
Molecular studies of lung neuroendocrine neoplasms uncover new concepts and entities
Lung neuroendocrine neoplasms (NENs) account for up to 25% of all lung cancers and can be subdivided into poorly differentiated, high-grade neuroendocrine carcinomas (NECs) including small cell lung carcinoma (SCLC, 20%) and large-cell neuroendocrine carcinomas (LCNEC, 3%), and well-differentiated neuroendocrine tumours (NETs, 2%) including the low- and intermediate-grade typical and atypical carcinoids, respectively (1). Each of these lung NEN subtypes shows different behaviour in terms of clinical presentation, prognosis and aetiology: whereas SCLC and LCNEC are smoking-related very aggressive, highly metastatic tumours with poor prognosis, typical and atypical carcinoids have thus far unclear association with smoking and are less aggressive, with longer survival expectancy (2). The distinction between lung NETs and NECs is crucial for the clinical management of these diseases; surgical resection is the treatment of choice for localized tumours—mostly NETs—while systemic treatment and, more recently, immunotherapy are preferred for disseminated NECs. However, cytotoxic agents are mostly ineffective in these tumours (3).
Molecular landscape of lung NENs
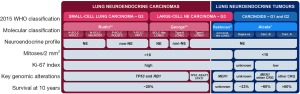
NECs and NETs have very distinct molecular characteristics and it is widely accepted that they are different diseases and not simply a continuum of neoplasms with a common pathogenesis (4,5): both typical and atypical pulmonary carcinoids show low mutation rate and frequent alterations in chromatin-remodelling genes (6,7) with very rare mutations in TP53 and RB1 genes. These latter two genes are, conversely, universally inactivated in SCLC (8,9). The molecular spectrum of LCNEC is more heterogeneous, with two well-distinguished large groups of samples: type-I LCNEC, with frequent alterations in TP53, STK11 and KEAP1, and type-II LCNEC, with alterations in both TP53 and RB1 (10,11) (Figure 1). While the nomenclature given by Rekhtman and colleagues (NSCLC-like and SCLC-like LCNEC for type-I and type-II, respectively) is accurate in terms of mutational patterns, it does not reflect the expression patterns: type-I LCNECs have the mutational characteristics of NSCLCs, but expression profiles similar to SCLC, while type-II LCNECs have mutational characteristics of SCLC but low neuroendocrine expression profiles similar to NSCLCs (10). Recent studies on molecular subtypes of SCLC point to distinct expression profiles marked by the differential expression of key transcription factors (13) (Figure 1). With regard to the strong similarities of LCNEC and SCLC, and the difficulties in distinguishing these cancer types in cell culture and by histology, it remains to be understood how many of these SCLC subtypes may rather reflect similarities to LCNEC.
Evidence for grade-3 carcinoids in the lung
The distinction between NECs and NETs is currently based on morphologic characteristics, necrosis, and mitotic count (>10 per 2 mm2 for NECs) (4). Ki-67 proliferation index is only recommended to separate lung NECs and NETs in non-resection specimens (14,15), with a threshold of 20% as the upper limit for NETs (4). Lung carcinoids correspond to the second most frequent type of NETs in the body (accounting for 30%), right after enteropancreatic NETs (50%) (16). In contrast to the lung, for the enteropancreatic system, a grade-3 category of NETs has recently been considered when the tumours present a Ki-67 index higher than 20%, or more than 20 mitoses/2 mm2 along with well-differentiated morphology (17). A recent study by Rekhtman and colleagues on a series of 132 stage IV lung carcinoids has shown that, in the metastatic setting, lung carcinoids frequently exceed the mitotic count and Ki-67 index ceiling criteria for grade-2 NETs, suggesting that these tumours might be the counter partners of grade-3 enteropancreatic NETs (18). The use of the Ki-67 index to stratify metastatic lung NETs in clinically relevant groups according to different treatments (somatostatin analogs, m-TOR inhibitors, peptide receptor radionuclide therapy, chemotherapy different from platin) has been previously proposed by Pelosi and collaborators (19-21). Rindi and collaborators have also proposed a grading system for lung NENs—from grade 1 to grade 3—combining mitotic count, necrosis and Ki-67 index applying lung-specific cut-off values (22).
The hypothesis that grade-3 lung NETs that have a relatively high Ki-67 index compared to other lung NETs might exist is also supported by molecular data. Interestingly, Rekhtman and colleagues identified a small subgroup of LCNECs (classified as such based on the high mitotic count) that presented carcinoid-like morphology and MEN1 mutations, characteristic of lung carcinoids (11); they called them carcinoid-like LCNEC (Figure 1). In another study, led by Simbolo and colleagues (12), using a 58-gene expression signature assessed in 35 atypical carcinoids and 32 LCNEC, the authors identified three clusters: C1 enriched for LCNEC, C2, which contained both LCNEC (C2a) and atypical carcinoids (C2b), and C3 enriched for atypical carcinoids. Based on their molecular characteristics, the C1 and C2a LCNEC-enriched clusters are likely to correspond to the previously described type-II and type-I LCNECs, respectively (10,11). Interestingly, atypical-carcinoid enriched cluster C3, in which MEN1 inactivation seem to play a major role, included four LCNEC samples, three of which also harboured MEN1 truncating mutations.
In a recent study, Alcala and colleagues performed integrative multi-omic analyses for a total of 257 lung NENs. Unsupervised Multi-Omics Factor Analysis (MOFA) (23) of expression and methylation data of lung carcinoids and LCNEC samples identified three robust clusters with different molecular and clinical characteristics: cluster Carcinoid A (enriched for typical carcinoids, 10-year overall survival above 80%), cluster Carcinoid B (enriched for atypical carcinoids, 10-year overall survival of 60%) and cluster LCNEC (10-year overall survival of 21%) (Figure 1). Interestingly, one of the LCNEC samples fell into one of the carcinoid clusters, suggesting that this LCNEC sample shared the molecular characteristics of lung carcinoids (7).
Although these LCNEC cases with molecular characteristics of lung carcinoids remain anecdotal (n=7 in total), they might indeed correspond to the grade-3 carcinoids of the lung. Indeed, Ki-67 index assessment of the two carcinoid-like LCNECs identified by Rekhtman and colleagues showed that, although the Ki-67 index was relatively high for carcinoids (around 35%), it was relatively low when compared to that of type-I and type-II LCNECs (60% and 90%, respectively) (11).
Supra-carcinoids: a new molecular entity of lung NENs
One unexpected finding in the MOFA performed by Alcala and colleagues was the identification of six morphologically classified atypical carcinoids that fell into the molecular cluster of LCNEC. In addition to sharing the molecular features of LCNEC, they also present a poor survival (10-year overall survival of 33%). These samples were named supra-carcinoids (Figure 1), and they represented 17% of the atypical carcinoids in that series. The supra-carcinoids also showed distinct features, such as high levels of immune checkpoint receptors and ligands, and high neutrophil content (7), which might represent potential diagnostic and therapeutic candidates for this group of aggressive lung NETs.
Supra-carcinoids might also have been identified in the study from Simbolo and colleagues (12). Indeed, four of their atypical carcinoids fell into clusters C2a and C1, which were both enriched for LCNECs. This represents 13% of atypical carcinoids in their series, which is consistent with the 17% that Alcala and colleagues identified (7).
The discovery of supra-carcinoids suggests that the molecular link between lung NETs and NECs might be subtler than initially thought, especially between atypical carcinoids and LCNEC. In fact, over the past years, Pelosi and colleagues have been advancing the innovative idea in both the lung and the thymus that the progression or transition from lung NET to NEC, possibly through the accumulation of genetic anomalies, might be possible (14,24,25). To formally prove this hypothesis, in vitro and in vivo experimental models are needed. Dr. Talya Dayton in the lab of Hans Clevers is developing an organoid-based model of lung NETs that, if successful, could provide the perfect setting to test this hypothesis (personal communication).
Open science to move forward with the study of these rare diseases: tumour maps
The overall limitation to performing molecular studies on lung carcinoids and LCNEC is the limited number of samples of suitable quality to perform comprehensive genomic analyses of these rare diseases. The generally low number of samples included in each study makes it difficult to produce data from which to draw meaningful and reproducible conclusions. Thankfully, the overall field of research is moving towards open data, even if restrictions are still often present (26) and barriers and reluctance still exist (27,28). This allows researchers with few samples to answer a specific research question by performing comparative analyses with data that have already been published. For example, this approach led Alcala and colleagues to the discovery of the supra-carcinoids (7), through the integration of molecular data generated for 257 lung NENs in four different studies (6,8-10).
One additional step in the direction of more useful open science is the generation of so-called “tumour maps”, which provide an interactive way to explore the molecular data and allow easy statistical interrogation, including generating new hypotheses, but also locating samples from smaller studies into the molecular landscape (29). The placement of the data from a specific patient’s sample in a region of the map with other samples that have similar molecular characteristics might also assist clinicians in their diagnosis, prognosis and clinical management of that patient. As an example, we have recently added our comprehensive lung NEN molecular map to the UCSC Tumor Map portal (https://tumormap.ucsc.edu/?p=RCG_lungNENomics/LNEN, Gabriel et al. in preparation). Simbolo and colleagues (12) seem to have identified equivalent molecular groups of lung NENs as in previous studies (7,10). However, the variability introduced by (1) the fact that their samples were formalin-fixed paraffin-embedded tissue while the ones used to generate the lung NEN molecular map were snap frozen, and (2), the studies were based on different sequencing techniques, hampers the integration of both datasets. Indeed, data standardization remains an issue in this kind of approach (30).
Conclusions
Considerable progress has been made in the molecular characterization of lung NENs. In the context of LCNEC and carcinoids, these studies have led to the suggestion that grade-3 enteropancreatic NETs might have their equivalent entity in the lung, and perhaps also in the thymus. Multi-omic integrative analyses have identified a new molecular entity, the supra-carcinoids, whose full characterization warrants further investigation, not only including comprehensive molecular studies but also a central pathology review with detailed analysis of proliferation metrics. Finally, the above-mention observations further support the suggested molecular link between lung NECs and NETs. Further studies are nevertheless needed to assess the clinical implications of all of these discoveries.
Acknowledgments
We thank Ian Cree, Talya Dayton, Julie George, Giuseppe Pelosi, Natasha Rekhtman, and Aldo Scarpa for reading and commenting the Editorial. We acknowledge Morena Sarzo for designing the infographic. This work is part of the LungNENomics project and Rare Cancers Genomics initiative (www.rarecancersgenomics.com).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
References
- Rindi G, Klimstra DS, Abedi-Ardekani B, et al. A common classification framework for neuroendocrine neoplasms: an International Agency for Research on Cancer (IARC) and World Health Organization (WHO) expert consensus proposal. Mod Pathol 2018;31:1770-86. [Crossref] [PubMed]
- Travis WD. Advances in neuroendocrine lung tumors. Ann Oncol 2010;21 Suppl 7:vii65-71. [Crossref] [PubMed]
- Wolin EM. Advances in the Diagnosis and Management of Well-Differentiated and Intermediate-Differentiated Neuroendocrine Tumors of the Lung. Chest 2017;151:1141-6. [Crossref] [PubMed]
- Travis WD, Brambilla E, Nicholson AG, et al. The 2015 World Health Organization Classification of Lung Tumors: Impact of Genetic, Clinical and Radiologic Advances Since the 2004 Classification. J Thorac Oncol 2015;10:1243-60. [Crossref] [PubMed]
- Swarts DR, Ramaekers FC, Speel EJ. Molecular and cellular biology of neuroendocrine lung tumors: evidence for separate biological entities. Biochim Biophys Acta 2012;1826:255-71. [PubMed]
- Fernandez-Cuesta L, Peifer M, Lu X, et al. Frequent mutations in chromatin-remodelling genes in pulmonary carcinoids. Nat Commun 2014;5:3518. [Crossref] [PubMed]
- Alcala N, Leblay N, Gabriel AAG, et al. Integrative and comparative genomic analyses identify clinically relevant pulmonary carcinoid groups and unveil the supra-carcinoids. Nat Commun 2019;10:3407. [Crossref] [PubMed]
- Peifer M, Fernandez-Cuesta L, Sos ML, et al. Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat Genet 2012;44:1104-10. [Crossref] [PubMed]
- George J, Lim JS, Jang SJ, et al. Comprehensive genomic profiles of small cell lung cancer. Nature 2015;524:47-53. [Crossref] [PubMed]
- George J, Walter V, Peifer M, et al. Integrative genomic profiling of large-cell neuroendocrine carcinomas reveals distinct subtypes of high-grade neuroendocrine lung tumors. Nat Commun 2018;9:1048. [Crossref] [PubMed]
- Rekhtman N, Pietanza MC, Hellmann MD, et al. Next-Generation Sequencing of Pulmonary Large Cell Neuroendocrine Carcinoma Reveals Small Cell Carcinoma-like and Non-Small Cell Carcinoma-like Subsets. Clin Cancer Res 2016;22:3618-29. [Crossref] [PubMed]
- Simbolo M, Barbi S, Fassan M, et al. Gene Expression Profiling of Lung Atypical Carcinoids and Large Cell Neuroendocrine Carcinomas Identifies Three Transcriptomic Subtypes with Specific Genomic Alterations. J Thorac Oncol 2019;14:1651-61. [Crossref] [PubMed]
- Rudin CM, Poirier JT, Byers LA, et al. Molecular subtypes of small cell lung cancer: a synthesis of human and mouse model data. Nat Rev Cancer 2019;19:289-97. [Crossref] [PubMed]
- Fabbri A, Cossa M, Sonzogni A, et al. Thymus neuroendocrine tumors with CTNNB1 gene mutations, disarrayed ss-catenin expression, and dual intra-tumor Ki-67 labeling index compartmentalization challenge the concept of secondary high-grade neuroendocrine tumor: a paradigm shift. Virchows Arch 2017;471:31-47. [Crossref] [PubMed]
- Pelosi G, Rodriguez J, Viale G, et al. Typical and atypical pulmonary carcinoid tumor overdiagnosed as small-cell carcinoma on biopsy specimens: a major pitfall in the management of lung cancer patients. Am J Surg Pathol 2005;29:179-87. [Crossref] [PubMed]
- Cancer.net. Neuroendocrine Tumor of the Lung. 2018. Available online: https://www.cancer.net/cancer-types/neuroendocrine-tumor-lung. Accessed Consulted on 24 October 2019.
- Nagtegaal ID, Odze RD, Klimstra D, et al. The 2019 WHO classification of tumours of the digestive system. Histopathology 2020;76:182-8. [Crossref] [PubMed]
- Rekhtman N, Desmeules P, Litvak AM, et al. Stage IV lung carcinoids: spectrum and evolution of proliferation rate, focusing on variants with elevated proliferation indices. Mod Pathol 2019;32:1106-22. [Crossref] [PubMed]
- Pelosi G, Pattini L, Morana G, et al. Grading lung neuroendocrine tumors: Controversies in search of a solution. Histol Histopathol 2017;32:223-41. [PubMed]
- Pelosi G, Bianchi F, Hofman P, et al. Recent advances in the molecular landscape of lung neuroendocrine tumors. Expert Rev Mol Diagn 2019;19:281-97. [Crossref] [PubMed]
- Pelosi G, Massa F, Gatti G, et al. Ki-67 Evaluation for Clinical Decision in Metastatic Lung Carcinoids: A Proof of Concept. Clin Pathol 2019;12:2632010x19829259.
- Rindi G, Klersy C, Inzani F, et al. Grading the neuroendocrine tumors of the lung: an evidence-based proposal. Endocr Relat Cancer 2013;21:1-16. [Crossref] [PubMed]
- Argelaguet R, Velten B, Arnol D, et al. Multi-Omics Factor Analysis-a framework for unsupervised integration of multi-omics data sets. Mol Syst Biol 2018;14:e8124. [Crossref] [PubMed]
- Pelosi G, Bianchi F, Dama E, et al. Most high-grade neuroendocrine tumours of the lung are likely to secondarily develop from pre-existing carcinoids: innovative findings skipping the current pathogenesis paradigm. Virchows Arch 2018;472:567-77. [Crossref] [PubMed]
- Dinter H, Bohnenberger H, Beck J, et al. Molecular Classification of Neuroendocrine Tumors of the Thymus. J Thorac Oncol 2019;14:1472-83. [Crossref] [PubMed]
- Amann RI, Baichoo S, Blencowe BJ, et al. Toward unrestricted use of public genomic data. Science 2019;363:350-2. [Crossref] [PubMed]
- van Panhuis WG, Paul P, Emerson C, et al. A systematic review of barriers to data sharing in public health. BMC Public Health 2014;14:1144. [Crossref] [PubMed]
- Longo DL, Drazen JM. Data Sharing. N Engl J Med 2016;374:276-7. [Crossref] [PubMed]
- Newton Y, Novak AM, Swatloski T, et al. TumorMap: Exploring the Molecular Similarities of Cancer Samples in an Interactive Portal. Cancer Res 2017;77:e111-4. [Crossref] [PubMed]
- Cieślik M, Chinnaiyan AM. Cancer transcriptome profiling at the juncture of clinical translation. Nat Rev Genet 2018;19:93-109. [Crossref] [PubMed]


