
Mechanisms of resistance to pemetrexed in non-small cell lung cancer
Introduction
Cancer is a major public health problem worldwide, and lung cancer has remained the most common cause of cancer death in recent years. Among patients with lung cancer, non-small cell lung cancer (NSCLC) represents 87% of all cases, including adenocarcinoma and squamous cell carcinoma as its main subtypes (1). However, advances in lung cancer treatment have always been challenging over the past three decades, and the current 5-year survival for lung cancer is only 18% (2). Even though multiple approaches are recommended for NSCLC patients such as surgery, chemotherapy, targeted therapy, immunotherapy, and radiotherapy, systemic chemotherapy is still the mainstay of treatment for NSCLC, especially used as palliative care for advanced patients (3). With respect to chemotherapy, platinum is not only regarded as the standard first-line regimen for advanced NSCLC, but it is also recommended for patients with earlier stages (4,5).
Besides platinum, pemetrexed, one of the antifolates, has also been extensively used to treat NSCLC. Regarding to its function, pemetrexed is an antagonist against folates. Because folates donate one-carbon units to the biosynthesis of purines, thymidine, and hence DNA, the interruption of folate metabolism by pemetrexed contributes to ineffective DNA synthesis and failure of growth in tumor cells finally (6). In resembling the metabolism of folates, there are three steps by which pemetrexed exerts its inhibitory potency. Three uptake receptors are responsible for the cell entry of pemetrexed, including the reduced folate receptor (RFC), the folate receptor-α (FR-α) and the proton-coupled folate receptor (7-9). Afterwards, the agent is modified to its pharmacologically active forms by folylpolyglutamate synthetase (FPGS) in the cytosol and mitochondria, to be better retained in cells and to gain a greater affinity than the parent form (10). This process competes with hydrolysis of accumulated glutamate tails on pemetrexed, which is catalyzed by another enzyme called γ-glutamyl hydrolase (GGH) in the lysosome, leading to elimination of the drug (11). Finally, four primary folate-dependent enzymes are inhibited: thymidylate synthase (TS), dihydrofolate reductase (DHFR), glycinamide ribonucleotide formyl-transferase (GARFT), and 5-aminoimidazole-4-carboxamide ribonucleotide formyl-transferase (AICARFT) (12,13). In clinical treatments, pemetrexed has shown efficacy not only with platinum as a first-line therapy but also as a single agent for second-line treatment among advanced patients (14,15). In addition, modest efficacy has also been reported when pemetrexed is used as further-line treatment options (16,17). However, despite the high efficacy of pemetrexed, NSCLC will develop chemoresistance that hinders clinical efficacy after long-term use. According to recent reviews, the molecular mechanisms of drug tolerance can be split into two categories: cancer cells and their interactions with anticancer agents (18). In this review, we will elaborate known mechanisms from these two categories that apply to the resistance of pemetrexed in NSCLC treatment, to provide a better understanding as well as a guidance on new directions in this field and to facilitate more efficient therapies for patients in clinical treatments. Understanding pemetrexed resistance or toxicity may provide accurate patient stratifications and appropriate drug administration by clinicians.
Mechanisms relevant to cancer cells
Oncogene and oncoprotein alterations
The ErbB receptor family, including EGFR, HER2 (ErbB2), HER3 (ErbB3), and HER4 (ErbB4), regulate cell proliferation, anti-apoptosis, and invasion in cancer (19). In lung cancer, EGFR mutations occur in about 40% of Asian patients, mostly in adenocarcinomas (20). Better outcomes were obtained in EFGR-mutated patients rather than wild-type patients when treated with pemetrexed, indicating the EGFR mutation as a positive factor for pemetrexed in NSCLC [progression-free survival (PFS): 3.9 vs. 2.3 months, P=0.030] (21). Furthermore, in terms of the types of mutations, pemetrexed efficacy was found to be significantly higher in the exon 21 (Leu858Arg) mutation group than that in the exon 19 (deletion) group in another study (PFS: 3.3 vs. 1.8 months, P=0.0080) (22). Besides, the expression of ErbB protein was determined in biomedical lung cancer studies. A group of authors found that ErbB was significantly upregulated in pemetrexed-resistant A549 adenocarcinoma cell lines (A549/PEM) when compared with parent cells, and the cellular response to pemetrexed of A549/PEM was increased via silencing both EGFR and HER3 (23). Apart from ErbB, anaplastic lymphoma kinase (ALK) and ROS proto-oncogene 1 (ROS1) frequently occur in NSCLC patients, with 2–7% and 1–2% of all cases respectively (24,25). Favorable clinical outcomes have been reported in ALK-positive NSCLC patients when treated with pemetrexed, and the outcome was more evident when pemetrexed was used as single agent (P<0.0010) (26,27). Moreover, compared to EGFR mutations, NSCLC patients with ALK rearrangements showed better outcomes than EGFR-mutant or wild-type patients who had been treated with pemetrexed [time to progression (TTP): 9.2 vs. 1.4 vs. 2.9 months, P=0.0010) (28). Furthermore, a recent study comparing the relationship between ROS1 and the efficacy of pemetrexed in NSCLC cases reported better survival among patients with ROS1 translocations than those without translocations (P=0.0030) (29). In addition to EGFR, ALK and ROS1, the viral RAS oncogene family (KRAS, NRAS, and HRAS) has been investigated in NSCLC where KRAS mutations are frequently found in 25% of lung adenocarcinomas, particularly in non-Asians while NRAS mutations are rarely found in NSCLC (30). A certain study has been conducted to determine the possible correlation between the drug efficacy of pemetrexed and several gene aberrations incorporating RAS. The results showed that advanced NSCLC patients with no matter which mutation had better clinical outcomes than those without any mutation after receiving pemetrexed as maintenance treatment [PFS, P=0.0060; overall survival time (OS), P=0.0010] (31). Apart from the abovementioned factors, one of the most common tumor suppressor genes, p53 together with its transcriptional target p21, is worth discussing. In NSCLC, a previous study reported notable elevations of p21 and p27 in A549 cells after receiving pemetrexed (32). Furthermore, knocking down the p53-regulated zinc finger protein (ZMAT3) was reported to sensitize the response of A549 cell lines to pemetrexed (P<0.050) (33).
Oncoprotein thyroid transcription factor 1 (TTF-1) is a tissue-specific transcription factor expressed mainly in the epithelial cells of the lungs. It is used as an important marker to classify NSCLC when no clear morphological features can be found, because the expression frequency of TTF-1 is very high in lung adenocarcinoma but rare in lung squamous cell carcinoma (34). It was reported in two studies that patients with TTF-1-positive NSCLC tumors, who underwent pemetrexed-based regimens, had significantly better outcomes than those with negative NSCLC tumors (OS: 25.4 vs. 14.2 months; hazard ratio: 0.55) (OS: 10.4 vs. 6.0 months, P<0.0010) (35,36).
Together, oncogene mutations, including EGFR, ALK, ROS1, KRAS, and NRAS as well as TTF-1 status are positive factors for pemetrexed in NSCLC patients, while tumor suppressor factors and exon 19 (deletion) in EGFR may signify resistance, but the results still need to be confirmed. A table summarizing the pemetrexed-resistance mechanisms relevant to oncogene and oncoprotein alterations in NSCLC is included in Table 1. Further detailed are needed to define the precise drug efficacy of pemetrexed for NSCLC patients harboring specific gene alterations.
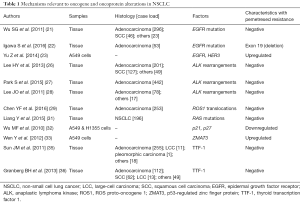
Full table
DNA synthesis and replication
Synthesis of purines and pyrimidines is the most outstanding factors in DNA synthesis and replication. Among the four target enzymes of pemetrexed, TS has most frequently been investigated. The correlation between low levels of TS expression and chemosensitivity to pemetrexed was first found in 61 human tumor samples, including lung cancer (37). Subsequently, this correlation was proven to be an inverse one on NSCLC patients (P=0.0060) (38). In addition, significantly beneficial outcomes were reported in NSCLC patients with low expression levels of TS in another clinical study (PFS: 5.6 vs. 3.5 months, P=0.013) (39). Views suggested by predecessors have been elucidated by relevant basic studies as described below. It was reported that the more TS is expressed by NSCLC cell lines, the weaker the sensitivity to pemetrexed (40). However, another study reported an opposite result, showing that TS mRNA was significantly reduced in a dose-dependent manner in lung adenocarcinoma cells treated with pemetrexed (32), so further studies are needed to resolve this controversy. On the gene level, many studies have searched for additional mechanisms associated with TS expression and pemetrexed efficacy in NSCLC. The most important observation has been the amplification of TS. Higher copy numbers of TS were found in pemetrexed-resistant lung cancer sublines when compared with parental cells (41). Moreover, after receiving pemetrexed, advanced NSCLC patients with amplified TS showed a worse response to pemetrexed when compared with those in the unamplified group (RR: 0 vs. 35.3%, P=0.054) (42). Another aspect investigated on the gene level has been the variant genotypes of TS. Of note, there are three different types of polymorphisms in TS. Two types of polymorphisms are in the promoter of TS: a 28-base-pair (bp) variable number tandem repeat (VNTR), including two and three repeats (2R/3R) located in the 5'-untranslated region (UTR) and a 6 bp deletion or insertion in the 3'-UTR. A third modality is the SNP, of which G > C is often the case studied (43). In this review, research conclusions relating to the 5'-UTR of TS will be described first. NSCLC patients with the 3R in the 5'-UTR were found to have superior clinical outcomes after receiving pemetrexed (RR: 100%; P=0.017) (44). Nonetheless, an opposite finding was reported among non-SCC patients treated with pemetrexed, in which the clinical outcomes of the 3R genotype carriers were insignificantly poorer than their counterparts, and a significant shorter survival was observed when there was a synchronous G nucleotide in the second repeat of VNTR (P=0.020) (45). Apart from the 5'-UTR of TS, there is still research with respect to genotypes in the 3'-UTR. NSCLC patients with 6bp deletion/6bp deletion polymorphisms have been reported to have a better prognosis than those with 6bp deletion/6bp insertion genotypes (PFS: 3.4 vs. 2.9 months; P=0.0010; OS: 14 vs. 12 months; P=0.000) (46). Apart from TS, other primary targets of pemetrexed, including DHFR and GARFT, have been investigated in recent years. While the association between the expression of DHFR and pemetrexed efficacy remains a controversial issue, higher GARFT status appears to play a pivotal role in NSCLC cell line resistance to pemetrexed (P=0.0080) (32,37,40).
Purines and pyrimidines are subsequently utilized for composing ribonucleotides that make up of strands of DNA during its replication, where ribonucleotide reductase subunit M1 (RRM1), one of the subunits of ribonucleotide reductase (RNR) plays an essential role (47). A survey among NSCLC patients with neoadjuvant chemotherapy using pemetrexed revealed that the tumor response rate was inversely correlated with the RRM1 expression level, suggesting that higher expression of RRM1 indicated resistance to pemetrexed (P≤0.0010) (38).
Additionally, given the role that folate plays in metabolism, folate is also a crucial factor worthy to be discussed in this issue. Methylenetetrahydrofolate reductase (MTHFR) is an enzyme that converts 5,10-methytetrahydrofolate (5,10-MTHF), used as one-carbon units, to 5-methytetrahydrofolate, which provides methyl groups for DNA methylation not used for synthetic processes (48). With regard to NSCLC, a report suggested that clinical outcomes for NSCLC patients with the single nucleotide polymorphism (SNP) 677T > T genotype were better than those harboring the 677C > C or 677C > T genotypes, all of whom had received pemetrexed combined with platinum (P=0.030) (49).
In general, primary targets of pemetrexed such as TS, DHFR, and GARFT together with RRM1 and MTHFR are predictive factors for NSCLC patients under pemetrexed-based therapy. We may conclude that patients with higher TS expression or gene copy numbers as well as certain kinds of genotypes of TS or MTHFR are more likely to show weaker responses to pemetrexed. The same results were observed with those harboring elevated levels of GARFT and RRM1.
DNA damage and repair
The base excision repair (BER) system is a vital factor worthy to be discussed in this review. One of the most vital components of the BER system is ceaseless aberrant uracil incorporation and reincorporation induced by TS (50). Deoxyuridine triphosphate nucleotidohydrolase (dUTPase) and uracil DNA glycosylase (UDG) are two regulators of this process (51). By silencing dUTPase, a greatly diminished survival ability was observed in cells treated with pemetrexed (52). Additionally, higher expression of UDG was demonstrated to be significantly associated with higher drug resistance in NSCLC cell lines treated with pemetrexed, whereas knockdown of UDG expression resulted in a seven-fold increased sensitivity to pemetrexed compared to that of parental cells (P<0.0001) (53).
Apart from the abovementioned studies, another important mechanism used to repair damaged DNA is nucleotide excision repair (NER), among which excision repair cross-complementation group 1 (ERCC1) is one of the responsible polypeptides (47). As reported, low ERCC1 expression was significantly associated with prolonged survival in patients with advanced lung adenocarcinoma treated with pemetrexed as first-line therapy (RR: 52.3 vs. 30.8%, P=0.015; PFS: 6.4 vs. 3.8 months, P=0.0040) (54). Of interest, patients harboring a variant-type polymorphism of 8092C > A were found to have shorter survival compared with wild-type carriers after receiving a combined therapy including pemetrexed (RR: 10 vs. 48%, P=0.0050; OS: 14.7 vs. 21.1 vs. 30.1 months, P=0.032) (55).
Checkpoint kinase 1 (CHK1), human MutS homolog 2 (MSH2), and Ku protein are also biomarkers associated with DNA repair systems. Poor differentiation has been shown in lung adenocarcinoma cells with higher CHK1 levels, indicating an increased dependence on the CHK1 pathway in response to pemetrexed (56). Moreover, by evaluating multiple NSCLC cell lines after treatment with pemetrexed, MSH2 levels were demonstrated to be increased and the cytotoxicity of pemetrexed was found to be enhanced by the knockdown of MSH2 (P<0.010) (57). Similar to MSH2, pemetrexed-treated A549 cell viability was found to be augmented by silenced Ku80, one of the two subunits of Ku protein (58).
Overall, a higher status of DNA repair systems, including BER, NER, as well as CHK1, MSH2 and Ku protein may contribute to resistance to pemetrexed in NSCLC. A table summarizing pemetrexed-resistance mechanisms relevant to DNA biosynthesis processes in NSCLC that we discussed above is included in Table 2.
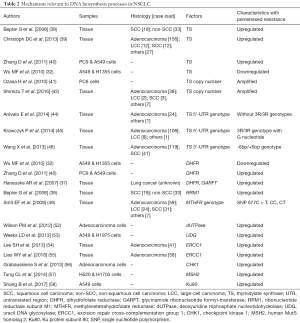
Full table
Other mechanisms relevant to cancer cells
Epithelial-to-mesenchymal transition (EMT) is a program causing epithelial cells to attain mesenchymal characteristics, enhancing tumor progression, and eventually, metastasis (59). An activated EMT pathway was found to be associated with resistance to pemetrexed in NSCLC cell lines, and the result was confirmed by the regression in pemetrexed efficacy after blocking the EMT pathway (P<0.050) (60). A further study not only achieved similar results to the previous study but pointed out that p38 and/or ERK signaling pathways might regulate the EMT process (61).
Beyond the EMT, cancer stem cells (CSCs) are emerging systems related to tumors, which have characteristics similar to embryonic stem cells (62). Using NSCLC cell lines and detecting CD166, one of the surface markers of stem cells, greater resistance to pemetrexed was found in CD166-postive cells rather than in CD166 positive cells, demonstrating a correlation between therapeutic resistance to pemetrexed and cancer cell stemness in NSCLC (P<0.010) (63). Furthermore, increased levels of stem cell factors were also found to be associated with resistance of NSCLC cell lines to pemetrexed (P<0.050) (60).
In addition, there are some other factors related to tumor cell migration and metastasis such as NME1 (NM23-H1), lipocalin 2 (LCN-2), cortactin (CTTN), and Src kinase. It was reported that after treatment with pemetrexed, NM23-H1 and LCN-2 were dramatically increased in A549 cell lines in a dose-dependent manner. Moreover, LCN-2-silenced cells appeared to be more responsive to pemetrexed, also in a dose-dependent manner (32). Besides, high expression of CTTN in A549 cells was also found to be correlated with resistance to pemetrexed and the sensitivity of A549 cells to pemetrexed was also augmented by knocking down CTTN (P<0.050) (33). The result was also the same with Src kinase in another clinical study indicating Src kinase as an adverse prognostic factor (PFS: 49.7 vs. 74.2 months, P=0.040) (64).
Apart from the abovementioned studies, three less studied aspects associated with resistance to pemetrexed in NSCLC, including cell apoptosis, microRNAs (miRNAs) expressions, and variations of mean corpuscular volume (MCV), are presented in the followings. First, enhanced prevention of cell apoptosis was strongly correlated with pemetrexed-induced resistance in A549 cells in terms of the overexpression of protectors against apoptosis such as flavin reductase (FR) and calreticulin (65). Secondly, in one study performed on advanced NSCLC patients, three miRNAs were implicated as predictive biomarkers for treatment efficacy of pemetrexed. In terms of clinical outcomes, it was pointed out that upregulation of miR-25 (PFS: 3.8 vs. 4.8 months, P=0.0070) or miR-210 (PFS: 3.7 vs. 4.9 months, P=0.0010) as well as downregulation of miR-145 (PFS: 3.8 vs. 5.0 months, P=0.0010) might be indicative of low pemetrexed drug efficacy (66). Last but not least, variations of MCV of erythrocytes were also shown to be potentially associated with drug efficacy in NSCLC. A study demonstrated that pemetrexed could induce a macrocytosis phenomenon in NSCLC patients. The study further evaluated the range of changes in MCV and found that patients with large variations of MCV, over 5 fL, had better clinical outcomes than those with less than 5-fL MCV (PFS: 7 vs. 3 months, P=0.0016; OS: 17 vs. 10 months, P=0.020) (67). However, the mechanism remains to be validated.
In summary, stronger driving forces for tumor migration and metastasis, for example, activate EMT, cancer cell stemness, and other changes in biomarkers are likely to induce insensitivity of NSCLC cells to pemetrexed. Besides, declined cell apoptosis, smaller variation ranges in MCV, as well as changed expressions of certain miRNAs may hinder the efficacy of pemetrexed for NSCLC patients. A table summarizing the aforementioned mechanisms of pemetrexed-resistance relevant to tumor cells in NSCLC is included in Table 3.
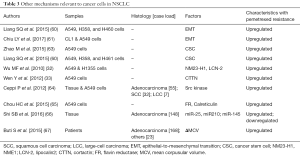
Full table
Mechanisms relevant to drug dynamics
Both of uptake and elimination processes affect the intracellular concentration of anticancer agents. RFC and FR-α are all transporters of pemetrexed. In one study, RFC was found to be markedly downregulated in certain pemetrexed-resistant NSCLC cells (P<0.050) (68). Additionally, two SNPs in RFC were evaluated in another study, including 2117C > T and 9148C > A, showing that patients with homozygous variant genotypes of SLC19A, for instance 2117T > T and 9148A > A, had a significantly better outcome in clinical events compared with their counterparts (2117C > T, OS: 16.7 vs. 14 vs. 8.9 months, P=0.030; 9148C > A OS: 22.7 vs. 10.3 vs. 9.4 months, P=0.045) (69). Furthermore, studies regarding the effects of FR-α expression on pemetrexed efficacy demonstrated that high membranous FR-α expression was significantly associated with prolonged patient survival, suggesting FR-α as a positive biomarker for prognosis of patients receiving pemetrexed (PFS: 5.6 vs. 3.7 months, P=0.031; OS: 22.1 vs. 11.5 months, P=0.013) (39). Regarding the elimination of anticancer agents, ATP-binding cassette (ABC) transporters are pivotal in this process. ABCC11, one of the seven subfamilies of ABC transporters, was found to be correlated with resistance to pemetrexed in NSCLC. The expression of ABCC11 was higher in pemetrexed-resistant lung adenocarcinoma PC6 cells than in parental cells (P<0.0001). Moreover, after assessing 538G > A of ABCC11, one of the functional SNPs, cells with 538A > A genotypes were more sensitive to pemetrexed than their counterparts (P=0.040) (70).
FPGS and its competitor GGH are both in charge of the balance of pemetrexed accumulation in cancer cells. In NSCLC, downregulated FPGS expression was found in some pemetrexed-resistant A549 cell lines (68). Consistently, another study showed the same results in PC6 cells and, furthermore, FPGS SNP 2572C > T was also discussed in this study. It was demonstrated that after treatment with pemetrexed together with platinum, 2572C > C carriers expressed significantly higher levels of FPGS protein (P=0.0022) and obtained better responses to pemetrexed compared to patients harboring 2572T > T or 2572C > T (RR 78.6% vs. 35.6%, P=0.0034) (71). Moreover, some studies observed the GGH SNP 1042C > T in NSCLC patients receiving pemetrexed in combination with gemcitabine and demonstrated that 1042C > T carriers had a longer clinical survival compared to patients with 1042T > T genotypes, but the result was not of significance (69).
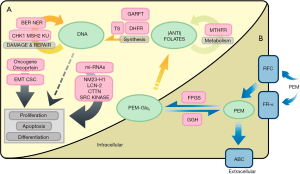
In conclusion, impaired cell entry and enhanced efflux of pemetrexed, as well as defective polyglutamylation all contribute to chemo-resistance to pemetrexed in NSCLC. A table summarizing the pemetrexed-resistance mechanisms relevant to drug dynamics and pharmacogenomics in NSCLC discussed above is included in Table 4. Moreover, a figure summarizing known mechanisms associated with pemetrexed-resistance in NSCLC that we discussed all above is available in Figure 1.
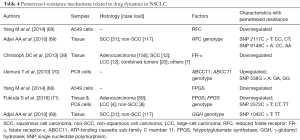
Full table
Search strategy and selection criteria
Data for this Review were identified by searches of PubMed, Web of Science, and references from relevant articles using the search terms “NSCLC”, “pemetrexed”, “resistance”, and “mechanism”. Abstracts and reports from meetings were included only when they related directly to previously published work. Only articles published in English between 2007 and 2018 were included.
Discussion
Pemetrexed has been approved for the treatment of NSCLC patients. In this article, we have focused on just two categories of mechanisms in association with pemetrexed resistance (biochemical changes in tumor cells and drug dynamics as well as pharmacogenomics). However, tissues and cell lines investigated in studies were not all treated with pemetrexed as a single agent, so the characteristics of resistance we have provided might be not only to pemetrexed alone. Besides, resistance to anticancer agents can be divided into two sections (intrinsic and acquired) (72), but hardly had any study reported explicitly whether the chemoresistance to pemetrexed were developed before or after the drug administration, remaining the nature of these mechanisms unclear. Therefore, to understand the intricate mechanisms and to individualize the adoption of pemetrexed in clinical use among NSCLC patients, there still exists the need of a full assessment of drug toxicities and resistance when patients are treated with combined regimens. Moreover, due to the wide range of practical use of pemetrexed, not only in lung cancer but also in various solid tumors such as breast cancer, gastric cancer, and head and neck cancer (73), some other novel biomarkers indicating chemoresistance to pemetrexed in NSCLC will be explored if drawing on the research carried on over the abovementioned tumors. Overall, a deeper and clearer understanding of resistance mechanisms to pemetrexed is required, which can potentially bring an exciting new facet to personalized medicine for lung cancer treatment.
Conclusions
Numerous clinical and molecule-level trials have been performed to determine the mechanisms of resistance to pemetrexed. Our review, taking all the results into consideration, showed that non-mutated status, higher expression of oncoprotein TTF-1 and pemetrexed target enzymes, enhanced DNA repair systems, activated EMT, and cancer stemness as well as impaired drug uptake, reinforced drug elimination, and defective polyglutamylation processes are correlated with development of resistance to pemetrexed in NSCLC. We have discussed some potential prospects of more precise diagnoses and individualized treatments for diverse patients. However, further detailed clinical studies on lung cancer and pemetrexed are still warranted in order to determine the precise drug efficacy of pemetrexed for NSCLC patients harboring the abovementioned or potential characteristics, which may provide accurate patient stratifications and appropriate drug administrations to be achieved by clinicians in the future.
Acknowledgments
The authors thank International Science Editing (http://www.internationalscienceediting.com) for editing this manuscript.
Funding: This work was supported by the National Natural Science Foundation of China (Grant Nos. 81672268, 81572295; www.nsfc.gov.cn).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
References
- Govindan R, Page N, Morgensztern D, et al. Changing epidemiology of small-cell lung cancer in the United States over the last 30 years: analysis of the surveillance, epidemiologic, and end results database. J Clin Oncol 2006;24:4539-44. [Crossref] [PubMed]
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin 2018;68:7-30. [Crossref] [PubMed]
- Lopci E, Rossi S. Tumor metabolism and prognostic role of EZH2 in non-small cell lung cancer. Transl Cancer Res 2017;6:S982-8. [Crossref]
- Schiller JH, Harrington D, Belani CP, et al. Comparison of four chemotherapy regimens for advanced non-small-cell lung cancer. N Engl J Med 2002;346:92-8. [Crossref] [PubMed]
- Nagasaka M, Gadgeel SM. Role of chemotherapy and targeted therapy in early-stage non-small cell lung cancer. Expert Rev Anticancer Ther 2018;18:63-70. [Crossref] [PubMed]
- Assaraf YG. Molecular basis of antifolate resistance. Cancer Metastasis Rev 2007;26:153-81. [Crossref] [PubMed]
- Chattopadhyay S, Wang Y, Zhao R, et al. Lack of impact of the loss of constitutive folate receptor alpha expression, achieved by RNA Interference, on the activity of the new generation antifolate pemetrexed in HeLa cells. Clin Cancer Res 2004;10:7986-93. [Crossref] [PubMed]
- Theti DS, Jackman AL. The role of alpha-folate receptor-mediated transport in the antitumor activity of antifolate drugs. Clin Cancer Res 2004;10:1080-9. [Crossref] [PubMed]
- Zhao R, Qiu A, Tsai E, et al. The proton-coupled folate transporter: impact on pemetrexed transport and on antifolates activities compared with the reduced folate carrier. Mol Pharmacol 2008;74:854-62. [Crossref] [PubMed]
- Shih C, Chen VJ, Gossett LS, et al. LY231514, a pyrrolo[2,3-d]pyrimidine-based antifolate that inhibits multiple folate-requiring enzymes. Cancer Res 1997;57:1116-23. [PubMed]
- Schneider E, Ryan TJ. Gamma-glutamyl hydrolase and drug resistance. Clin Chim Acta 2006;374:25-32. [Crossref] [PubMed]
- Joerger M, Omlin A, Cerny T, et al. The role of pemetrexed in advanced non small-cell lung cancer: special focus on pharmacology and mechanism of action. Curr Drug Targets 2010;11:37-47. [Crossref] [PubMed]
- Racanelli AC, Rothbart SB, Heyer CL, et al. Therapeutics by cytotoxic metabolite accumulation: pemetrexed causes ZMP accumulation, AMPK activation, and mammalian target of rapamycin inhibition. Cancer Res 2009;69:5467-74. [Crossref] [PubMed]
- Scagliotti GV, Parikh P, von Pawel J, et al. Phase III study comparing cisplatin plus gemcitabine with cisplatin plus pemetrexed in chemotherapy-naive patients with advanced-stage non-small-cell lung cancer. J Clin Oncol 2008;26:3543-51. [Crossref] [PubMed]
- Hanna N, Shepherd FA, Fossella FV, et al. Randomized phase III trial of pemetrexed versus docetaxel in patients with non-small-cell lung cancer previously treated with chemotherapy. J Clin Oncol 2004;22:1589-97. [Crossref] [PubMed]
- Sun JM, Lee KW, Kim JH, et al. Efficacy and toxicity of pemetrexed as a third-line treatment for non-small cell lung cancer. Jpn J Clin Oncol 2009;39:27-32. [Crossref] [PubMed]
- Chang MH, Ahn JS, Lee J, et al. The efficacy of pemetrexed as a third- or fourth-line therapy and the significance of thymidylate synthase expression in patients with advanced non-small cell lung cancer. Lung Cancer 2010;69:323-9. [Crossref] [PubMed]
- Gonen N, Assaraf YG. Antifolates in cancer therapy: structure, activity and mechanisms of drug resistance. Drug Resist Updat 2012;15:183-210. [Crossref] [PubMed]
- Scagliotti GV, Selvaggi G, Novello S, et al. The biology of epidermal growth factor receptor in lung cancer. Clin Cancer Res 2004;10:4227s-32s. [Crossref] [PubMed]
- Shigematsu H, Lin L, Takahashi T, et al. Clinical and biological features associated with epidermal growth factor receptor gene mutations in lung cancers. J Natl Cancer Inst 2005;97:339-46. [Crossref] [PubMed]
- Wu SG, Yang CH, Yu CJ, et al. Good response to pemetrexed in patients of lung adenocarcinoma with epidermal growth factor receptor (EGFR) mutations. Lung Cancer 2011;72:333-9. [Crossref] [PubMed]
- Igawa S, Sato Y, Ishihara M, et al. EGFR Mutation Genotype Impact on the Efficacy of Pemetrexed in Patients with Nonsquamous Nonsmall Cell Lung Cancer. Asian Pac J Cancer Prev 2016;17:3249-53. [PubMed]
- Yu Z, Li XM, Liu SH, et al. Downregulation of both EGFR and ErbB3 improves the cellular response to pemetrexed in an established pemetrexedresistant lung adenocarcinoma A549 cell line. Oncol Rep 2014;31:1818-24. [Crossref] [PubMed]
- Shaw AT, Yeap BY, Mino-Kenudson M, et al. Clinical features and outcome of patients with non-small-cell lung cancer who harbor EML4-ALK. J Clin Oncol 2009;27:4247-53. [Crossref] [PubMed]
- Bergethon K, Shaw AT, Ou SH, et al. ROS1 rearrangements define a unique molecular class of lung cancers. J Clin Oncol 2012;30:863-70. [Crossref] [PubMed]
- Lee HY, Ahn HK, Jeong JY, et al. Favorable clinical outcomes of pemetrexed treatment in anaplastic lymphoma kinase positive non-small-cell lung cancer. Lung Cancer 2013;79:40-5. [Crossref] [PubMed]
- Park S, Park TS, Choi CM, et al. Survival Benefit of Pemetrexed in Lung Adenocarcinoma Patients With Anaplastic Lymphoma Kinase Gene Rearrangements. Clin Lung Cancer 2015;16:e83-9. [Crossref] [PubMed]
- Lee JO, Kim TM, Lee SH, et al. Anaplastic lymphoma kinase translocation: a predictive biomarker of pemetrexed in patients with non-small cell lung cancer. J Thorac Oncol 2011;6:1474-80. [Crossref] [PubMed]
- Chen YF, Hsieh MS, Wu SG, et al. Efficacy of Pemetrexed-Based Chemotherapy in Patients with ROS1 Fusion-Positive Lung Adenocarcinoma Compared with in Patients Harboring Other Driver Mutations in East Asian Populations. J Thorac Oncol 2016;11:1140-52. [Crossref] [PubMed]
- Dearden S, Stevens J, Wu YL, et al. Mutation incidence and coincidence in non small-cell lung cancer: meta-analyses by ethnicity and histology (mutMap). Ann Oncol 2013;24:2371-6. [Crossref] [PubMed]
- Liang Y, Wakelee HA, Neal JW. Relationship of Driver Oncogenes to Long-Term Pemetrexed Response in Non--Small-Cell Lung Cancer. Clin Lung Cancer 2015;16:366-73. [Crossref] [PubMed]
- Wu MF, Hsiao YM, Huang CF, et al. Genetic determinants of pemetrexed responsiveness and nonresponsiveness in non-small cell lung cancer cells. J Thorac Oncol 2010;5:1143-51. [Crossref] [PubMed]
- Wen Y, Gamazon ER, Bleibel WK, et al. An eQTL-based method identifies CTTN and ZMAT3 as pemetrexed susceptibility markers. Hum Mol Genet 2012;21:1470-80. [Crossref] [PubMed]
- Rekhtman N, Ang DC, Sima CS, et al. Immunohistochemical algorithm for differentiation of lung adenocarcinoma and squamous cell carcinoma based on large series of whole-tissue sections with validation in small specimens. Mod Pathol 2011;24:1348-59. [Crossref] [PubMed]
- Sun JM, Han J, Ahn JS, et al. Significance of thymidylate synthase and thyroid transcription factor 1 expression in patients with nonsquamous non-small cell lung cancer treated with pemetrexed-based chemotherapy. J Thorac Oncol 2011;6:1392-9. [Crossref] [PubMed]
- Grønberg BH, Lund-Iversen M, Strøm EH, et al. Associations between TS, TTF-1, FR-alpha, FPGS, and overall survival in patients with advanced non-small-cell lung cancer receiving pemetrexed plus carboplatin or gemcitabine plus carboplatin as first-line chemotherapy. J Thorac Oncol 2013;8:1255-64. [Crossref] [PubMed]
- Hanauske AR, Eismann U, Oberschmidt O, et al. In vitro chemosensitivity of freshly explanted tumor cells to pemetrexed is correlated with target gene expression. Invest New Drugs 2007;25:417-23. [Crossref] [PubMed]
- Bepler G, Sommers KE, Cantor A, et al. Clinical efficacy and predictive molecular markers of neoadjuvant gemcitabine and pemetrexed in resectable non-small cell lung cancer. J Thorac Oncol 2008;3:1112-8. [Crossref] [PubMed]
- Christoph DC, Asuncion BR, Hassan B, et al. Significance of folate receptor alpha and thymidylate synthase protein expression in patients with non-small-cell lung cancer treated with pemetrexed. J Thorac Oncol 2013;8:19-30. [Crossref] [PubMed]
- Zhang D, Ochi N, Takigawa N, et al. Establishment of pemetrexed-resistant non-small cell lung cancer cell lines. Cancer Lett 2011;309:228-35. [Crossref] [PubMed]
- Ozasa H, Oguri T, Uemura T, et al. Significance of thymidylate synthase for resistance to pemetrexed in lung cancer. Cancer Sci 2010;101:161-6. [Crossref] [PubMed]
- Shimizu T, Nakagawa Y, Takahashi N, et al. Thymidylate synthase gene amplification predicts pemetrexed resistance in patients with advanced non-small cell lung cancer. Clin Transl Oncol 2016;18:107-12. [Crossref] [PubMed]
- Lurje G, Manegold PC, Ning Y, et al. Thymidylate synthase gene variations: predictive and prognostic markers. Mol Cancer Ther 2009;8:1000-7. [Crossref] [PubMed]
- Arévalo E, Castanon E, Lopez I, et al. Thymidylate synthase polymorphisms in genomic DNA as clinical outcome predictors in a European population of advanced non-small cell lung cancer patients receiving pemetrexed. J Transl Med 2014;12:98. [Crossref] [PubMed]
- Krawczyk P, Kucharczyk T, Kowalski DM, et al. Polymorphisms in TS, MTHFR and ERCC1 genes as predictive markers in first-line platinum and pemetrexed therapy in NSCLC patients. J Cancer Res Clin Oncol 2014;140:2047-57. [Crossref] [PubMed]
- Wang X, Wang Y, Wang Y, et al. Association of thymidylate synthase gene 3'-untranslated region polymorphism with sensitivity of non-small cell lung cancer to pemetrexed treatment: TS gene polymorphism and pemetrexed sensitivity in NSCLC. J Biomed Sci 2013;20:5. [Crossref] [PubMed]
- Besse B, Olaussen KA, Soria JC. ERCC1 and RRM1: ready for prime time? J Clin Oncol 2013;31:1050-60. [Crossref] [PubMed]
- Crider KS, Yang TP, Berry RJ, et al. Folate and DNA methylation: a review of molecular mechanisms and the evidence for folate's role. Adv Nutr 2012;3:21-38. [Crossref] [PubMed]
- Smit EF, Burgers SA, Biesma B, et al. Randomized phase II and pharmacogenetic study of pemetrexed compared with pemetrexed plus carboplatin in pretreated patients with advanced non-small-cell lung cancer. J Clin Oncol 2009;27:2038-45. [Crossref] [PubMed]
- Webley SD, Welsh SJ, Jackman AL, et al. The ability to accumulate deoxyuridine triphosphate and cellular response to thymidylate synthase (TS) inhibition. Br J Cancer 2001;85:446-52. [Crossref] [PubMed]
- Van Triest B, Pinedo HM, Giaccone G, et al. Downstream molecular determinants of response to 5-fluorouracil and antifolate thymidylate synthase inhibitors. Ann Oncol 2000;11:385-91. [Crossref] [PubMed]
- Wilson PM, LaBonte MJ, Lenz HJ, et al. Inhibition of dUTPase induces synthetic lethality with thymidylate synthase-targeted therapies in non-small cell lung cancer. Mol Cancer Ther 2012;11:616-28. [Crossref] [PubMed]
- Weeks LD, Fu P, Gerson SL. Uracil-DNA glycosylase expression determines human lung cancer cell sensitivity to pemetrexed. Mol Cancer Ther 2013;12:2248-60. [Crossref] [PubMed]
- Lee SH, Noh KB, Lee JS, et al. Thymidylate synthase and ERCC1 as predictive markers in patients with pulmonary adenocarcinoma treated with pemetrexed and cisplatin. Lung Cancer 2013;81:102-8. [Crossref] [PubMed]
- Liao WY, Ho CC, Tsai TH, et al. Combined effect of ERCC1 and ERCC2 polymorphisms on overall survival in non-squamous non-small-cell lung cancer patients treated with first-line pemetrexed/platinum. Lung Cancer 2018;118:90-6. [Crossref] [PubMed]
- Grabauskiene S, Bergeron EJ, Chen G, et al. CHK1 levels correlate with sensitization to pemetrexed by CHK1 inhibitors in non-small cell lung cancer cells. Lung Cancer 2013;82:477-84. [Crossref] [PubMed]
- Tung CL, Chiu HC, Jian YJ, et al. Down-regulation of MSH2 expression by an Hsp90 inhibitor enhances pemetrexed-induced cytotoxicity in human non-small-cell lung cancer cells. Exp Cell Res 2014;322:345-54. [Crossref] [PubMed]
- Shang B, Jia Y, Chen G, et al. Ku80 correlates with neoadjuvant chemotherapy resistance in human lung adenocarcinoma, but reduces cisplatin/pemetrexed-induced apoptosis in A549 cells. Respir Res 2017;18:56. [Crossref] [PubMed]
- Thiery JP, Acloque H, Huang RY, et al. Epithelial-mesenchymal transitions in development and disease. Cell 2009;139:871-90. [Crossref] [PubMed]
- Liang SQ, Marti TM, Dorn P, et al. Blocking the epithelial-to-mesenchymal transition pathway abrogates resistance to anti-folate chemotherapy in lung cancer. Cell Death Dis 2015;6:e1824. [Crossref] [PubMed]
- Chiu LY, Hsin IL, Yang TY, et al. The ERK-ZEB1 pathway mediates epithelial-mesenchymal transition in pemetrexed resistant lung cancer cells with suppression by vinca alkaloids. Oncogene 2017;36:242-53. [Crossref] [PubMed]
- Gangemi R, Paleari L, Orengo AM, et al. Cancer stem cells: a new paradigm for understanding tumor growth and progression and drug resistance. Curr Med Chem 2009;16:1688-703. [Crossref] [PubMed]
- Zhao M, Zhang Y, Zhang H, et al. Hypoxia-induced cell stemness leads to drug resistance and poor prognosis in lung adenocarcinoma. Lung Cancer 2015;87:98-106. [Crossref] [PubMed]
- Ceppi P, Rapa I, Lo Iacono M, et al. Expression and pharmacological inhibition of thymidylate synthase and Src kinase in nonsmall cell lung cancer. Int J Cancer 2012;130:1777-86. [Crossref] [PubMed]
- Chou HC, Chen JY, Lin DY, et al. Identification of Up- and Down-Regulated Proteins in Pemetrexed-Resistant Human Lung Adenocarcinoma: Flavin Reductase and Calreticulin Play Key Roles in the Development of Pemetrexed-Associated Resistance. J Proteome Res 2015;14:4907-20. [Crossref] [PubMed]
- Shi SB, Wang M, Tian J, et al. MicroRNA 25, microRNA 145, and microRNA 210 as biomarkers for predicting the efficacy of maintenance treatment with pemetrexed in lung adenocarcinoma patients who are negative for epidermal growth factor receptor mutations or anaplastic lymphoma kinase translocations. Transl Res 2016;170:1-7. [Crossref] [PubMed]
- Buti S, Bordi P, Tiseo M, et al. Predictive role of erythrocyte macrocytosis during treatment with pemetrexed in advanced non-small cell lung cancer patients. Lung Cancer 2015;88:319-24. [Crossref] [PubMed]
- Yang M, Fan WF, Pu XL, et al. Significance of thymidylate synthase expression for resistance to pemetrexed in pulmonary adenocarcinoma. Oncol Lett 2014;7:227-32. [Crossref] [PubMed]
- Adjei AA, Salavaggione OE, Mandrekar SJ, et al. Correlation between polymorphisms of the reduced folate carrier gene (SLC19A1) and survival after pemetrexed-based therapy in non-small cell lung cancer: a North Central Cancer Treatment Group-based exploratory study. J Thorac Oncol 2010;5:1346-53. [Crossref] [PubMed]
- Uemura T, Oguri T, Ozasa H, et al. ABCC11/MRP8 confers pemetrexed resistance in lung cancer. Cancer Sci 2010;101:2404-10. [Crossref] [PubMed]
- Fukuda S, Oguri T, Kunii E, et al. A folylpoly-gamma-glutamate synthase single nucleotide polymorphism associated with response to pemetrexed treatment combined with platinum for non-small cell lung cancer. Lung Cancer 2016;102:15-20. [Crossref] [PubMed]
- Kim ES. Chemotherapy Resistance in Lung Cancer. Adv Exp Med Biol 2016;893:189-209. [Crossref] [PubMed]
- Jarmuła A. Antifolate inhibitors of thymidylate synthase as anticancer drugs. Mini Rev Med Chem 2010;10:1211-22. [Crossref] [PubMed]


