkpoint in cancer. Immunol Rev 2017;276:145-64. [Crossref] [PubMed]
- Weiskopf K, Jahchan NS, Schnorr PJ, et al. CD47-blocking immunotherapies stimulate macrophage-mediated destruction of small-cell lung cancer. J Clin Invest 2016;126:2610-20. [Crossref] [PubMed]


