
Measuring tumor mutation burden in non-small cell lung cancer: tissue versus liquid biopsy
Introduction
Over the past decades, important progresses have been achieved in the treatment of advanced non-small cell lung cancer (NSCLC), especially following the development of targeted therapies for tumors carrying specific actionable mutations, including epidermal growth factor receptor-tyrosine kinase inhibitors (EGFR-TKIs) and anaplastic lymphoma kinase (ALK) inhibitors. However, these approaches are unfeasible to treat tumors that do not carry these molecular alterations, which represent the majority of NSCLCs. In addition, the inevitable TKI-treatment resistance does further outline the need for alternative therapeutic options that can improve outcomes in advanced NSCLC patients. For a long time, NSCLC has been considered a non-immunogenic disease, because of its ability to escape the immune checkpoints and suppress immune response (1). The latest technological improvements, combined with an increase in the knowledge of tumor immunology mechanisms in NSCLC, have led to an increased interest in cancer immunotherapy. The introduction of immune checkpoint inhibitors (IOs) caused relevant changes in the treatment of advanced NSCLC, demonstrating improvements in patients’ outcomes and quality of life. Atezolizumab, nivolumab and pembrolizumab are immune-oncology compounds approved in the second-line treatment of metastatic NSCLC (2-4). These three molecules target programmed death-ligand 1 (PD-L1)/programmed death-1 (PD-1) axis and disrupt inhibition of the immune response, leading to T-cell activation and thereby restoring anti-tumour immunity (5-8). These IOs demonstrated improved survival as compared to chemotherapy in second line after failure of platinum doublet therapy in lung cancer (9-11). In addition, following the KEYNOTE-024 clinical trial results, in which pembrolizumab showed an impressive 30.2-month overall survival (OS) compared with 14.2 months for chemotherapy, this compound received the Food and Drug Administration (FDA) and European Medicines Agency (EMA) approval as first-line therapy in NSCLC patients whose tumors express ≥50% of the PD-L1 (12). However, the identification of the patients who will benefit from immunotherapy is still imprecise. PD-L1 expression is the only predictive biomarker currently used for patient selection, analysed by means of immunohistochemistry (IHC). However, it has several limitations, including the lack of standardization among the assays and the platforms used, which can determine a variation in the results, and it is not sufficient to fully explain clinical outcomes, because of the responses registered also in some patients with low PD-L1 expression (13,14). Thus, the need for biomarkers to stratify patients for immunotherapy is still unmet.
Tumor mutation burden (TMB) as predictive biomarker of response to immunotherapy
TMB has newly emerged as a possible independent biomarker to predict patient responses to immunotherapy in different tumor types, including lung cancer (2,3,15-22). TMB measures the number of somatic mutations within a tumor and thus is defined as the total number of somatic mutations per coding area of a tumor genome (5-8,23). In general, the accumulation of somatic mutations in the cancer genome is a multifactorial process, caused by factors that can be intrinsic—which include deficiencies or alterations in DNA repair genes or genes involved in DNA replication such as the polymerase epsilon (POLE)—and extrinsic—environmental factors, such as smoking status or ultraviolet (UV) exposure (24-27). The presence of high numbers of somatic mutations can lead to modifications of the proteins that are encoded by the mutated genes. The modified proteins are recognized by the immune system as “non-self” and may therefore represent tumor-specific neoantigens capable of activating specific anti-tumor immune responses (23,28,29). Indeed, the accumulation of somatic mutations and the neoantigen load have been demonstrated to correlate with response to IOs also in NSCLC (2). For all these reasons the possibility to use TMB measured by means of whole exome sequencing (WES) has been used as a proxy to deduce the neoantigen load of the tumors, introducing the rationale for its use as a biomarker for treatment with immunotherapies (20). Considering the complexity of the mechanisms that cause the accumulation of somatic mutations, the TMB value can differ among different tumor types and within the same tumor histotype (25,26,30). In lung cancer, for example, although the level of somatic mutations is generally high (25,26), it is significantly reduced in tumors carrying driver mutations (i.e., alterations in EGFR, ALK, ROS genes), while it seems not affected by the presence of KRAS or BRAF alterations (31). Because EGFR, ALK, and ROS1 alterations are more frequent among never or light smokers, this correlation further strengthens the connection between smoke and TMB (32). In this scenario, pharmaceutical companies have started to incorporate TMB assays into their clinical trials, to clinically evaluate and validate its role in the prediction of response to immunotherapies, not only in NSCLC, but also in other tumors such as melanoma and urothelial carcinomas (16-18).
TMB validation in clinical trials
WES studies
Considering the wide range of alterations that can be accumulated during cancer growth, WES is the golden standard method for measuring TMB, allowing the detection of somatic mutations present within the entire exome, which are likely to produce neoantigens (Table 1) (16,41). In a cohort of 34 NSCLC patients who received pembrolizumab, a median number of somatic nonsynonymous mutations of 299 was detected by WES in the group registering improved clinical benefits in terms of progression free survival (PFS), overall response rate (ORR) and durable clinical benefits (DCB), while a median of 127 alterations was present in the other group (P=0.0008) (2). Within the CheckMate-026 trial, comparing nivolumab versus platinum-based chemotherapy as first line therapy in NSCLC, an exploratory analysis was conducted on 312 patients to assess the effects of the TMB on treatment outcomes. Patients were divided in three groups according to TMB measured by WES (low TMB: <100 mutations; medium TMB: between 100 and 242 mutations, high TMB: >243 mutations). In the group with high TMB, the PFS was improved [9.7 vs. 5.8 months, HR 0.62 (95% CI: 0.38–1.00)] and response rate was higher (47% vs. 28%), when patients received nivolumab (3). Since in this study patients were originally stratified according to PD-L1 expression, these two biomarkers were compared. In the group with both high TMB and PD-L1 expression ≥50%, a higher response rate was registered (75%), while the group with the lower results for both parameters had the worst outcome (16%) (3). Recently, as part of the CheckMate-012 study, WES was used to analyze 75 patients with NSCLC treated with nivolumab plus ipilimumab, an IO targeting the cytotoxic T-lymphocyte-associated antigen 4 (CTLA-4). The final efforts were to improve clinical responses by using the combination of different immunotherapies and to evaluate whether the combination can enlarge the number of responders, thus limiting the role of TMB. Again, high TMB (>median, 158 mutations) demonstrated to be predictive of improved PFS, ORR and DCB compared to low TMB (median PFS 17.1 versus 3.7 months, HR 0.41; 95% CI: 0.23–0.73, log rank P=0.0024; ORR 51% versus 13%, Fisher’s exact P=0.0005; DCB 65% versus 34%, Fisher’s exact P=0.011). In addition, the negative predictive role of low TMB could not be overcome by the combination immunotherapy (22). In the paper, the authors highlighted the problems which are related to the use of WES approaches in clinical routine. The use of WES in clinical practice results difficult, especially in NSCLC, due to required gDNA input, minimum coverage needed and turnaround time. For this reason, they tried to estimate TMB using only the genes included in two targeted next generation sequencing (NGS) panels approved by FDA, the Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT) and FoundationOne panel (42,43). These two panels were able in silico to infer TMB similarly to WES (22).
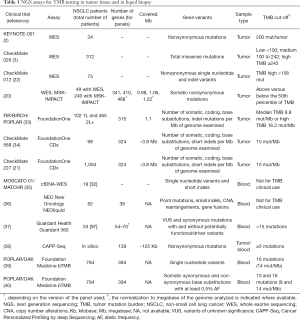
Full table
Targeted NGS studies
Targeted NGS constitutes the most reasonable approach to be translated into the clinics, because it has the major advantage of being less expensive and complex than WES. This application has become quite popular in clinical oncology, since it is a flexible method and easily adaptable to different uses (Table 1). Correlations between TMB results measured by targeted sequencing assay and by WES have been performed to understand the optimal number of genes to be covered to correctly deduce TMB. Evidence suggest that, more important than the number of genes, is the genomic area covered by the panel, demonstrating that TMB can be correctly inferred targeting at least 1Mb of the genome (44). TMB evaluated by means of targeted panels remains effective in predicting the response to immunotherapies in NSCLC, as demonstrated by different clinical trials. Rizvi et al. used three subsequent versions of the MSK-IMPACT NGS gene panel (covering 0.98, 1.06, and 1.22 Mb in the 341-, 410-, and 468-gene panel versions, respectively) to analyse 240 NSCLC patients treated with IOs. Using TMB data from the NGS panel to perform clinical correlations, higher TMB (median 8.5 vs. 6.6 single nucleotide variants/Mb) was present in patients with DCB, defined as partial response/stable disease for more than 6 months. In addition, MSK-IMPACT was compared to WES in a subgroup of 49 patients, confirming the good correlation between targeted NGS and WES when a minimum area of 1 Mb is sequenced (20). Pre-treatment tumor specimens from 102 first line and 465 second line or subsequent NSCLC patients enrolled in the POPLAR, BIRCH and FIR trials, all evaluating the role of atezolizumab as monotherapy, were analysed with the FoundationOne panel covering 315 cancer-related genes, corresponding to 1.1 Mb of genomic area. Atezolizumab produced improved outcomes in patients with a TMB value superior to the established cut-offs, defined by the 75th (high) and 50th (median) percentile. Thus, the obtained cut-offs were not univocal. TMB and PD-L1 expression were also independently associated with improved atezolizumab efficacy (33). Within the phase II CheckMate 568 trial, which evaluated the combination nivolumab plus ipilimumab in NSCLC, a TMB ≥10 mutations per megabase was identified as an effective cut-off to select patients most likely to have a clinical response. NGS analysis was conducted using FoundationOne CDx Assay, which covers 324 cancer-related genes for approximately 0.8 Mb of genomic area covered (34,45). The subsequent phase III CheckMate 227 examined PFS with the combination nivolumab plus ipilimumab versus chemotherapy among naïve NSCLC patients with a high TMB as defined by the CheckMate 568 (>10 mutations/Mb). This study confirmed findings from the CheckMate 568 trial: TMB is a robust predictive biomarker for the identification of patients responsive to immunotherapy, with a median PFS in patients receiving nivolumab and ipilimumab versus chemotherapy of 7.2 vs. 5.4 months, (HR 0.58; 97.5% CI: 0.41–0.81, P<0.001) and an ORR of 45.3% versus 26.9%, respectively. In addition, TMB was described as an independent biomarker from PD-L1 expression (21). Despite the fact that this panel covers <1 Mb of genome area, it has been clinically validated and has received FDA approval. In general, larger panels would have a better concordance with WES, however, the importance of the panel validation within clinical trials remains essential and can induce its introduction in the clinical practice. A comprehensive study conducted across 27 cancer types including NSCLC, evaluated the objective response rate for anti-PD-1 or anti-PD-L1 therapy against the TMB value and demonstrated a significant correlation between objective response rate and TMB (15). All the studies conducted up to now do confirm the importance and robustness of TMB as predictive biomarker of response to immunotherapy.
TMB analysis on liquid biopsy samples
Although primary tumor sample, obtained as formalin-fixed paraffin-embedded (FFPE), fine needle aspiration biopsy (FNAB) or cytological sample, is considered the standard starting material to perform NGS analyses, it can constitute a limiting factor especially in NSCLC, because it is often not available or insufficient and because the quality of the obtained DNA can negatively influence downstream testing. Liquid biopsy is becoming a common alternative approach to perform molecular analyses, since it is less invasive than tumor biopsy and it is easily repeatable. Circulating cell free DNA (cfDNA) derived from blood is currently being evaluated to assess TMB, allowing the parallel development of the methodologies on this alternative source of tumor DNA (Table 1). cfDNA could be used to evaluate TMB before treatment but also to monitor clinical responses. The use of gene panels to evaluate TMB seems more feasible when it comes to cfDNA testing (Table 2). However, TMB assessment on liquid biopsy must face the problem that the circulating DNA deriving from tumor cells [circulating tumor DNA (ctDNA)] is often only a small fraction of the cfDNA. A study by Koeppel et al. assessed TMB in cfDNA isolated from blood of 32 metastatic patients with various cancer types, including 19 NSCLC, enrolled in the MOSCATO 01 and MATCHR trials. They described a sensitivity of 53% when cfDNA-WES was compared to tissue WES. All the cases for which great differences in TMB were present between cfDNA and tissue-derived DNA, were supposed to be either ctDNA negative or mildly positive samples (considered as a percentage of detected variants 5 times smaller in cfDNA than in tissue), suggesting that ctDNA amount can highly influence sequencing results. When comparing WES and targeted NGS performed with the Cancer Hotspot Panel v2 from Thermo Fisher on cfDNA, a concordance rate of 92% was obtained, considering the regions covered by targeted NGS panel (35). An exploratory analysis performed with the Cancer Personalized Profiling by deep Sequencing (CAPP-Seq) approach evaluated the relationship between the number of mutations detected with the CAPP-Seq panel and with WES (38). The CAPP-Seq is an ultrasensitive NGS approach optimized for low DNA input and is able to enrich 139 recurrently mutated genes, in total covering ~125 kb (46). The number of mutations identified in plasma was used to infer the expected number of nonsynonymous mutations identified by CAPP-Seq on tumor tissue. The CAPP-Seq TMB correlated with WES data (r=0.93). The estimated WES-TMB was 41 times the mutations detected by CAPP-Seq and a TMB of about 200 mutations, considered as the standard for high TMB measured by WES, corresponded to ≥5 mutations detected with CAPP-Seq. To validate the CAPP-Seq/WES ratio, these methods were performed on 5 tumor samples from lung cancer patients, confirming the obtained results (38). CAPP-Seq based panels are commercially available, although data on TMB calculation from cfDNA have not been reported yet. A comprehensive hybrid capture-based NGS assay called NEOliquid was developed for the analysis of 39 clinically relevant genes in plasma samples (36). This method was compared with routine diagnostic testing performed on tissue from 82 NSCLC patients, with a concordance of 97.6%, a sensitivity of 70.8% and a specificity of 100%. Despite the potential utility of this approach and its validity when used on cfDNA, the use of this panel for TMB evaluation should be further investigated to ensure the concordance with WES and its ability to predict clinical responses (36). Another attempt to compare TMB from cfDNA with tissue was performed in a preliminary study conducted in a cohort of 97 patients, of which 53 with NSCLC. For tissue, the FoundationOne panel was used, while the Guardant360 was used for cfDNA testing. In this study, the obtained correlation was rather disappointing, due to a lack of concordance between tissue and cfDNA TMB. The authors suggest that a possible reason for the unsatisfying results may be due to differences in sequencing length, in mutation detection and in the methods for TMB measurement (37). Indeed, data on concordance between tissue and cfDNA vary among the different studies available up to now, both because of the analytical challenges that the use of cfDNA poses and of the limited datasets we have at disposal in this moment. All the studies comparing TMB analyses on cfDNA and tissue underline that further steps in the increase of sensitivity should be done, although the TMB performed with WES on cfDNA better correlates with tissue results (35,47). For sure, further evaluation on tissue-plasma concordance will be needed to ensure the feasibility of targeted NGS panels on plasma samples to determine TMB.

Full table
Clinical use of cfDNA for TMB evaluation
Despite these contrasting results on tissue and cfDNA concordance, the clinical utility of blood TMB (bTMB) was first assessed in a study conducted on 69 patients with different tumor types, including NSCLC, receiving IOs; cfDNA NGS analysis was performed with the Guardant360 panel (47). The ORR, PFS and OS registered in the group with high alterations number [defined as a total number of variants of unknown significance (VUS) >3 mutations or as a total number of alterations ≥6] were greater than in the group with low alterations number (47). A bTMB assay was recently developed by Foundation Medicine (48). This panel demonstrated its analytical validity by using a combination of 500 cell line DNA and clinical samples. The assay counts somatic mutations present at low allele frequency (0.5%) across 394 genes, starting from as little as 1% tumor content in at least 20 ng of cfDNA. Accuracy was established by comparing the bTMB assay and the previously validated Foundation Medicine TMB assay on the same samples. The obtained average positive percent agreement (PPA), negative percent agreement (NPA) and positive predictive values (PPV), calculated on the basis of the bTMB cut-offs of 10 and 16, were 95.2%, 100.0%, and 100.0% (48). The bTMB assay is being used to enroll patients for a phase II/III clinical trial evaluating atezolizumab versus chemotherapy as first line therapy in NSCLC (BFAST clinical trial NCT03178552). Analysis of bTMB within a bigger study using the 394-gene panel validated by Fabrizio et al. (48) demonstrated a significant association between PFS and TMB in patients treated in second line with atezolizumab. The study involved 211 samples from the POPLAR study and 583 samples from the OAK trial (39). In the POPLAR test cohort, a bTMB ≥16 corresponded to increased PFS and OS when patients received atezolizumab compared to docetaxel. In the validation cohort from the OAK study, the bTMB ≥16 population was the 27% of all evaluable patients. A significant benefit in terms of PFS in patients receiving atezolizumab was observed in the group with a bTMB ≥16 (HR 0.65; 95% CI: 0.47–0.92). However, the PPA between tissue and bTMB test was only 64%, highlighting the difficulties to standardize TMB testing on cfDNA. Recently, Fabrizio et al. compared the 394-gene panel analysis performed on blood samples with the TMB results obtained with both the FoundationOne test performed on tissue and a CLIA-validated assay for cfDNA, the Foundation FACT, covering 62 genes. Analyses conducted in a subgroup of 259 patients enrolled in the POPLAR and OAK study demonstrated an overall agreement and a PPA of 81.5% and 63.6%, respectively, between the 394-gene blood panel and the FoundationOne panel on tissue. When using the bTMB calculated by the CLIA assay, the PPA decreased to 17%, evidencing the importance of having a minimum genomic area covered to ensure a robust bTMB measurement. In addition, clinical validation was also performed on the entire cohort of patients enrolled in the two clinical trials. Cases with a bTMB ≥16 total mutations, corresponding to 14 mutations/Mb, were considered as high. Within POPLAR, the PFS HR for bTMB high patients was 0.57. The clinical utility of the panel was validated applying the bTMB cutoff of 14 mut/Mb in the OAK population, and obtaining a PFS HR of 0.65 (40). Taken together, the data suggest that the hypermutated state, also when assessed in cfDNA, can help identifying responders. However, high grade of variability in the identification of the bTMB values are still registered in the different clinical trials. At the present, the data available for TMB analysis in liquid biopsy are few and do not allow to reach a univocal conclusion. Two important clinical trials evaluating tumor mutational burden in the blood of patients with first-line NSCLC are currently ongoing and will probably provide more evidence on the feasibility of cfDNA in measurement of TMB. The B-F1RST (ClinicalTrials.gov identifier: NCT02848651) is a phase II, open-label, prospective, multicenter study designed to evaluate the efficacy and safety of single-agent atezolizumab as a first-line therapy NSCLC and it will evaluate the association between TMB measured in the blood and efficacy in predict clinical outcome in patients. The B-FAST (Blood-First Assay Screening Trial; ClinicalTrials.gov identifier: NCT03178552) is a phase II/III global, multicenter, open-label trial for the evaluation of the safety and efficacy of atezolizumab (or alectinib) in patients with unresectable, advanced/metastatic NSCLC. Treatment decision is performed on the basis of the presence of actionable somatic mutations or on the TMB score, both measured in the blood. The obtained data could help to clarify the role of liquid biopsy as a noninvasive, predictive biomarker for checkpoint inhibitor response in NSCLC.
Conclusions
In the research for new biomarkers predictive of response to immunotherapy in NSCLC patients, the importance of having blood based-approaches is evident, since it would allow to expand the cohort of patients who can receive a molecular characterization, as already happened with the EGFR mutation analysis in liquid biopsy. However, while TMB seems a very promising tool in immuno-oncology, there are different challenges to overcome for its adoption into clinical practice, both when using tissue and blood. First, both tissue and blood do present their own advantages and disadvantages. FFPE material is for sure the most convenient and reliable material, between the two, since tumor load can be measured and eventually micro-dissected to ensure a high percentage of neoplastic cell content. However, it displays quality problems mainly caused by formalin fixation, which could produce a highly fragmented and low quality-DNA. The damaged DNA can negatively influence downstream interpretation, since false positive calls due to deamination may be included in the TMB calculation. In addition, as already stated, in NSCLC patients it can be sometimes difficult to obtain tissue material, because NSCLC is often diagnosed at advanced stages. Liquid biopsy is a less invasive method to obtain tumor derived-DNA, it has patient’s compliance and is also easily repeatable, if needed. It can also ideally be representative of tumor heterogeneity, especially in advanced disease. On the other side, cfDNA may not contain sufficient ctDNA for NGS approaches, which generally require higher amounts of DNA input, or may contain no tumor DNA at all. Data on concordance between cfDNA and tissue need further evaluations, to ensure the consistency of the results obtained from blood (Table 2). The lack of standardization in the approaches used for TMB evaluation makes difficult to compare the results. The targeted panels cover different megabases of the genome. In addition, the type of somatic mutations (synonymous, non-synonymous and drivers’ alterations) included in TMB calculation differs among the methods used in the studies, as well as the cut-offs used to stratify patients. The importance of having pre-defined cut-off would simplify the interpretation of clinical outcomes and would of course help clinicians identify the optimal treatment approach. In this respect, some projects are ongoing to harmonize this analysis: the Quality Assurance Initiative Pathology (QuIP) initiative of the German Society of Pathology and the Federal Association of German Pathologists, The Friends of Cancer Research consortium, and the International Quality Network for Pathology (IQNPath) and European Society of Medical Oncology (ESMO) joint project on TMB are working to provide standardization of the procedures and harmonization of the results. For the first time, an attempt to provide harmonization of biomarker analyses is made prior to its clinical introduction in clinical practice. As the immunotherapy value increases and the use of TMB as biomarker is investigated, the number of results that will be collected will grow in parallel. This will allow to characterize TMB in a robust manner and will also allow to better understand how this biomarker works and how to better determine treatment decisions. The opportunity provided by the recent improvements both in the technologies available, including NGS, and in the use of liquid biopsy for molecular profiling of the tumor has for sure improved the management of NSCLC and offered a proof of how far research has come in personalizing the available therapies according to patient’s needs.
Acknowledgements
None.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Raez LE, Cassileth PA, Schlesselman JJ, et al. Allogeneic vaccination with a B7.1 HLA-A gene-modified adenocarcinoma cell line in patients with advanced non-small-cell lung cancer. J Clin Oncol 2004;22:2800-7. [Crossref] [PubMed]
- Rizvi NA, Hellmann MD, Snyder A, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 2015;348:124-8. [Crossref] [PubMed]
- Carbone DP, Reck M, Paz-Ares L, et al. First-Line Nivolumab in Stage IV or Recurrent Non-Small-Cell Lung Cancer. N Engl J Med 2017;376:2415-26. [Crossref] [PubMed]
- Steuer CE, Ramalingam SS. Tumor Mutation Burden: Leading Immunotherapy to the Era of Precision Medicine? J Clin Oncol 2018;36:631-2. [Crossref] [PubMed]
- Kim JM, Chen DS. Immune escape to PD-L1/PD-1 blockade: seven steps to success (or failure). Ann Oncol 2016;27:1492-504. [Crossref] [PubMed]
- Liontos M, Anastasiou I, Bamias A, et al. DNA damage, tumor mutational load and their impact on immune responses against cancer. Ann Transl Med 2016;4:264. [Crossref] [PubMed]
- Sharma P, Allison JP. The future of immune checkpoint therapy. Science 2015;348:56-61. [Crossref] [PubMed]
- Giannakis M, Mu XJ, Shukla SA, et al. Genomic Correlates of Immune-Cell Infiltrates in Colorectal Carcinoma. Cell Rep 2016;15:857-65. [Crossref] [PubMed]
- Horn L, Spigel DR, Vokes EE, et al. Nivolumab Versus Docetaxel in Previously Treated Patients With Advanced Non-Small-Cell Lung Cancer: Two-Year Outcomes From Two Randomized, Open-Label, Phase III Trials (CheckMate 017 and CheckMate 057). J Clin Oncol 2017;35:3924-33. [Crossref] [PubMed]
- Herbst RS, Baas P, Kim DW, et al. Pembrolizumab versus docetaxel for previously treated, PD-L1-positive, advanced non-small-cell lung cancer (KEYNOTE-010): a randomised controlled trial. Lancet 2016;387:1540-50. [Crossref] [PubMed]
- Fehrenbacher L, Spira A, Ballinger M, et al. Atezolizumab versus docetaxel for patients with previously treated non-small-cell lung cancer (POPLAR): a multicentre, open-label, phase 2 randomised controlled trial. Lancet 2016;387:1837-46. [Crossref] [PubMed]
- Reck M, Rodriguez-Abreu D, Robinson AG, et al. Pembrolizumab versus Chemotherapy for PD-L1-Positive Non-Small-Cell Lung Cancer. N Engl J Med 2016;375:1823-33. [Crossref] [PubMed]
- Ilie M, Hofman V, Dietel M, et al. Assessment of the PD-L1 status by immunohistochemistry: challenges and perspectives for therapeutic strategies in lung cancer patients. Virchows Arch 2016;468:511-25. [Crossref] [PubMed]
- Grigg C, Rizvi NA. PD-L1 biomarker testing for non-small cell lung cancer: truth or fiction? J Immunother Cancer 2016;4:48. [Crossref] [PubMed]
- Yarchoan M, Hopkins A, Jaffee EM. Tumor Mutational Burden and Response Rate to PD-1 Inhibition. N Engl J Med 2017;377:2500-1. [Crossref] [PubMed]
- Van Allen EM, Miao D, Schilling B, et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science 2015;350:207-11. [Crossref] [PubMed]
- Snyder A, Makarov V, Merghoub T, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med 2014;371:2189-99. [Crossref] [PubMed]
- Rosenberg JE, Hoffman-Censits J, Powles T, et al. Atezolizumab in patients with locally advanced and metastatic urothelial carcinoma who have progressed following treatment with platinum-based chemotherapy: a single-arm, multicentre, phase 2 trial. Lancet 2016;387:1909-20. [Crossref] [PubMed]
- Le DT, Uram JN, Wang H, et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med 2015;372:2509-20. [Crossref] [PubMed]
- Rizvi H, Sanchez-Vega F, La K, et al. Molecular Determinants of Response to Anti-Programmed Cell Death (PD)-1 and Anti-Programmed Death-Ligand 1 (PD-L1) Blockade in Patients With Non-Small-Cell Lung Cancer Profiled With Targeted Next-Generation Sequencing. J Clin Oncol 2018;36:633-41. [Crossref] [PubMed]
- Hellmann MD, Ciuleanu TE, Pluzanski A, et al. Nivolumab plus Ipilimumab in Lung Cancer with a High Tumor Mutational Burden. N Engl J Med 2018;378:2093-104. [Crossref] [PubMed]
- Hellmann MD, Nathanson T, Rizvi H, et al. Genomic Features of Response to Combination Immunotherapy in Patients with Advanced Non-Small-Cell Lung Cancer. Cancer Cell 2018;33:843-52.e4. [Crossref] [PubMed]
- Schumacher TN, Schreiber RD. Neoantigens in cancer immunotherapy. Science 2015;348:69-74. [Crossref] [PubMed]
- Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature 2009;458:719-24. [Crossref] [PubMed]
- Lawrence MS, Stojanov P, Polak P, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 2013;499:214-8. [Crossref] [PubMed]
- Alexandrov LB, Nik-Zainal S, Wedge DC, et al. Signatures of mutational processes in human cancer. Nature 2013;500:415-21. [Crossref] [PubMed]
- Lin EI, Tseng LH, Gocke CD, et al. Mutational profiling of colorectal cancers with microsatellite instability. Oncotarget 2015;6:42334-44. [Crossref] [PubMed]
- Brown SD, Warren RL, Gibb EA, et al. Neo-antigens predicted by tumor genome meta-analysis correlate with increased patient survival. Genome Res 2014;24:743-50. [Crossref] [PubMed]
- Anagnostou V, Smith KN, Forde PM, et al. Evolution of Neoantigen Landscape during Immune Checkpoint Blockade in Non-Small Cell Lung Cancer. Cancer Discov 2017;7:264-76. [Crossref] [PubMed]
- Vogelstein B, Papadopoulos N, Velculescu VE, et al. Cancer genome landscapes. Science 2013;339:1546-58. [Crossref] [PubMed]
- Spigel DR, Schrock AB, Fabrizio D, et al. Total mutation burden (TMB) in lung cancer (LC) and relationship with response to PD-1/PD-L1 targeted therapies. J Clin Oncol 2016;34:9017. [Crossref]
- Govindan R, Ding L, Griffith M, et al. Genomic landscape of non-small cell lung cancer in smokers and never-smokers. Cell 2012;150:1121-34. [Crossref] [PubMed]
- Kowanetz M, Zou W, Shames D, et al. Tumor mutation burden (TMB) is associated with improved efficacy of atezolizumab in 1L and 2L+ NSCLC patients. J Thorac Oncol 2016;12:S321-2. [Crossref]
- Ramalingam S, Hellmann M, Awad MM, et al. Tumor mutational burden (TMB) as a biomarker for clinical benefit from dual immune checkpoint blockade with nivolumab (nivo) + ipilimumab (ipi) in first-line (1L) non-small cell lung cancer (NSCLC): identification of TMB cutoff from CheckMate 568. Cancer Res 2018;78:Abstract nr CT078.
- Koeppel F, Blanchard S, Jovelet C, et al. Whole exome sequencing for determination of tumor mutation load in liquid biopsy from advanced cancer patients. PLoS One 2017;12:e0188174. [Crossref] [PubMed]
- Müller JN, Falk M, Talwar J, et al. Concordance between Comprehensive Cancer Genome Profiling in Plasma and Tumor Specimens. J Thorac Oncol 2017;12:1503-11. [Crossref] [PubMed]
- Davis AA, Chae YK, Agte S, et al. Comparison of tumor mutational burden (TMB) across tumor tissue and circulating tumor DNA (ctDNA). J Clin Oncol 2017;35:e23028. [Crossref]
- Chaudhuri AA, Chabon JJ, Lovejoy AF, et al. Early Detection of Molecular Residual Disease in Localized Lung Cancer by Circulating Tumor DNA Profiling. Cancer Discov 2017;7:1394-403. [Crossref] [PubMed]
- Gandara DR, Kowanetz M, Mok TSK, et al. Blood-based biomarkers for cancer immunotherapy: Tumor mutational burden in blood (bTMB) is associated with improved atezolizumab (atezo) efficacy in 2L1 NSCLC (POPLAR and OAK). Ann Oncol 2017;28:460-1. [Crossref]
- Fabrizio D, Malboeuf C, Lieber D, et al. A Blood-Based Next Generation Sequencing Assay to Determine Tumor Mutational Burden (bTMB) Is Associated with Benefit to an Anti-PD-L1 Inhibitor, Atezolizumab. Cancer Res 2018;78:Abstract nr 5706.
- Rooney MS, Shukla SA, Wu CJ, et al. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell 2015;160:48-61. [Crossref] [PubMed]
- FDA. FDA announces approval, CMS proposes coverage of first breakthrough-designated test to detect extensive number of cancer biomarkers. 2017. Available online: https://www.fda.gov/NewsEvents/Newsroom/PressAnnouncements/ucm587273.htm
- FDA. FDA unveils a streamlined path for the authorization of tumor profiling tests alongside its latest product action. 2017. Available online: https://www.fda.gov/NewsEvents/Newsroom/PressAnnouncements/ucm585347.htm
- Chalmers ZR, Connelly CF, Fabrizio D, et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med 2017;9:34. [Crossref] [PubMed]
- FDA. FoundationOne CDx: Summary of Safety and Effectiveness Data (SSED). 2017. Available online: https://www.accessdata.fda.gov/cdrh_docs/pdf17/P170019B.pdf
- Newman AM, Bratman SV, To J, et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med 2014;20:548-54. [Crossref] [PubMed]
- Khagi Y, Goodman AM, Daniels GA, et al. Hypermutated Circulating Tumor DNA: Correlation with Response to Checkpoint Inhibitor-Based Immunotherapy. Clin Cancer Res 2017;23:5729-36. [Crossref] [PubMed]
- Fabrizio D, Malboeuf C, Lieber D, et al. Analytic validation of a next generation sequencing assay to identify tumor mutational burden from blood (bTMB) to support investigation of an anti-PD-L1 agent, atezolizumab, in a first line non-small cell lung cancer trial (BFAST). Ann Oncol 2017.28.


