
Prognostic model based on circular RNA circPDK1 for resected lung squamous cell carcinoma
Introduction
Lung squamous cell carcinoma (LSCC) is the second most common pulmonary malignancy worldwide, accounting for 20–30% of non-small cell lung cancer (NSCLC) cases (1,2). Although the poor prognosis of LSCC has raised the concern of researchers for decades, it was still unable to be predicted accurately by now (3).
Serum tumor markers have long been investigated as prognostic factors because they are simple parameters to measure. The used lung cancer markers for diagnosis and prognosis include carcinoembryonic antigen (CEA), Cytokeratin 19 fragment (CK19), Carbohydrate antigen 125 (CA125), Serum gastrin-releasing peptide precursor (ProGRP), SCCAg and Neuron-specific enolase (NSE) (4-9). However, the sensitivity and specificity of theses markers were always too low to predict lung malignancies. Besides, the serum level of these markers fluctuates in different duration of disease. Therefore, more stable, and accurate biomarkers were needed for LSCC.
Circular RNAs are a group of circular-stated transcripts associated with the progression of multiple human malignancies and diseases (10). Compared with linear mRNAs, these circular products are highly stable in vivo and more suitable as biomarkers (11). So far, circular RNA has been revealed as a potential biomarker in multiple malignancies, including gastric, colorectal, pancreatic and breast cancer (12-16). However, few studies have revealed the relationship between LSCC and circular RNAs.
In this work, we identified a circular RNA, circPDK1, which is overexpressed in LSCC and associated with good prognosis of resected LSCC patients. We aimed to build a prognostic model including circPDK1 and other clinicopathological factors, thereby revealing its potential as a prognostic biomarker for LSCC.
Methods
Next-generation sequencing
Ten cases of LSCC tumor tissues and their adjacent normal tissues were collected intraoperatively for transcriptome-wide associated sequencing. The sequencing was performed by BioMarker Technologies (Beijing, China).
Study population and follow-up
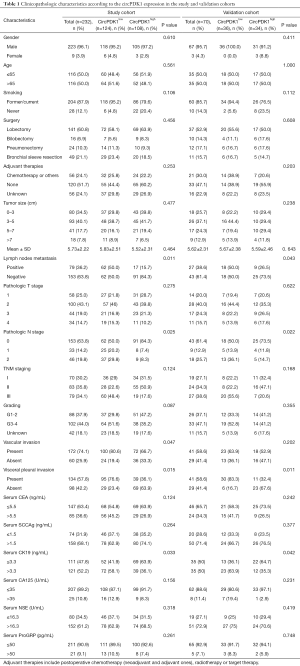
Three hundred two cases of LSCC samples and their adjacent normal tissues were collected from four Chinese medical centers during October 2013 and December 2014. All the individuals were primary lung squamous cell cancer patients underwent R0 resection and systematic lymph node dissection. The patients who had received preoperative radiotherapy, chemotherapy, or other adjuvant therapies, or those with recurrence and distant metastasis were excluded. Normal lung tissues were collected from a distance more than 5 cm from the tumor margin and used as negative controls. All collected tissue samples were immediately stored at −80 °C until use. The clinicopathological information of the patients was retrospectively collected from the electronic medical records. The serum levels of tumor markers (CEA, SCCAg, NSE, CK19, CA125, and ProGRP) were routinely measured preoperatively. The cutoff value of these tumor markers were set according to other literatures: CEA, 5.5 ng/mL; SCCAg, 1.5 ng/mL; NSE, 16.3 U/mL, CK19, 3.3 ng/mL; CA125, 35 U/mL; ProGRP, 50 pg/mL (8,17,18). These cutoff values were also following the values showed in Survival status was collected via outpatient clinic revisit records or telephone call. The mean follow-up period of the patients was 62.2 months (range, 56–70 months). The patient loss to follow-up was excluded. For all patients, the postsurgical pathological staging was determined according to the eighth edition TNM classification. Individuals were divided into study cohort (n=232) and validation cohort (n=70) randomly. The baseline information and clinicopathologic characteristics of the two cohorts were seen in Table 1. Informed consents have been obtained from all the patients or their families.
Full table
RNA extraction and quantitative real-time polymerase chain reaction (QRT-PCR)
Total RNA was extracted from tissues with TRIZOL reagent (Solarbio, China). The SuperScript® III First-Strand Synthesis System for RT-PCR (Thermo Fisher Scientific, USA) was used to reverse-transcribe RNA into cDNA. QRT-PCR was performed on an Applied Biosystems 7500 Real-Time PCR system (Applied Biosystems, USA) with SYBR® Green PCR Master Mix (Thermo Fisher Scientific, USA). The primers were as follows: circPDK1 forward: 5'-TCGAAAACACATTGGAAGCA and reverse: 3'-TGCCGCAGAAACATAAATGA. Relative RNA levels were calculated with 2–ΔCT [where ΔCt = Ct(gene) – Ct(β-actin)]. mRNA PDK1 forward: 5'-CAGCAGTCCTTGGCATTTGT and reverse: 3'-ATACGTCTCAGGCTCTGCTC. Additionally, the fold change of RNA expression in tumor tissues vs. adjacent normal tissues was calculated with the 2–ΔΔCT method. GAPDH was used as an internal control.
Prognostic analysis
Overall survival (OS) was defined as the period from after operation to patient death or the last follow-up. Relapse-free survival (RFS) was defined as the period from surgery to tumor recurrence or patient death from a non-cancerous cause. The univariate and multivariate survival analyses were performed by Cox proportional hazards regression models to analyze the potential prognostic factors. The 5-year survival was analyzed with Kaplan-Meier plotter (K-M plot) survival curve. The prognostic model was built based on the results of multivariate analysis. Nomograms (19) was built to calculate the 5-year RFS and OS. C-index and calibration curves were used to achieve internal validation. External validation was performed in the validation cohort.
Statistical analysis
All data were analyzed with the statistical package SPSS 22.0 (SPSS Inc., Chicago, IL, USA) and R language (version 3.5.1, the “survival” and “rms” packages). Data are presented as mean ± SD. Graphs were made with GraphPad Prism 6 (GraphPad Software, CA, USA). Student’s t-test, chi-square test, multiple logistic regression, and Pearson correlation analysis were used. P values <0.05 were considered statistically significant.
Results
CircPDK1 was identified as a prognostic biomarker of LSCC
Through next-generation sequencing, we identified a unique circular RNA, circPDK1, which was highly expressed in LSCC tissues but was scarcely detected in normal tissues (Figure 1A,B).
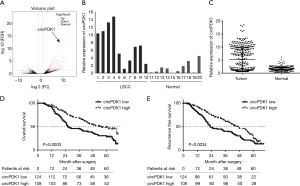
Next, we detected the relative expression of circPDK1 in each LSCC specimen. The results of qRT-PCR proved that circPDK1 was significantly higher in LSCC tissues than that in normal tissues (5.05 folds change, P<0.01) (Figure 1C). However, the expression of circPDK1 fluctuated among different patients.
To explore the correlation between circPDK1 and 5-year RFS/OS of LSCC patients, we divided the cases into circPDK1high and circPDK1low groups according to its mean expression (2–ΔΔCT =3.7), then, K-M plot analysis was performed in the study cohort. The OS of circPDK1low and circPDK1high was 29.0% and 48.2%, respectively (P=0.0003, median survival time (MST) was 26 and 55 months) (Figure 1D). The RFS of circPDK1low and circPDK1high was 17.7% and 25.9%, respectively [P=0.0034, median survival time (MST) was 23 and 45 months] (Figure 1E). To our data, elevated circPDK1 indicated better RFS and OS of resected LSCC.
Correlations between circPDK1 expression and clinicopathological characteristics
The study cohort included 223 males and 9 females with a median age of 65 years old (range, 47–82). Most patients had a smoking history (204 patients; 87.9%). According to the 8th TNM staging, 70 patients (30.2%) were in stage I, 83 ones (35.8%) were in stage II, and 79 ones (34.1%) were in stage III. The number of patients who underwent lobectomy, bilobectomy, pneumonectomy, and bronchial sleeve resection was 141 (60.8%), 16 (6.9%), 24 (10.3%) and 49 (21.1%), respectively. Fifty-six patients (24.1) received adjuvant therapies postoperatively, while 120 ones (51.7%) were not. The detail clinicopathological features among the enrolled patients are listed in Table 1.
In the study cohort, circPDK1low showed more lymph nodes metastasis (50.0% vs. 15.7%, P=0.025), more VI (80.6% vs. 66.7%, P=0.047), more VPI (76.6% vs. 36.1%, P=0.015) and more cases with serum CK19 higher than 2.2 ng/mL (58.1% vs. 36.1%, P=0.033) than circPDK1high. In the validation cohort, the results were similar, except for VI (P=0.202) (Table 1).
Survival analysis in the study cohort
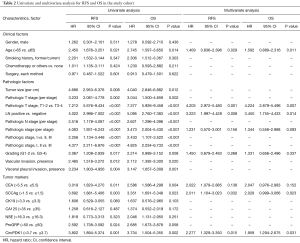
According to univariate analysis in study cohort with the Cox proportional hazards regression model, we found multiple clinicopathologic factors were significant to both RFS and OS, including older than 65 years (P=0.021 and 0.014, respectively), larger tumor size (P=0.008 and 0.012, respectively), more advanced pathologic stage (P<0.001, respectively), more advanced pathologic T stage (P<0.001, respectively), more advanced pathologic N stage (P<0.001, respectively), LN metastasis status (P<0.001, respectively), higher tumor grading (P=0.017 and 0.008, respectively), presence of VPI (P=0.004 and 0.001, respectively), less circPDK1 expression in tumor (P=0.001 and 0.002, respectively), higher level of serum CEA (P=0.011 and 0.004, respectively) and SCCAg (P=0.003 and 0.023, respectively). Also, the presence of VI was significantly associated with RFS (P=0.012) but not to OS (P=0.220). At the multivariate analysis, age 65 years or older (P=0.029 and 0.011, respectively), pathologic T stage III and IV (P=0.001 and 0.007, respectively), LN metastasis status (P=0.008 and 0.014, respectively), SCCAg >1.5 ng/mL (P=0.032 and 0.023, respectively) and relative circPDK1 expression >3.7 (P=0.015 and 0.031, respectively) were significant independent predictors for shorter DFS and OS (Table 2).
Full table
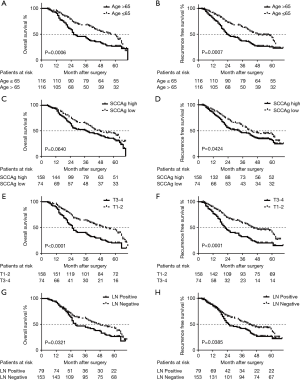
The prognostic value of circPDK1 has been proved via K-M plot analysis before (Figure 1D,E). The survival rates according to the presence of other four independent predictors were analyzed by K-M plot, either. To the results, patients older than 65 years showed significantly poorer OS and RFS than the patients 65 years old or younger (P<0.001, respectively) (Figure 2A,B). While to the patients with higher serum SCCAg (>1.5 ng/mL), a significantly lower curve was observed in RFS (P=0.0424), but not in OS (P=0.0640) (Figure 2C,D). Furthermore, T3-4 patients had a significantly poorer OS and RFS than T1-2 ones (P<0.0001, respectively) (Figure 2E,F). Finally, the LN positive patients also showed significantly poorer OS and RFS than negative ones (P=0.0321 and 0.0385, respectively) (Figure 2G,H).
A prognostic model based on circPDK1 and multiple clinicopathological characteristics
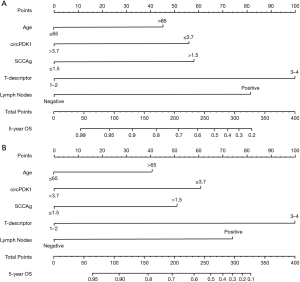
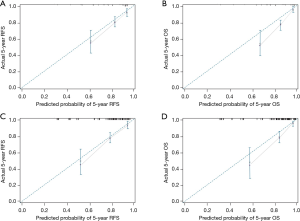
According to the outcomes of the multivariate analysis, a prognostic model was built with circPDK1, T staging, LN status, age, and SCCAg. The nomograms were performed to predict the 5-year RFS and OS (Figure 3). The internal validation of the accuracy to predict RFS and OS has been shown with calibration curves (Figure 4A,B). According to the curves, there was a great correlation between the nomogram-estimated prediction and the actual observed result. The C-index of our model to 5-year RFS and OS was 0.8214 and 0.8359, respectively.
Predictive accuracy of the model in the validation cohort
To perform external validation, the predictive accuracy of this prognostic model to 5-year OS and RFS of resected LSCC was evaluated in the validation cohort, either. In the validation cohort, the C-index of our model for predicting 5-year RFS and OS was 0.8098 and 0.7924, respectively, which showed the excellent discriminative ability of prognosis. Finally, the nomograms were confirmed in the validation cohort, and the calibration curves proved high accuracy of prognosis prediction (Figure 4C,D).
Discussion
In this work, we identified a circular RNA, circPDK1, which is overexpressed in LSCC tumor tissues and exhibited well susceptibility and specificity for LSCC prognosis. Eventually, a new prognostic model was built with circPDK1 and several clinicopathologic factors (T staging, LN status, age, and SCCAg) and verified to be efficient for 5-year RFS/OS prediction of resected LSCC patients.
Nomograms model were used to calculate the probability of 5-year OS and RFS in this work, which has been validated as a reliable method for prognostic prediction (19). Our model consisted of five factors: circPDK1, T staging, LN status, age, and SCCAg that were independent prognostic factors according to the multivariate analysis. According to the hazard ratio (HR) of multivariate analysis, we integrated these clinicopathologic factors into the nomograms, thereby providing an individualized prognostic assessment for each individual. For example, a 68-year-old LSCC patient who underwent R0 resection and systematic LN dissection showed the clinicopathologic features of T3 without mediastinal LN metastasis; his preoperative serum SCCAg concentration is 1.3 ng/mL; the relative expression of circPDK1 in his tumor tissues is 2.7. The nomograms demonstrate that his 5-year RFS and OS is 67% and 72%, respectively. It is indicated that after 5 years, the recurrence risk of this patient is 33%, and the cancer-related mortality risk is 28%. C-index shows the probability that the predicted results are consistent with the observed ones, which is always used to assess the predictive capability of the prognostic model (19). In the study cohort, the C-index of the nomograms for RFS and OS was 0.8214 and 0.8098, respectively. While in the validation cohort, the C-index for RFS and OS was 0.8359 and 0.7924, respectively. Hence, our model that based on circPDK1, T staging, LN status, age, and SCCAg could accurately predict the 5-year RFS and 5-year OS of resected LSCC.
Similar methods have been used in other studies. In the year 2015, Pilotto et al. built a risk stratification model with age, T staging, LN metastasis and tumor grading for prognostic prediction of resected LSCC and performed the external multicenter validation of their model in the year 2017. Moreover, Wang et al. exhibited their nomograms model based on CEA, CK19, TNM staging, and Karnofsky performance status (KPS) for 1-, 3-, and 5-year OS of resected LSCC in 2018. Recently, Liu et al. verified the prognostic value of the tumor spread through air spaces (STAS) in lung adenocarcinoma (LUAD) and then set up a prognostic model based on STAS, VPI, VI, and histological subtype (8,20-22). Compared with those studies, there were more clinicopathological characteristics in our data, such as postoperative adjuvant therapies and multiple serum tumor markers. Besides, our model firstly included a circular RNA, which exhibited excellent potential to be a prognostic marker of LSCC. In considering the stability and abundance in tissues and blood serum, circular RNAs would become the clinical frequently-used biomarkers in the future.
In earlier studies, many serum lung cancer markers have been showed the prognostic value in LSCC (7-9,23). However, the views about the biomarkers’ prediction efficiency were inconsistent. In the year 2013, Nagashima et al. (24) proposed that older age, high serum CEA level, and nodal metastasis are significant prognostic factors to LSCC. While in 2014, Kinoshita et al. (17) reported that high serum SCCAg (>5.5 ng/mL) indicated the poorer prognosis of early-stage LSCC when combined with VPI status and VI status. However, the prognostic role of CEA was not observed in the latter study. One hand, the cut off values of tumor markers in different studies were inconsistent, that had a significant impact on the diagnostic usability of the test results (4,9,25-27). In our institution, the standard cut off values of CEA, SCCAg, NSE, CK19, CA125, ProGRP were 3.4 ng/mL, 1.5 ng/mL, 17 ng/mL, 3.3 ng/mL, 35 U/mL and 63 pg/mL, respectively. However, to enhance the rigor and persuasiveness of this work, we set the cut off values according to several pieces of authoritative literature similar to our study (CEA, 5.5 ng/mL; NSE, 16.3 U/mL and ProGRP, 50 pg/mL) (8,17,18). In the other hand, there were inevitable confounding factors in retrospective data that lead to statistical bias. CEA and pathologic stage have been reported as independent risk factors of LSCC by multiple studies. To our data, no statistical significances were observed in CEA (RFS and OS: P=0.138 and 0.152, respectively) and pathologic stage (RFS and OS: P=0.156 and 0.093, respectively) to the multivariate analysis. In considering that the P values of these items were close to 0.05; thus, the outcomes might be interfered by some confounding factors.
According to the next-generation sequencing data, circPDK1 was transcribed by pyruvate dehydrogenase kinase 1 (PDK1) gene (28). The function of this circular RNA has never been revealed in NSCLC before. To our data, elevated circPDK1 was associated with fewer lymph nodes metastasis, less VI and VPI presence and lower serum concentration of CK19. These results indicated an antitumor function of it in LSCC. However, although transcribed from the same gene, linear PDK1 and circPDK1 exert distinct functions in lung cancer. According to previous studies, PDK1 promotes cancer malignant behaviors, whereas inhibition of PDK1 increases the anti-cancer effect of EGFR tyrosine kinase inhibitors in non-small cell lung cancer (29). We also detected the expression of mRNA PDK1 in LSCC, while there was no significant difference between the tumor and normal tissues (Figure S1A). In addition, we observed a negative correlation between circPDK1 and linear PDK1 in p-LSCC (r=−0.71, P=0.009; Figure S1B). We assumed that, when more circPDK1 was transcribed, the production of linear PDK1 decreased. We have also analyzed the role of linear RNA PDK1 in LSCC in public database (Data source: K-M plot database, http://kmplot.com), and found that its association with LSCC prognosis was not as strong as that of circPDK1 (Figure S1C,D,E). All these findings were worthy of further studies.

This study has some limitations. First, although it is a multicenter study, the sample size was still small for a prognostic model. However, to detect the expression of circPDK1, we must get well-preserved LSCC tissues of all the study objects, while most of the LSCC specimens were poor preserved or not collected 5 years ago. Second, the function of circPDK1 in LSCC has never been reported by previous studies. Hence, it is unconvincing to build a prognostic model with this RNA. However, in our analysis, we have confirmed its potential as a prognostic marker in LSCC. We should reveal the function of circPDK1 in LSCC and further explore its underling molecular biological mechanisms in next work.
Acknowledgments
Funding: This study was funded by Key R & D programs in Shandong Province (grant number.: 2018GSF118119).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The medical ethics for this research were approved by the Ethic Committee of the Affiliated Hospital of Qingdao University (No. QYFYKY 2018-10-11-2).
References
- Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2009. CA Cancer J Clin 2009;59:225-49. [PubMed]
- Lopci E, Rossi S. Tumor metabolism and prognostic role of EZH2 in non-small cell lung cancer. Transl Cancer Res 2017;6:S982-8. [Crossref]
- Martínez-Terroba E, Behrens C, Agorreta J, et al. 5 protein-based signature for resectable lung squamous cell carcinoma improves the prognostic performance of the TNM staging. Thorax 2019;74:371-9. [PubMed]
- Pollan M, Varela G, Torres A, et al. Clinical value of p53, c-erbB-2, CEA and CA125 regarding relapse, metastasis and death in resectable non-small cell lung cancer. Int J Cancer 2003;107:781-90. [PubMed]
- Saintigny P, Coulon S, Kambouchner M, et al. Real-time RT-PCR detection of CK19, CK7 and MUC1 mRNA for diagnosis of lymph node micrometastases in non small cell lung carcinoma. Int J Cancer 2005;115:777-82. [PubMed]
- Watanabe S, Oda M, Go T, et al. Should mediastinal nodal dissection be routinely undertaken in patients with peripheral small-sized (2 cm or less) lung cancer? Retrospective analysis of 225 patients. Eur J Cardiothorac Surg 2001;20:1007-11. [PubMed]
- Johnson DH, Marangos PJ, Forbes JT, et al. Potential utility of serum neuron-specific enolase levels in small cell carcinoma of the lung. Cancer Res 1984;44:5409-14. [PubMed]
- Wang J, Jiang W, Zhang T, et al. Increased CYFRA 21-1, CEA and NSE are Prognostic of Poor Outcome for Locally Advanced Squamous Cell Carcinoma in Lung: A Nomogram and Recursive Partitioning Risk Stratification Analysis. Transl Oncol 2018;11:999-1006. [PubMed]
- Hatzakis KD, Froudarakis ME, Bouros D, et al. Prognostic value of serum tumor markers in patients with lung cancer. Respiration 2002;69:25-9. [PubMed]
- Salzman J. Circular RNA Expression: Its Potential Regulation and Function. Trends Genet 2016;32:309-16. [PubMed]
- Barrett SP, Salzman J. Circular RNAs: analysis, expression and potential functions. Development 2016;143:1838-47. [PubMed]
- Vo JN, Cieslik M, Zhang Y, et al. The Landscape of Circular RNA in Cancer. Cell 2019;176:869-81.e13. [PubMed]
- Hansen TB, Kjems J, Damgaard CK. Circular RNA and miR-7 in cancer. Cancer Res 2013;73:5609-12. [PubMed]
- Chen J, Li Y, Zheng Q, et al. Circular RNA profile identifies circPVT1 as a proliferative factor and prognostic marker in gastric cancer. Cancer Lett 2017;388:208-19. [PubMed]
- Chen B, Huang S. Circular RNA: An emerging non-coding RNA as a regulator and biomarker in cancer. Cancer Lett 2018;418:41-50. [PubMed]
- Li Z, Yanfang W, Li J, et al. Tumor-released exosomal circular RNA PDE8A promotes invasive growth via the miR-338/MACC1/MET pathway in pancreatic cancer. Cancer Lett 2018;432:237-50. [PubMed]
- Kinoshita T, Ohtsuka T, Hato T, et al. Prognostic factors based on clinicopathological data among the patients with resected peripheral squamous cell carcinomas of the lung. J Thorac Oncol 2014;9:1779-87. [PubMed]
- Lang D, Horner A, Brehm E, et al. Early serum tumor marker dynamics predict progression-free and overall survival in single PD-1/PD-L1 inhibitor treated advanced NSCLC-A retrospective cohort study. Lung Cancer 2019;134:59-65. [PubMed]
- Iasonos A, Schrag D, Raj GV, et al. How to build and interpret a nomogram for cancer prognosis. J Clin Oncol 2008;26:1364-70. [PubMed]
- Pilotto S, Sperduti I, Novello S, et al. Risk Stratification Model for Resected Squamous-Cell Lung Cancer Patients According to Clinical and Pathological Factors. J Thorac Oncol 2015;10:1341-8. [PubMed]
- Pilotto S, Sperduti I, Leuzzi G, et al. Prognostic Model for Resected Squamous Cell Lung Cancer: External Multicenter Validation and Propensity Score Analysis exploring the Impact of Adjuvant and Neoadjuvant Treatment. J Thorac Oncol 2018;13:568-75. [PubMed]
- Liu A, Hou F, Qin Y, et al. Predictive value of a prognostic model based on pathologic features in lung invasive adenocarcinoma. Lung Cancer 2019;131:14-22. [PubMed]
- Kulpa J, Wojcik E, Reinfuss M, et al. Carcinoembryonic antigen, squamous cell carcinoma antigen, CYFRA 21-1, and neuron-specific enolase in squamous cell lung cancer patients. Clin Chem 2002;48:1931-7. [PubMed]
- Nagashima T, Sakao Y, Mun M, et al. A clinicopathological study of resected small-sized squamous cell carcinomas of the peripheral lung: prognostic significance of serum carcinoembryonic antigen levels. Ann Thorac Cardiovasc Surg 2013;19:351-7. [PubMed]
- Yang DW, Zhang Y, Hong QY, et al. Role of a serum-based biomarker panel in the early diagnosis of lung cancer for a cohort of high-risk patients. Cancer 2015;121 Suppl 17:3113-21. [PubMed]
- Sigari N, Mohsenpour B, Nikkhoo B, et al. Determination of the best prognostic value of serum tumor markers in patients with suspected lung cancer in an Iranian population. Clin Lab 2014;60:23-7. [PubMed]
- Ishibashi N, Maebayashi T, Aizawa T, et al. Serum tumor marker levels at the development of intracranial metastasis in patients with lung or breast cancer. J Thorac Dis 2019;11:1765-71. [PubMed]
- Sradhanjali S, Reddy MM. Inhibition of Pyruvate Dehydrogenase Kinase as a Therapeutic Strategy against Cancer. Curr Top Med Chem 2018;18:444-53. [PubMed]
- Yang Z, Zhang SL, Hu X, et al. Inhibition of pyruvate dehydrogenase kinase 1 enhances the anti-cancer effect of EGFR tyrosine kinase inhibitors in non-small cell lung cancer. Eur J Pharmacol 2018;838:41-52. [PubMed]


