
Frontiers of ctDNA, targeted therapies, and immunotherapy in non-small-cell lung cancer
Overview of non-small-cell lung cancer (NSCLC)
Lung cancer, the most common type of cancer worldwide, has a morbidity rate of 11.6% and a mortality rate of 18.4% (1). Approximately 85% of lung cancers are within a group of histological subtypes collectively known as NSCLC. NSCLC is thought to originate in lung epithelial cells and comprises diverse histological subtypes, of which lung adenocarcinoma and lung squamous cell carcinoma are the most common (2). The most common genetic alterations in NSCLC are mutations in the receptor tyrosine kinases (RTKs) epidermal growth factor receptor (EGFR), Kirsten rat sarcoma (KRAS), tumor suppressor p53 and liver kinase B1 (LKB1); anaplastic lymphoma kinase (ALK) gene fusion; ROS1 gene fusion; and MET amplification (3,4). Over decades, treatment strategies for lung cancer have changed from chemotherapy to personalized medicine, such as tyrosine kinase inhibitors (TKIs), which specifically target mutations based on individual patients. Immunotherapy, a developing treatment, has already proven effective in treating NSCLC patients. Through controlling the statuses of programmed cell death-1 (PD-1), programmed cell death ligand-1 (PD-L1) and cytotoxic T-lymphocyte antigen-4 (CTLA-4) by antibodies called immune checkpoint inhibitors (ICIs), patients can have a prolonged life with improved quality. Circulating tumor DNA (ctDNA) quantification is often used in TKI-based targeted therapy to facilitate more precise clinical decisions and prognoses. The efficiency of three generations of EGFR- or ALK-TKIs can be evaluated by monitoring the ctDNA level of corresponding EGFR and ALK mutant genes, respectively.
Activating mutations in EGFR, such as exon 19 del and L858R on exon 21, sensitize the majority of NSCLC tumor cells to the first-generation EGFR-TKIs gefitinib and erlotinib and the second-generation EGFR-TKIs afatinib and dacomitinib. Among patients with these tumors, 50% develop acquired resistance due to the EGFR T790M mutation, which was the source of major concern until the development of the third-generation EGFR-TKIs osimertinib and rociletinib (5-7). With the widespread use of osimertinib, the EGFR C797S resistance mutation appeared as well (8). In addition to secondary EGFR mutations, bypass mechanisms such as MET or ERBB2 amplification, Hippo pathway inhibition, and insulin-like growth factor 1 receptor (IGF1R) activation also contribute to resistance to EGFR-TKIs (9-12). EML4-ALK gene fusion is found in 3–7% of NSCLC patients (13-15). Similar to the resistance to EGFR, resistance to each of the three generations of ALK-TKIs occurs.
KRAS mutations, which are found in approximately 30% of lung adenocarcinomas and 3% of lung squamous cell carcinomas, are not as targetable as EGFR and ALK mutations. KRAS mutations account for 90% of RAS mutations found in lung adenocarcinoma (16). Among all mutations detected in NSCLC patients, mutations in KRAS and EGFR constitute more than 60% of the mutations found in lung adenocarcinoma (4,17,18). KRAS and EGFR mutations, however, are usually mutually exclusive, but when these mutations coexist, KRAS mutations may result in tumors that are drug-refractory to EGFR-TKIs and do not respond to anti-EGFR monoclonal antibodies (19,20). Activated KRAS activates downstream pathways, including the BRAF/MEK/ERK and PI3K/AKT/mTOR pathways. Potential targeted therapies for KRAS-mutant lung cancer have focused on inhibiting the downstream effectors of these signaling pathways instead of mutated KRAS. Unlike EGFR-TKIs, which have evolved into the third generation, the development of clinically effective small molecule drugs for KRAS has met with great obstacles over the past decades. Recently, the association between PD-(L)1 and KRAS has been discussed in several studies, and some have noted that PD-1 expression is significantly associated with the presence of KRAS mutations (21-24). However, more investigation is needed to increase understanding of immunotherapy so that developments in KRAS treatment no longer remain stagnant.
ctDNA aids diagnosis and treatment tracking
Tissue biopsy plays an important role in analyzing tumor properties but remains invasive and may cause harm to patients. In addition, tissue biopsy is not always feasible, and it cannot fully account for the temporal and spatial heterogeneity of cancer cells (25). Liquid biopsy, on the other hand, is noninvasive and provides a dynamic view of tumors with overall heterogeneity (26). Because liquid biopsy is highly sensitive to ctDNA, it can be utilized as an early detection method for cancer and remains useful during treatment procedures to evaluate treatment response (27,28).
ctDNA
ctDNA molecules produced by tumors have certain properties not possessed by normal cell-free DNAs (cfDNAs) (27). A wide variation in length is the most distinctive feature of ctDNAs. ctDNAs are typically highly fragmented; thus, their size varies between about 70 and 200 bp. Some studies reported that the most frequently observed ctDNAs are 180 bp in size with a classic ladder pattern that correlates with the caspase-activated DNase digestion of chromatin in apoptotic tumor cells (29), and ctDNA fragments larger than 10 kb might originate from necrosis (30). Two recent studies pointed out that the modal size of ctDNA for many cancer types is about 166 bp, which is the length of DNA wrapped around the chromatosome, and the result of protection from enzymatic degradation due to histone binding to nuclear DNA (31,32). Furthermore, an enrichment in mutated ctDNA fragments longer than 167 bp were also reported, notably around 250–320 bp (32). However, shorter length of ctDNA fragments compared to wild-type allele was found in human hepatocellular carcinoma (134–144 vs. 167 bp), melanoma (132–145 vs. 165 bp), lung cancer, renal cell carcinoma, and colorectal cancer (33-35). In ctDNA analysis, an additional DNA fragment length cutoff could be applied for mutation identification to increase overall sensitivity (36). It is likely that due to different origins, various fragment sizes are generated. The mechanisms behind ctDNA fragmentomics still require further research.
Because of the low concentration of ctDNA in blood, it is difficult to detect ctDNA using nucleic acid detection methods suitable for the qualitative detection of a large amount of DNA. However, with the development of highly sensitive DNA detection techniques with a low false-positive rate, such as quantitative polymerase chain reaction (qPCR), digital polymerase chain reaction (dPCR), next-generation sequencing (NGS), NGS-based techniques and other methods, the detection of ctDNAs at low concentrations became possible (37). Mutations in ctDNA molecules from tumor tissues can clearly reflect tumoral mutations and tumor heterogeneity (28,38), thus enabling the detection of tumors by detecting ctDNA mutations.
qPCR has been used to detect cfDNA from blood taken from lung cancer patients (39-41). A significant correlation between NSCLC severity and the levels of cfDNA was observed, and high cfDNA concentrations indicated more severe disease (41). cfDNA detection is currently more often used as a supplementary measure for diagnosis and to monitor NSCLC patients.
Detecting EGFR mutations by ctDNA
EGFR mutations can cause resistance to EGFR-TKIs in NSCLC patients (38,42). Sensitizing EGFR mutations are present in 14% to 38% of NSCLC patients depending on the location of the mutation and the ethnicity of the patient (43). In recent years, commonly used techniques to detect ctDNA EGFR mutations have been mainly the amplification refractory mutation system (ARMS) approach, droplet digital PCR (ddPCR) and NGS-based methods (44-46). These techniques differ in their sensitivity, accuracy, specificity and coverage in detecting EGFR mutations (37).
ARMS is based on qPCR that uses specific probes to identify EGFR mutant sequences. As ARMS has an overall sensitivity of 0.1–1% (47,48), the abundance of the mutated DNA must be above this threshold to depress the false-negative rate. However, the false-negative rate in plasma samples, which is approximately 30%, is still relatively high compared with that in tumor tissue. The sensitivity and specificity of plasma EGFR mutation detection are 92% and 100%, respectively, compared with those of tumor EGFR mutation detection (49). There are two subcategories of ARMS: ADx-ARMS and cobas-ARMS. Cobas-ARMS exhibits superior sensitivity in detecting the EGFR T790M mutation (0.1%) and sufficient sensitivity (90%) (44). The obvious advantage of ARMS is its use of simple workflows and consequent rapid turnaround time. Furthermore, ARMS is normally performed at initial tumor biopsy and therefore offers first-hand information about tumors (44). However, flaws in this method exist as cobas-ARMS can detect only EGFR variants, and studies have shown that ARMS cannot replace tumor tissue biopsy to detect EGFR mutation status (50). In addition, the sensitivity of ARMS is unmatched with that of ddPCR, which is approximately 0.01% and thus ten times greater than that of ARMS (48). The increased precision and accuracy of ddPCR used to detect plasma T790M status are also advantages of ddPCR over ARMS.
ddPCR, which exhibits an unparalleled sensitivity of 0.001–0.4%, can detect EGFR, KRAS, ALK and other mutations with greater precision than other methods (51-53). Due to the small amount of ctDNA present in the serum of lung cancer patients, a highly sensitive method such as ddPCR is essential. ddPCR costs less and requires shorter turnaround time; besides, ddPCR is highly accessible without complex bioinformatics support for data analysis. And it was shown by studies (54-56) that the sensitivity of ddPCR for the detection of EGFR or KRAS mutations in lung cancer can be as sensitive and highly concordant as NGS, sometimes even more. However, ddPCR requires specific primers, and we can detect only one locus per reaction, which limits its use in multiplex tests.
NGS-based methods are the most accepted and recognized method used to detect EGFR mutations with both high and low allele frequencies, especially those with low allele frequency. In a study performed by Xu et al. (44) cobas-ARMS, ddPCR and NGS all detected mutations with low allele frequencies well, and ddPCR and NGS also yielded excellent positive coincidence rates. Overall, the NGS platform demonstrates sensitivity equivalent to that of ddPCR, the most sensitive platform for EGFR mutational profiling. A sensitivity of 100% (30/30) for amplicon-based NGS was achieved compared to 87% (26/30) for ddPCR in the detection of EGFR activating mutations in advanced NSCLC (57). Similar studies generated data of 100% (18/18) NGS vs. 94% (17/18) ddPCR (54), and 83% (30/36) NGS vs. 69% (25/36) ddPCR (58). What makes NGS-based techniques truly stand out is their ability to detect a wide range of mutations in EGFR and other clinically important genes, such as ALK and RAS, which ddPCR is unable to do because it can detect one only locus in each reaction. Researchers believe that ctDNA can be used to confirm all EGFR primary driver mutations, along with additional mutations in other disease-relevant genes. These advantages have made NGS relatively costly compared to other methods. One setback of NGS is its high false-positive rate. It is recommended that the results of both NGS and ddPCR are combined or repeatedly compared with an NGS library to validate the detected mutation. For example, the combination of NGS to discover EGFR primary mutations and other mutations with the specificity of the ddPCR assay was shown to be the optimal workflow for T790M analysis (37). In summary, NGS analysis is an ideal method for detecting EGFR primary mutations that avoids the possible harm caused by tumor biopsy. Furthermore, ddPCR can be used as an additional reassurance during NGS analysis, especially when detecting mutations with low allele frequencies (<0.1%). When the amount of cfDNA is limited, NGS is a better option than dPCR and qPCR because it requires only 10 ng of cfDNA per analysis, while dPCR and qPCR require 15 and 40 ng, respectively, according to Bartels et al. (37).
In recent years, different branches of NGS-based methods such as amplicon-based NGS (54,57,59) and ultra-deep NGS (55,60) have shown transcendent sensitivity and specificity in the detection of mutations in NSCLC. Amplicon-based NGS demonstrated high accuracy for point mutations and indels, and it can also detect chromosomal rearrangement and fusion genes in ctDNA. This barcoded NGS in which PCR was used to enrich target genes provided excellent sensitivity by limiting sequencing artifacts compared to hybrid capture-based NGS, and further lowed the required amount of ctDNA sample (61). A shallow sequencing depth was a problem for NGS method, however, ultra-deep NGS assays can overcome this and achieved a 75% sensitivity in detecting lung cancer oncogenic driver mutations, according to Li et al. (55).
ctDNA assists clinical decision making
EGFR-activating mutations are the most important mutations among the many driver oncogenes that play critical roles in NSCLC (62). Initial patient responses to EGFR-TKI treatment are often very positive. As ctDNA is a convenient and precise parameter collected in real-time, it can show sudden outbreaks that appears as spikes in data that correlate with the apoptosis of tumor cells within 1 week after first-time TKI use (63,64). Riediger et al. (65) reported an 11-fold acute increase in ctDNA levels only 26 hours after therapy started. After the initial outbreak, the amount of ctDNA continues to decrease over the following weeks. However, after a partial response (PR) or even complete response (CR) for a short time, the efficiency of TKIs decreases, which may lead to a stable disease (SD) condition and in some cases prevent TKIs from countering the effect of the mutation, finally eliminating its efficacy. The state in which specific TKIs no longer function is called a resistant state. Median progression-free survival (PFS) is 10–16 months among patients treated with EGFR-TKIs, and almost half of acquired TKI resistance is caused by secondary EGFR T790M mutation (5,66).
Tracking ctDNA dynamics helps determine disease state
Tracking changes in ctDNA levels is informative for determining a patient’s disease state and capturing dynamic changes during TKI treatment (67). Changes in ctDNA levels also reflect the different sensitivities of heterogeneous cancer cell clones because of the different responses shown by EGFR ctDNA to EGFR-TKIs. All of the advantages of ctDNA monitoring are crucial for clinical intervention. ctDNA levels show a good correlation with radiologic CR/PR but exhibit a poor correlation with patients in SD/progressive disease (PD) states (68). Cancer cell clones in radiologic SD/PD patients with good ctDNA responses may be a mixture of cancer cells with wild-type EGFR and EGFR with activating mutations. Poor ctDNA responses indicate that the tumor may carry de novo resistance to EGFR-TKIs (69). Notably, early (often within one week) ctDNA responses are not a good predictor of radiologic response because the sudden increase in ctDNA results from rapid cancer cell apoptosis (63).
Differences in ctDNA levels before and after treatment
Husain et al. observed peaks in the level of ctDNA, followed by median decreases of 86% and 81% from weeks one to two, respectively, indicating EGFR L858R and exon 19 del (64). Because patients who do not undergo therapy cannot exhibit patterns such as temporal spikes in their ctDNA profiles, instantaneous changes in ctDNA profiles before and after treatment reveal a significant quantitative rise in the number of EGFR copies after therapy, which reflects cell apoptosis within days of exposure to the drug.
In a drug-sensitive state, the continuous decline in ctDNA levels in T790M-positive patients continues for a median of 6 months (70). Urine ctDNA levels decline to a greater extent than widely used plasma ctDNA levels. Urine ctDNA detection also has a remarkable 88% overall concordance rate with the detection of ctDNA in tissue samples. The concordance rate with plasma ctDNA detection is 98%. This shows the feasibility of replacing plasma ctDNA detection with urinal ctDNA detection, which exhibits a similar sensitivity and improved convenience (70).
Mechanisms of TKI resistance
EGFR
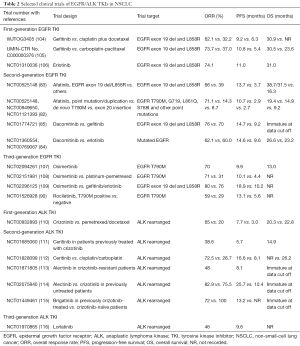
Three generations of TKIs have been designed to target EGFR mutations. As ctDNA has aided the detection and verification of resistance to various drugs, the mechanisms behind resistance to these drugs have been studied for more than a decade. Three generations of EGFR-TKIs are summarized in Table 1 (77,82-85,90). In the following section, we focus on several widely accepted resistance mechanisms along with less discussed novel resistance. Figure 1 shows most of the mechanisms of resistance to different inhibitors through signaling pathways mentioned in this review. Hopefully, the study of these mechanisms will shed light on the clinical treatment of NSCLC patients.

Full table
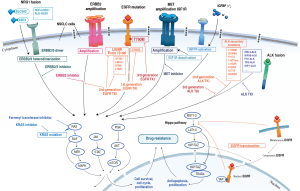
EGFR T790M and C797S
EGFR T790M, the most common mutation against first-generation TKIs, accounts for 50–60% of acquired resistance to TKIs. When the third-generation TKI osimertinib was developed, the response rate to osimertinib was 62.5%, and 52% of patients with the EGFR T790M mutation exhibited a PFS of 12 months (6). Though osimertinib is a revolutionary drug, there are still cases of osimertinib resistance. Novel osimertinib resistance mechanisms can be divided into different types: (I) the acquisition of tertiary mutations that restore EGFR signaling pathways and (II) bypass mechanisms, such as MET or ERBB2 amplification, Hippo pathway inhibition, or IGF1R signaling activation.
EGFR C797S is sensitive to first-generation TKIs but resistant to second- and third-generation TKIs. The second-generation EGFR-TKI dacomitinib directly induced secondary EGFR T790M or C797S mutations in Ba/F3 cells transfected with EGFR 19 del, L858R, or G719A mutants, and there was no significant difference in the timing of the emergence of the T790M and C797S mutations (8,78). The C797S mutation occurred in 11% of L858R mutant clones (4 of 35) and 32% of G719A mutant clones (12 of 38) established with low-dose dacomitinib (8). Afatinib, another second-generation EGFR-TKI, also induced the C797S mutation as a resistance mechanism (78). First-generation EGFR-TKIs exert their effects independent of the cysteine at position 797 (94). Gefitinib or erlotinib can be used to overcome resistance to the C797S mutation. Erlotinib is particularly effective in treating EGFR C797S-positive tumors (78-80). Tumors with T790M and the trans C797S mutation responded to a combination of erlotinib and osimertinib. In patients with acquired osimertinib resistance, gefitinib monotherapy was shown to successfully shrink T790M-negative/C797S-positive tumors (77,95). By studying the T790M and C797S mutations in lung adenocarcinoma, NSCLC containing T790M/cis-C797S mutations was shown to be more aggressive than that containing T790M/trans-C797S mutations (95). Tumoral heterogeneity plays an important role in the mechanism of dual cis/trans resistance.
Tumor heterogeneity reveals resistance potential
Overcoming tumor heterogeneity is a major challenge for the personalized treatment of cancer. The most effective treatment protocol can be designed only after tumor DNA profiling, which reveals the heterogeneity of each individual tumor.
Cancer cells simultaneously carry several types of activating EGFR mutations even before EGFR-TKI treatment begins (68). If these cells possess the ability to develop multiple resistance mechanisms and are more adaptable to TKIs, resistance can emerge earlier (96). There may be an intrinsic minor population of T790M-positive cancer cells in EGFR-mutated tumor clones carrying multiple mutations (69). In addition, T790M-negative drug-tolerant cells may persist in patients with drug-resistant EGFR T790M-positive tumors and undergo further evolution to acquire resistance to subsequent therapies (97). Emergence of the T790M mutation accounts for 50% of acquired resistance. Furthermore, the T790M mutation was more frequent among patients with the activating EGFR 19 del mutation than those with the L858R mutation (6). By understanding the emergence of EGFR T790M mutation heterogeneity, better clinical decisions can be made during the period of disease resistance.
EGFR T790M heterogeneity
The presence of resistance mutations, such as the T790M mutation, does not necessarily lead to an overall resistant phenotype in tumor cells. In some cases, despite the selective pressure by TKIs in the treatment process, cancer cells with the T790M mutation still fail to dominate (68). The heterogeneity within tumors plays a much more important role than previously estimated. For instance, EGFR T790M-positive clones emerge from not only pre-existing clones but also initially EGFR T790M-negative drug-tolerant cells through what is called the de novo acquisition of T790M (97). This mixed tumor evolution eventually determines the overall response to TKIs although a wide variety of tumor cells originating from different parental cells are contained within the tumor.
Heterogeneous EGFR-mutated cancer cells undergo convergence and divergence in response to EGFR-TKIs. Minor clones are eliminated first, and mutations such as T790M then arise and induce acquired resistance (68). For instance, the ratio of T790M to exon 19 del mutations changed significantly and fluctuated during treatment. While most EGFR alleles with exon 19 del seem to carry the T790M mutation at the first and second T790M DNA peaks, this does not appear to continue during the development of acquired resistance because exon 19 del alleles may have amplified more rapidly than T790M alleles over time (68).
Several studies have shown that the T790M mutation can be spatiotemporally heterogeneous in a patient because of selective pressure from EGFR-TKIs (66,69,97-99). This heterogeneity also reflects the dynamics within resistant tumor cell clones, which are likely made up of TKI-sensitive and TKI-resistant cells. Upon TKI withdrawal, the ratio of TKI-sensitive to TKI-resistant cells will increase as a result of the repopulation of TKI-sensitive cells due to the absence of selective pressure, thus leading to the regain of tumoral TKI sensitivity (100). In conclusion, long TKI-free intervals may reduce TKI-resistant clones and induce restoration of EGFR-TKI sensitivity. Elimination of the T790M mutation is the result of a significant reduction in TKI resistance, such as that observed in T790M-positive clones. T790M heterogeneity should be taken into consideration when making clinical decisions to apply TKI therapy in patients after the development of acquired resistance. Furthermore, by using T790M-positive clones as a predictive marker during TKI treatment, a better TKI rechallenge scheme with designed “on and off” TKI exposure can maximize elimination of the T790M-positive clones in the tumor.
The heterogeneity of T790M-positive patients is the result of a mixture of T790M-positive and T790M-negative cells that may have been present in heterogeneous tumors before TKI treatment. TKI pressure plays a selective role and controls the ratio between these two types of cells, thus determining the total tumor status (69). In contrast, the selective pressure exerted by TKIs can never induce T790M-positive cells in a T790M-negative cell clone.
EAI045 is the first allosteric TKI engineered thus far to overcome L858R/T790M and C797S mutations (93). EAI045 is 1000-fold more selective for mutant EGFR than for wild-type EGFR. Cetuximab, a monoclonal antibody that can block EGFR dimerization by preventing EGF ligand binding, synergizes with EAI045 by converting the inhibitor-resistant receiver population into a monomeric form that is remarkably sensitive to EAI045 (92,93). EGFR mutations at position C797 do not affect the efficacy of EAI045, as C797 is far from the allosteric binding pocket (91). In addition, EAI045 in combination with cetuximab potently inhibited L858R/T790M/C797S in Ba/F3 cells (93). More studies are needed to determine the clinical efficacy of EAI045 and shed light on its application.
T790M loss and C797S
The two main mechanisms of resistance to osimertinib are loss of the T790M mutation and emergence of the C797S mutation. These two mutations cause more than 60% of resistance (88,101). T790M loss is a common mechanism of resistance in patients treated with osimertinib. Half of the patients in a progressive state exhibited T790M loss during osimertinib treatment (86,88,102). Twenty-six percent of the patients were observed to harbor EGFR C797 and L792 mutations, and these mutations were exclusive to T790M-preserved cases (86). The C797S mutation was detected in approximately 40% of EGFR-mutated NSCLC patients with T790M who developed acquired resistance to osimertinib (88). These results reveal that there are different resistance patterns in cases in which T790M is preserved or lost. In most T790M-preserved cases, resistance was associated with continued EGFR activation through known tertiary mutations that cause resistance, such as C797S, or activation of bypass signaling pathways, whereas resistance in T790M-loss cases occurred through diverse and predominantly EGFR-independent alternative competing mechanisms, such as MET amplification and small cell transformation (86).
Patients who develop early resistance to osimertinib are likely to have competing resistance mechanisms in other tumor subclones, while patients who develop late resistance to osimertinib are more likely to have maintained T790M and acquired the C797S mutation (87). Although both T790M-loss and T790M-preserved patients had decreased T790M levels after 1 to 3 weeks of osimertinib treatment, repeated testing for T790M is still required to distinguish between these two biologically distinct types of osimertinib resistance.
Other novel mutations occur at a relatively low frequency. Among these mutations, L792F was detected in 1.76% (6 of 340) of patients with lung adenocarcinoma treated with osimertinib (89). The L792F mutation results in a level of resistance between that to both first- and third-generation EGFR-TKIs due to the T790M and C797S mutations (89). Mutations in cis with T790M cannot be inhibited by cetuximab or EAI045 (89). However, L792F-positive tumors were also found to be much less resistant to second-generation TKIs, especially dacomitinib (78). More precise treatment strategies and additional combinational approaches are required for patients with the EGFR L792F mutation.
ALK
Rearrangements of ALK, which are mutually exclusive with mutations in EGFR or KRAS, account for 3–7% of mutations (13-15). The most common ALK rearrangement occurs in the echinoderm microtubule-associated protein-like 4 (EML4) gene, producing an EML4-ALK fusion (103). To treat ALK fusion, TKIs of ALK, such as crizotinib, ceritinib, alectinib and brigatinib, have been developed. We examined several clinical trials of ALK-TKIs and EGFR-TKIs and illustrate the clinical efficacies of ALK-TKIs and EGFR-TKIs in Table 2 (111-116). Crizotinib is a competitive ATP inhibitor of ALK and MET tyrosine kinases that received Food and Drug Administration (FDA) approval in 2011 (117,118). A phase I clinical trial of crizotinib demonstrated a high overall response rate (ORR) of 60.8% and a median PFS of 9.7 months. Subsequent phase III trials demonstrated the prolonged PFS and increased ORR of crizotinib compared to those of standard first- or second-line chemotherapy (110,119).
Full table
Resistance to first- and second-generation ALK-TKIs
Acquired resistance to the first-generation ALK-TKI crizotinib has been identified in approximately 30% to 40% of patients. The occurrence of resistance may be attributed to three causes: (I) the acquisition of secondary resistant mutations that were reported to occur in 22–36% of patients (120); (II) ALK copy number alterations; and (III) the upregulation of bypass signaling pathways leading to ALK-independent growth, such as the activation of the EGFR, MET, KIT, IGF1R, and/or other pathways (120-122).
Secondary ALK mutations in NSCLC are distributed throughout the kinase domain. The mutations L1196M, G1269A, F1174, 1151Tins, L1152R, S1206Y, I1171T and G1202R have been determined to confer major resistance to crizotinib (123-126). L1196M is a mutation of the gatekeeper residue, and G1269A is a mutation of a residue in the ATP-binding pocket that, upon its mutation, can cause changes that prevent crizotinib binding (127).
To counter resistance to crizotinib, the second-generation ALK inhibitors ceritinib, alectinib and brigatinib were developed. Approximately 40–50% of crizotinib-resistant patients were shown to respond to these TKIs, which exhibit a median PFS of 7–12 months (128-130). Secondary mutations at I1171 confer resistance to alectinib, and the I1171N and I1171T mutations destabilize ALK inhibitor binding while stabilizing the tyrosine kinase in its activated conformation (131,132). Ceritinib, however, was found to be capable of overcoming the I1171T mutation as well as several crizotinib-resistant ALK mutations, such as L1196M, G1269A and S1206Y, in preclinical models (133).
Resistance to third-generation ALK-TKIs
The third-generation ALK-TKI lorlatinib is an ATP-competitive inhibitor of recombinant ALK and ROS1 kinases. Lorlatinib has the ability to penetrate the brain and central nervous system, where frequent lung cancer metastasis occurs. In preclinical settings, lorlatinib was shown to have low nanomolar potency against wild-type ALK and be highly effective against all clinically acquired ALK mutations, including the highly resistant G1202R mutant, which is resistant to both first- and second-generation ALK inhibitors, by impairing drug binding through steric hindrance (126,134). Lorlatinib can also potently inhibit wild-type ROS1 and the G2032R ROS1 mutant in vitro and in vivo (135).
The antitumor efficacy of lorlatinib has been shown in two recent clinical trials (116,136). In a phase I trial of lorlatinib in NSCLC patients with ALK or ROS1 rearrangement, lorlatinib showed a high response rate and a median duration of response of 11.7 months in 42% of patients (11 of 26) previously treated with first- and second-generation ALK-TKIs. Then, patients with different medical histories were involved in a phase II trial. Among these patients, the treatment-naïve group of ALK-positive NSCLC patients showed the best response rate; 87% (26 of 30) of the patients in this group showed a PR, and only 1 of 30 patients remained at disease progression. Consistent with those in the phase I trial, patients previously treated with crizotinib showed a good response rate that was second to the response of the treatment-naïve group. In all patients previously treated with at least one ALK-TKI, responses were rapid with a median of 1.4 months and durable. Lorlatinib is a new option for patients whose disease has progressed after treatment with crizotinib or second-generation ALK inhibitors.
Bypass mechanisms
Bypass mechanisms for EGFR-TKI
Amplification of MET and ERBB2
MET gene amplification and the hyperactivation of MET are mechanisms of resistance to both first- and third-generation EGFR-TKIs, such as erlotinib, rociletinib and osimertinib, in patients that exhibit multiple mechanisms of resistance, such as those in whom tumor cells have undergone epithelial-mesenchymal transition (EMT) or those with small cell lung cancer (SCLC) transformation (10,38). An inverse correlation between EGFR T790M and MET amplification was observed (137). EGFR and MET have been shown to act simultaneously to activate downstream effectors, such as PI3K/AKT/mTOR and RAS/RAF/ERK, and ultimately regulate tumor cell proliferation. MET signaling activation likely serves as a compensatory pathway for the loss of the EGFR-driven signaling cascade (138).
ERBB2 overexpression accounts for approximately 3–26% of acquired resistance to first-generation EGFR-TKIs and 5% of acquired resistance to third-generation TKIs in NSCLC patients (38,74-76). By activating the EGFR-independent phosphorylation of ERBB3 and downstream activation of the PI3K/AKT pathway, a bypass mechanism is created even in the presence of first-generation EGFR inhibitors (9,10). The sensitivity of the third-generation EGFR-TKIs rociletnib and osimertinib decreased when they were used at nanomolar concentrations in a PC9/GR NSCLC cell line overexpressing ERBB2 (139). Shi et al. (10) demonstrated that ERBB3 phosphorylation in both HCC827/ER and HCC827/AR cells was minimally inhibited by osimertinib alone and could be fully suppressed only when osimertinib was combined with a MET inhibitor. Hence, the sensitivity to third-generation TKIs is restored by MET inhibition resulting from the suppression of ERBB3 phosphorylation.
Tumor resistance caused by the activation of accessory pathways can be theoretically overcome by a combination of EGFR inhibitors and other involved molecules, which serves as a potential strategy to counter acquired resistance often observed during the treatment of EGFR-mutated NSCLC. Dual inhibition of MET and ERBB has also been performed to determine the intratumor heterogeneity and plasticity in acquired resistance (140). The combination of capmatinib (MET inhibitor) and afatinib was shown to be more effective than afatinib as a single agent. This drug combination completely suppressed tumor growth in a patient-derived xenograft (PDX) mouse model that showed the necessity of MET amplification to lung cancer cell survival (140). KRAS G12C mutant clones emerged upon the blocking of two upstream activating components of the MAPK pathway, EGFR and MET, which suggests that the development of resistance in NSCLC cells is a flexible process.
IGF1R activation
IGF1R stimulates cell proliferation primarily through the PI3K/AKT and RAS/MAPK signaling pathways. Activation of IGF1R enhances PI3K/AKT signaling, giving rise to resistance to EGFR-TKIs in PC9 cells resistant to either PF299804 or WZ4002 (11). IGF1R upregulation as an immediate response to erlotinib was also observed in erlotinib-resistant HCC827 cells with additional acquired features of EMT, whereas MET overexpression and secondary EGFR mutations were absent (71,81). Exogenous IGF1 also activated IGF1R in the PC9 and H460 cell lines (72). Targeting IGF1R with AG-1024 and inhibiting EGFR with gefitinib exerted antiproliferative effects in the H1975 cell line via a reduction in AKT phosphorylation and the subsequent upregulation of BCL-2-interacting mediator of cell death (BIM) (141,142).
The loss of IGF binding protein 3 (IGFBP3) has been reported to induce IGF1R activation and EGFR-TKI resistance. Overexpression of IGFBP3 or inhibition of IGF1R increased the sensitivity of NSCLC cell lines to 3rd-generation EGFR-TKIs (11,73,143). A recent study found that both increased or decreased IGFBP expression induced the activation of IGF1R in response to TKI and served as a bypass mechanism in cells with MET amplification. In addition, the activation of IGF/IGF1R signaling was found in cell lines resistant to both the MET-TKI PHA665752 and the EGFR-TKI gefitinib (144). IGF1R knockout enhanced MET amplification, resulting in resistance to erlotinib. In addition, IGF1R knockdown attenuated EMT, which involved a decrease in E-cadherin expression and an increase in vimentin, snail, and nuclear β-catenin expression in PC9/GR and H460/ER cells (72). However, tumoral clones with MET amplification do not always exhibit great advantages. IGF1R hyperactivation and heterogeneous EMT features, but not MET amplification, led to resistance to high-dose erlotinib in the HCC827 cell line. MET amplification tends to emerge instead of EMT as the resistance mechanism in HCC827 cells after their exposure to low concentrations of EGFR inhibitors (145).
TGFβ1 pathways
EGFR inhibition can result in an autocrine transforming growth factor β1 (TGFβ1) pathway loop and stimulate the downstream SMAD pathway (145). Hematopoietic pre-B-cell leukemia transcription factor (PBX)-interacting protein (HPIP/PBXIP1) silencing can suppress TGFβ1 secretion by inhibiting SMAD2 activation (146). Only continuous TGFβ1 secretion can promote and maintain mesenchymal transition and EGFR-TKI resistance; thus, this induced mesenchymal transition was reversible upon the removal of TGFβ1 (145). The reversal of TGFβ1-induced EMT by E-cadherin overexpression in resistant cells can also restore TKI sensitivity (72). The expression of EMT-related markers and TGFβ1/SMAD2 was higher in cells transfected with miRNA-132 inhibitor (147). Furthermore, miRNA-138 knockdown cells exhibited mesenchymal phenotypes (148). These results imply that miRNA-132 inhibits EMT by regulating TGFβ1/SMAD2 in NSCLC cells, while TGFβ1 downregulates miRNA-138, contributing to an EMT phenotype. This reversibility is clinically significant because the relief of EGFR inhibition could deplete TGFβ1 to reverse EMT, which in turn might resensitize tumors to EGFR-TKIs, thus prolonging the duration of EGFR-TKI therapy.
Hippo pathway inhibition
The Hippo pathway consists of a large network of proteins that include neurofibromin-2 (NF2), core kinase cassette containing mammalian STE20-like protein kinase 1/2 (MST1/2), large tumor suppressor 1/2 (LATS1/2), adaptor proteins Salvador homologue 1 (SAV1) and MOB kinase activator 1 (MOB1). These proteins limit tissue growth by promoting LATS1/2-dependent phosphorylation of the oncoproteins Yes-associated protein (YAP) and transcriptional coactivator with the PDZ-binding motif (TAZ) (149-151). YAP and TAZ promote cell proliferation by regulating the activity of different transcription factors such as TEADs and SMADs (152). The association between Hippo pathway inhibition and the development of EGFR-TKI resistance was discussed in several recent studies (12,153-157).
Translocation of EGFR from the plasma membrane to the cytoplasm and nuclear membrane inhibits the Hippo pathway. EGF could stimulate the translocation of membranous EGFR (mEGFR) into the cytoplasm (cEGFR) and nucleus (nEGFR) by binding to importin-β through its nuclear localization sequence or binding with YAP (12,158,159). As a result, the expression of mEGFR decreased, and the expression of cEGFR increased. cEGFR interacted with SIK2 and enhanced its ability to bind to SAV1, which inhibited the interaction between LATS1 and MST1. Furthermore, downstream YAP phosphorylation was inhibited, thus increasing the nuclear translocation of YAP and ultimately inhibiting the Hippo pathway by binding with the transcription factor TEAD (12). Therefore, resistance against the first-generation EGFR-TKIs gefitinib and erlotinib is associated with inhibition of the Hippo pathway and enhanced YAP activity (155,156). Furthermore, the combination of the YAP inhibitor verteporfin with erlotinib sensitized the erlotinib-resistant H1975 cell line (155).
Bypass mechanisms for ALK-TKI
EGFR activation
Multiple bypass mechanisms could induce EGFR-TKI resistance, and the emergence of bypass signaling during ALK-TKI treatment also contribute to resistance in ALK-positive NSCLC. EGFR activation as a bypass mechanism for ALK-TKI crizotinib, alectinib, and ceritinib was found in several cell models and one mouse model in recent studies (160-162). The activation of EGFR pathway induced by TGFα contributed to the resistance to alectinib, and upon TGFα knockdown, the sensitivity of alectinib in H3122-alecinib resistant NSCLC cells was restored (162). Epidermal growth factor (EGF) was also found to induce resistance to alectinib by activating EGFR signaling (161). Furthermore, through dual targeting of ALK and EGFR with alectinib and afatinib in mouse xenograft model, EGFR downstream signals to PI3K/AKT and MAPK were inhibited, and tumor volume decreased significantly (162). Similar study also discussed acquired resistance to ceritinib through EGFR bypass signaling activation in H3122 cells (160). Besides EGFR signaling, increased expression levels of other members of ERBB family such as ERBB2/3 induced by EGF were reported to be mechanisms contributing to resistance to ALK-TKI in EML4-ALK positive H3122 cells (163). Dual inhibition of ALK/ERBB family by shRNA and dacomitinib showed further antiproliferative effects in DFCI076 NSCLC cells that are resistant to both ALK inhibitor crizotinib and TAE684 (164). Similarly, crizotinib plus afatinib that inhibited ALK, EGFR, and ERBB2 signaling was able to inhibit the growth of H3122-crizotinib resistant cells (165), confirming EGFR activation as a resistance mechanism for ALK-TKI. However, if the cell line is resistant to both of the inhibitor, then even a combined inhibition might not work. In a comprehensive study by Katayama et al. (120), H3122 CR3 cells which were resistant to both crizotinib and gefitinib remained less sensitive to the combination of crizotinib plus gefitinib compared to crizotinib alone.
MET activation
Although mentioned in a smaller number of studies, MET activation also induces resistance to ALK-TKI. Hepatocyte growth factor (HGF) induced MET activation and triggered resistance to crizotinib and TAE684. HGF stimulated the phosphorylation of MET and its adaptor protein, GAB1, and activated downstream AKT and ERK1/2 pathways thus finally cause resistance to TAE684 (166). Another study also reported that HGF induced resistance to alectinib in H3122 and H2228 cell lines (167). MET activation but not gene amplification was observed in tissues from patients with ALK rearrangement (168). However, in another study, circulating tumor cells and ctDNA were analyzed by targeted NGS, and MET amplification up to sevenfold was detected after initiating crizotinib treatment (169). Further studies are still required to demonstrate whether MET amplification contribute to resistance to ALK-TKI.
Activation of KIT or IGF1R
Besides the activation of EGFR or MET signaling, a few studies discussed KIT or IGF1R signaling, and how they contributed to resistance to ALK-TKI. ALK positive H3122 cells with KIT overexpression showed sensitive to crizotinib. However, in the presence of stroma-derived stem cell factor (SCF), KIT-overexpressing H3122 cells exhibited high resistance to crizotinib through activation of downstream intermediates ERK and AKT (120). In another study that involved patients receiving crizotinib treatment, a significant decrease in PFS was correlated with high phosphorylation level of KIT in ALK-positive patients (170). Activating KIT mutation D816G was also identified in crizotinib-resistant cells, however, until now, only in ROS1-positive cell lines (171).
Through a special case of a patient with ALK-fusion who significantly responded to IGF1R-specific antibody, the combination of ALK plus IGF1R inhibitors was investigated in H3122 cell model and confirmed an enhanced antiproliferative response (122). This synergistic effect was verified by subsequent study in NSCLC (172). Furthermore, cellular dependence on ALK decreased because of increased IGF1R signaling induced by stimulation. IGF1R/insulin receptor substrate 1 (IRS1) signaling in the presence of ALK inhibitor therefore became a mechanism by which cells evade ALK blockade (122). In vitro experiments have confirmed that both ALK and IGF1R activation are inhibited after treatment with ceritinib or TAE684 (173), however, the clinical utility of these two ALK-TKIs in inhibiting ALK/IGF1R still remains to be defined.
Other novel resistance mechanisms
Neuregulin 1 (NRG1) fusion
CD98hc (SLC3A2, solute carrier family 3 member 2) is the heavy chain of CD98 and forms large neutral amino acid transporter LAT1 (SLC7A5) in cells. Overexpression of SLC3A2 occurs widely in cancer cells and is associated with poor clinical prognosis (174). SLC3A2 is upregulated in human osteosarcoma and promotes tumor growth through the PI3K/AKT signaling pathway (175).
The NRG1 gene encodes the growth factor NRG1; members of the NRG1 family are structurally related to EGF and stimulate ERBB3 RTKs (176). The EGF-like domain of NRG1 in the SLC3A2-NRG1 chimeric protein was shown to be critical for NSCLC proliferation and tumorigenesis (177). A SLC3A2-NRG1 fusion protein activated the formation and phosphorylation of the ERBB2-ERBB3 heterocomplex and downstream PI3K/AKT/mTOR signaling pathway. Inhibition of both ERBB2 and ERBB3 blocked the downstream signaling intermediates AKT and ERK (178). Therefore, the dual inhibition of ERBB2/3 might be a suitable strategy to block the signals activated by SLC3A2-NRG1.
CD74 is the most common NRG1 fusion partner, and CD74-NRG1 fusion occurs in approximately 1.7% of patients with lung adenocarcinomas (179). CD74-NRG1 increases the expression and phosphorylation of the EGF-like domain of NRG1 III-β3 and leads to the heterodimerization of ERBB3 and ERBB2, subsequently activating the downstream PI3K/AKT pathway (179). An increase in the NRG1 ligand level was directly related to resistance to crizotinib treatment (180). Furthermore, the treatment of NSCLC cells with second-generation ALK inhibitors activated EGFR family pathways through activation of the NRG1-ERBB3-EGFR axis (181,182). Therefore, activation of the NRG1/ERBB3 pathway is a potential mechanism of TKI resistance.
Invasive mucinous adenocarcinoma (IMA) is a highly malignant type of lung adenocarcinoma that is mainly caused by KRAS mutations. However, an aberrant, novel tumor driver, the CD74-NRG1 fusion gene, was also found to contribute to KRAS-negative IMA tumorigenesis (179). CD74-NRG1 and KRAS mutations are mutually exclusive. Expression of the CD74-NRG1 protein not only induced sphere formation in vitro but also enhanced tumor initiation in vivo. The CD74-NRG1 protein activates the PI3K/AKT/NFKB signaling pathway, leading the IGF2 autocrine/paracrine circuit to initiate and maintain cells with cancer stem cell properties. IGF1R, the CD74-NRG1 receptor, was enhanced in an NFKB-dependent manner in cells expressing CD74-NRG1 (183).
Other NRG1 fusion proteins such as SDC4-NRG1 and ALK-NGR1 have been reported in some studies (184,185). Although SDC4-NRG1 fusion displays a rapid and durable PR to afatinib and NRG1 has been shown to respond to EGFR and ERBB2/3 inhibitors in the preclinical setting (184,186,187), targeting downstream of NRG1 through the direct inhibition of ERBB3 and other molecules in this pathway is thought to be a better strategy for clinical application than the use of a broad EGFR/ERBB family inhibitor to target mutant tyrosine kinases (183,184).
Overexpression of MET and BCL-2
AC0010 is a pyrrolopyrimidine-based, irreversible, third-generation EGFR inhibitor that selectively inhibits EGFR-activating and T790M mutations with an up to 298-fold increase in potency compared with its inhibition of wild-type EGFR (188). A phase I study of AC0010 suggested that AC0010 has a well-tolerated safety profile and shows promising antitumor activity in NSCLC patients with acquired resistance to a first-generation EGFR-TKIs (189). However, AC0010 cannot overcome resistance caused by the overexpression of MET and BCL-2. The BCL-2 inhibitor navitoclax inhibited cell growth in the AC0010-resistant cell line H1975-AVR1 (97,138). Navitoclax together with gefitinib showed an enhanced ability to eradicate NSCLC cells (190). Combination treatment with AC0010 and crizotinib inhibited the growth of H1975-P1-R1 cells, and synergistic effects with a 73.5% inhibitory rate at a nontoxic dose were observed (138). Thus, the overexpression of BCL-2 and MET is responsible for acquired resistance to AC0010 in NSCLC.
Immunotherapy
Tumors can evade immune detection by exploiting inhibitory immune checkpoints, such as the PD-1/PD-L1 pathway. PD-1 signaling, which is driven primarily by the adaptive expression of PD-L1 within the tumor, represses the ability of T lymphocytes to recognize tumor-specific antigens, resulting in tumor progression and metastasis (191). The PD-1/PD-L1 expression level is mediated directly by EGFR, ALK, and the presence of TKIs that exert their effects by influencing the expression level of EGFR and ALK and downstream signaling cascades. Studies have shown that the expression level of PD-L1 is significantly upregulated in NSCLC cell lines expressing an EGFR driver mutation and the EML4-ALK fusion protein (192,193). The EGFR-TKIs gefitinib and erlotinib both increased the expression of PD-L1 in NSCLC cell lines with mutant EGFR (194).
PD-1/PD-L1 ICIs
The introduction of ICIs, such as monoclonal antibodies that target CTLA-4 and PD-1/PD-L1, has shed light on a new strategy to treat NSCLC patients. Currently, three widely used agents (two PD-1 inhibitors, nivolumab and pembrolizumab, and one PD-L1 inhibitor, atezolizumab) are approved as standard treatment options for pretreated NSCLC patients. Clinical trials of durvalumab and avelumab are underway.
A phase III study of nivolumab revealed the increased overall survival (OS) of patients with advanced-stage NSCLC who exhibited disease progression after cytotoxic therapy treated with nivolumab (12.2 months) compared to the OS of those treated with docetaxel (9.4 months) for the first time (195). Other studies have also shown the increased OS of nivolumab-treated NSCLC patients (17%) compared to that of docetaxel-treated NSCLC patients (8%) (196). Pembrolizumab and atezolizumab also showed greater efficacy than platinum-based cytotoxic therapy, with a PFS of 10.3 months observed in pembrolizumab-treated patients compared to 6.0 months in a chemotherapy group. A significant improvement in OS was also observed (197). A similar comparison of atezolizumab and docetaxel also showed that atezolizumab can increase OS in patients, although how the effects of atezolizumab treatment on the ORR and PFS differ from those of docetaxel still needs further investigation (198,199). ICIs combined with chemotherapy can act synergistically to improve treatment effects. The combination of pembrolizumab and pemetrexed/platinum-based drugs significantly improved OS and PFS in patients with untreated metastatic nonsquamous NSCLC with no EGFR or ALK mutations (200).
Durvalumab and avelumab were developed more recently than the three other aforementioned ICIs, so an investigation of their efficacy compared to that of regular chemotherapy is underway. Some evidence already suggests the utility of durvalumab to treat NSCLC patients, as a significantly longer PFS (16.8 months with durvalumab vs. 5.6 months with placebo) was observed in patients with stage III NSCLC without disease progression after platinum-based chemoradiotherapy (201,202). Another phase III trial showed that avelumab did not increase OS compared to that following docetaxel treatment in PD-L1-positive NSCLC patients previously treated with platinum-based chemoradiotherapy. However, patients undergoing avelumab treatment had fewer adverse reactions, showing the favorable safety profile of avelumab (203).
Combining anti-PD-(L)1 with anti-CTLA-4
The expression of CTLA-4 delivers an inhibitory signal into T-cells, inhibiting T-cell activation (204). Thus, the dual targeting of PD-(L)1 and CTLA-4 may produce a greater and more durable tumor response than PD-(L)1 inhibition alone. One critical study showed that nivolumab monotherapy (1-year PFS rate of 29%) was less beneficial in patients with a higher tumor mutational burden (TMB), while the combination of nivolumab and ipilimumab (1-year PFS rate of 42%) was significantly effective (205). Nivolumab plus ipilimumab was also shown to induce a strong and durable response and exhibited a tolerable safety profile in clinical trials (206). However, combination therapy with pembrolizumab plus ipilimumab was associated with increased toxicity in 51 patients with advanced NSCLC, despite the high antitumor activity of the two drugs (207).
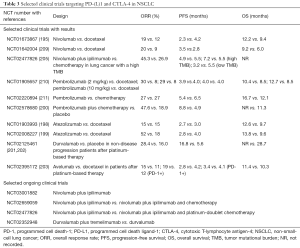
When tremelimumab, another CTLA-4 antibody, was used with the PD-L1 inhibitor durvalumab in a phase I study, the complete suppression of PD-L1 was observed in most patients, indicating effective targeting by durvalumab. Additionally, increased peripheral T-cell activation and proliferation were observed in patients treated with durvalumab plus tremelimumab compared with that in patients treated with durvalumab monotherapy, even when tremelimumab was administered at the lowest dose (208), demonstrating the synergistic effect of dual PD-L1 and CTLA-4 inhibition. Another clinical trial (NCT02352948) of durvalumab plus tremelimumab treatment is ongoing. However, how dual PD-(L)1 and CTLA-4 blockade improves the response rate compared with that following PD-(L)1 blockade monotherapy remains unclear. Table 3 contains information on finished and ongoing anti-PD(L)1/CTLA-4 clinical trials for further reference (209-211). Recent studies (197,212-215) showed significant correlation between PD-(L)1/CTLA-4 expression levels and the effects of immunotherapy, low PD-(L)1 expression levels were linked with poor responses.
Full table
TMB predicts responses to immunotherapy
Immunotherapy has changed the landscape of NSCLC treatment. However, just like ctDNA as a biomarker to predict the response of TKI therapy, novel biomarkers are being evaluated for their potential to provide precise and dynamic prediction of the response to immunotherapy. Although this evaluation is only at its early stage, biomarkers such as PD-(L)1 expression level, the presence of EGFR/ALK mutations, TMB, and microsatellite instability were all associated with the responses to immunotherapy (216,217). The dynamics of TMB were shown to be independent of PD-L1 expression (218,219), and greater benefit was consistently observed with high TMB and PD-L1 expression, suggesting the potential of TMB and PD-(L)-1 expression level as independent biomarkers (220). In patients with stage IV or recurrent NSCLC with a high TMB, the 1-year PFS rate was 43% with nivolumab plus ipilimumab treatment and 13% with chemotherapy. Notably, in patients with a low TMB, the PFS with nivolumab plus ipilimumab treatment was similar to that with chemotherapy, suggesting TMB as a biomarker (205). Several other studies also noted that higher TMB predicts favorable outcomes such as duration and survival of patients receiving anti-PD-1/PD-L1 therapy in NSCLC (216,219,221). Furthermore, TMB was significantly lower among patients with EGFR, ALK, or ROS1 mutation compared to wild-type (216). Unsolved questions on the mechanism of combined PD-(L)1 and CTLA-4 blockade and the synergy of this treatment regime with high TMBs remain.
Significant links between ctDNA, TKI targeted therapy, and immunotherapy
ctDNAs as prognostic markers to determine acquired resistance and the effectiveness of TKIs
Even when patients exhibit an initial disease progression event observed by the monitoring of ctDNA levels, continued TKI treatment [osimertinib (86), afatinib (222)] can remain effective. In a phase III trial, continued afatinib treatment after disease progression doubled the patient PFS (5.6 months with afatinib treatment vs. 2.8 months with chemotherapy) (222). When a drug continues to suppress the majority of tumor cells, compensatory pathways may be upregulated. Upon drug withdrawal, dramatic disease progression called “tumor flare” might occur (223). However, treatment should be based on each patient’s mutation profile; for instance, a patient with the EGFR T790M mutation might exhibit disease progression after rociletinib treatment for a certain duration (7). In this case, a change in EGFR-TKI from rociletinib to osimertinib seems to be a better strategy than continued rociletinib treatment. A prolonged PR to osimertinib after PD with rociletinib treatment was observed in EGFR T790M-positive NSCLC patients (224,225).
The ctDNA levels in EGFR T790M-positive patients exhibited a specific, similar, and sudden spike that developed at different time points during routine ctDNA profiling after TKI treatment, but this pattern was not observed in EGFR T790M-negative patients (70). This result illustrates the possibility of predicting cancer progression by analyzing ctDNA profiles and determining the possible mutation type. Analysis of ctDNA shedding in samples taken before and after disease progression showed a positive correlation between the level of ctDNA shedding and tumor burden, suggesting that disease in these non-shedding patients is entirely controlled at progression. The emergence of clinically important resistance may correlate with the presence of detectable ctDNA (226). Resistance caused by the EGFR T790M mutation has irreversible outcomes but can be detected by early ctDNA analysis, which can identify the mutation even before it has a large impact on patients (64).
EGFR T790M ctDNA profiling also provides solid evidence of the effectiveness of TKIs and indications for drug switching strategies. EGFR T790M was strongly correlated with large increases in mutant DNA levels. This strongly suggests switching from first-generation TKIs, which no longer effectively control disease progression, to third-generation TKIs with the need of intervention at an earlier time (67). Therefore, EGFR T790M has emerged as a favorable prognostic marker to determine acquired resistance and an important predictive marker for the effectiveness of TKIs.
ctDNAs as predictive markers for immunotherapy to assess pseudoprogression and blood TMB (bTMB)
ctDNAs are widely used as an efficient clinical tool to monitor the effects of chemotherapy and targeted therapy. Recently, more and more studies focused on the application of ctDNA assays in monitoring responses of immunotherapy by ICIs.
ctDNA levels correlated with tumor progression in patients receiving anti-CTLA-4 and anti-PD-L1 therapy, and pseudoprogression was revealed by undetectable ctDNA 3 weeks prior to clinical improvement (227). Other studies (228-230) also proved the efficiency of ctDNA, decreased ctDNA levels were observed in response to treatment several weeks after initiation. Guibert et al. (231) monitored responses to anti-PD-1 treatment of KRAS mutated lung adenocarcinoma by ddPCR on plasma ctDNA and discriminated pseudo from true progression. Patients with pseudoprogression often began with undetectable ctDNA at baseline or detectable ctDNA at baseline followed by a greater than 10-fold decrease in ctDNA level in response to treatment (232); these patients with low or undetected ctDNA level at the beginning of and during therapy often have better responses to immunotherapy (233). Patients with more than 50% decrease compared to ctDNA baseline levels showed superior PFS and OS than those by less than 50% (234). Baseline ctDNA level, and ctDNA level post-treatment, could be prognostic factors in patients receiving ICIs.
ctDNA has been investigated as a predictive marker that reveals TMB in response to immunotherapy (235-237). TMB assessed by targeted NGS was significantly associated with improved benefit among patients with NSCLC treated with ICIs (238). Recent studies (239-241) investigated bTMB profiled with ctDNA sequencing and whether it predicts responses to immunotherapy. bTMB quantified by NGS were highly concordant with tissue TMB (tTMB), so that TMB can be accurately measured in plasma. bTMB also associated with clinical benefit such as longer PFS in patients treated with atezolizumab (240). Although there are only a few relative studies, researchers positively concluded that bTMB quantified by ctDNA had the potential to be a novel biomarker for prognosis in patients with NSCLC treated by immunotherapy. However, due to the limitation of sequencing depth, in order to quantify TMB by ctDNA, mutations with a minimum allele frequency must be above 1% in ctDNA for a bTMB score to be valid (240).
Combining TKI therapy and immunotherapy
Many studies have pointed out, EGFR/ALK status in NSCLC cell lines were correlated with the expression level of PD-(L)1. For example, EGFR activating mutations, EGFR T790M, EML4-ALK fusion, and MET overexpression were significantly associated with increased expression of PD-L1 (192,193). However, the number of studies investigating how TKI treatment affects immunotherapy is still limited.
Although EGFR mutations or ALK rearrangements were associated with low ORRs to PD-1/PD-L1 inhibitors (242), and EGFR T790M upregulated PD-L1 level (243); It is contradictory to our current understanding that Haratani et al. (244) found out that 25 patients with EGFR mutation but negative for T790M benefited more from nivolumab treatment after EGFR-TKI therapy, possibly because of higher PD-L1 expression levels in those T790M-negative patients. A few nivolumab responders with high levels of PD-L1 expression also experienced copy number gain for MET. Prospective clinical trials are required to confirm the efficacy of anti-PD-1 treatment for EGFR T790M-negative patients. Another recent study (245) focused on combining erlotinib/gefitinib with pembrolizumab as first-line therapy for NSCLC patients with EGFR sensitizing mutations. ORRs were 41.7% and 14.3% in pembrolizumab plus erlotinib and pembrolizumab plus gefitinib, respectively. However, there were no improvement of ORRs compared to previous monotherapies, and 5 in 7 patients receiving pembrolizumab plus gefitinib were observed to have drug-related liver toxicity. Patients received pembrolizumab plus erlotinib were observed to have adverse events similar to monotherapy, demonstrating the safety profile of this combination. Combining immunotherapy with EGFR-TKI therapy might be reasonable, nevertheless, due to the small size of cohort in this study, further evaluation is still required. In another clinical trial (NCT02013219) (246), atezolizumab plus erlotinib demonstrated a manageable safety profile and achieved ORR of 75%, a great improvement from previous atezolizumab monotherapy. Trial (NCT02088112) (247) on durvalumab plus gefitinib achieved ORR of 78.9%. Trials ongoing (NCT02013219, NCT02088112, NCT02364609, and NCT02143466) all involve the combination of EGFR/ALK-TKI therapy with immunotherapy, a novel treatment strategy that is still at an early stage of exploration.
Future prospects
Decades have passed since researchers first found that mutations in the KRAS, EGFR and ALK genes were related to lung cancer tumorigenesis. Lung cancer treatment has evolved from regular chemotherapy to targeted therapy using RTK inhibitors that diminish only cancer cells with certain gene mutations and to immunotherapy that utilizes our immune system to efficiently and precisely attack tumor cells through regulating PD-(L)1 and CTLA-4 or the genetic engineering of T-cells to perform chimeric antigen receptor T-cell immunotherapy (CAR-T). Each treatment revolution has helped us tackle the world’s second largest disease, which causes many deaths. Lots of ongoing studies continue to discuss the potential of TMB as a novel biomarker for immunotherapy, and using ctDNA to calculate bTMB as a more convenient and dynamic approach. Furthermore, ongoing trials on the combination of targeted therapy and immunotherapy might shed light on new strategies for NSCLC treatment.
Genetic profiling is the first step in targeted treatment, and serum or urine ctDNA sampling has greatly simplified the whole biopsy process, making it simpler than tissue biopsy. ctDNA sampling also enables the noninvasive, sensitive and dynamic capture of DNA information from heterogeneous tumors. ctDNA is often used in targeted therapy as a tool to aid in determining treatment efficacy and predicting prognosis.
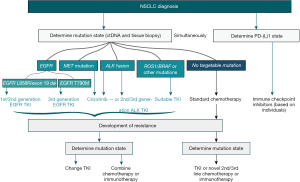
Although targeted therapy is effective, there are still many cases of developed resistance. Three generations of TKIs have been developed to treat the clinically crucial RTKs in NSCLC EGFR and ALK; however, different TKI resistance mechanisms, such as secondary RTK mutations, gene fusion or signal activation bypass, occur for each drug despite our increasing understanding of the biology and treatment of NSCLC. Figure 2 describes the clinical strategies used to manage resistance in NSCLC. NSCLC patients still face many challenges, such as relatively poor survival rates and frequent metastasis. We hope that the rapid development of targeted therapy and immunotherapy will facilitate the continuous development of new drugs and effective treatment strategies for NSCLC patients.
Acknowledgments
Funding: This research was supported by the Fundamental Research Funds of Zhejiang Sci-Tech University; National Natural Science Foundation of China (81728012); Zhejiang Xinmiao Talents Program (2018R406031); National Undergraduate Training Program for Innovation and Entrepreneurship (201910338028), China.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Travis WD, Brambilla E, Nicholson AG, et al. The 2015 World Health Organization classification of lung tumors: impact of genetic, clinical and radiologic advances since the 2004 classification. J Thorac Oncol 2015;10:1243-60. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014;511:543-50. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature 2012;489:519-25. [Crossref] [PubMed]
- Kobayashi S, Boggon TJ, Dayaram T, et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med 2005;352:786-92. [Crossref] [PubMed]
- Remon J, Caramella C, Jovelet C, et al. Osimertinib benefit in EGFR-mutant NSCLC patients with T790M-mutation detected by circulating tumour DNA. Ann Oncol 2017;28:784-90. [Crossref] [PubMed]
- Piotrowska Z, Niederst MJ, Karlovich CA, et al. Heterogeneity underlies the emergence of EGFRT790 wild-type clones following treatment of T790M-positive cancers with a third-generation EGFR inhibitor. Cancer Discov 2015;5:713-22. [Crossref] [PubMed]
- Kobayashi Y, Fujino T, Nishino M, et al. EGFR T790M and C797S mutations as mechanisms of acquired resistance to dacomitinib. J Thorac Oncol 2018;13:727-31. [Crossref] [PubMed]
- Engelman JA, Zejnullahu K, Mitsudomi T, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science 2007;316:1039-43. [Crossref] [PubMed]
- Shi P, Oh YT, Zhang G, et al. Met gene amplification and protein hyperactivation is a mechanism of resistance to both first and third generation EGFR inhibitors in lung cancer treatment. Cancer Lett 2016;380:494-504. [Crossref] [PubMed]
- Cortot AB, Repellin CE, Shimamura T, et al. Resistance to irreversible EGF receptor tyrosine kinase inhibitors through a multistep mechanism involving the IGF1R pathway. Cancer Res 2013;73:834-43. [Crossref] [PubMed]
- Rong X, Liang Y, Han Q, et al. Molecular mechanisms of tyrosine kinase inhibitor resistance induced by membranous/cytoplasmic/nuclear translocation of epidermal growth factor receptor. J Thorac Oncol 2019;14:1766-83. [Crossref] [PubMed]
- Gainor JF, Varghese AM, Ou SH, et al. ALK rearrangements are mutually exclusive with mutations in EGFR or KRAS: an analysis of 1,683 patients with non-small cell lung cancer. Clin Cancer Res 2013;19:4273-81. [Crossref] [PubMed]
- Rikova K, Guo A, Zeng Q, et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell 2007;131:1190-203. [Crossref] [PubMed]
- Takeuchi K, Choi YL, Soda M, et al. Multiplex reverse transcription-PCR screening for EML4-ALK fusion transcripts. Clin Cancer Res 2008;14:6618-24. [Crossref] [PubMed]
- Pylayeva-Gupta Y, Grabocka E, Bar-Sagi D. RAS oncogenes: weaving a tumorigenic web. Nat Rev Cancer 2011;11:761-74. [Crossref] [PubMed]
- Dogan S, Shen R, Ang DC, et al. Molecular epidemiology of EGFR and KRAS mutations in 3,026 lung adenocarcinomas: higher susceptibility of women to smoking-related KRAS-mutant cancers. Clin Cancer Res 2012;18:6169-77. [Crossref] [PubMed]
- Herbst RS, Morgensztern D, Boshoff C. The biology and management of non-small cell lung cancer. Nature 2018;553:446-54. [Crossref] [PubMed]
- Pao W, Wang TY, Riely GJ, et al. KRAS mutations and primary resistance of lung adenocarcinomas to gefitinib or erlotinib. PLoS Med 2005;2:e17. [Crossref] [PubMed]
- Linardou H, Dahabreh IJ, Kanaloupiti D, et al. Assessment of somatic k-RAS mutations as a mechanism associated with resistance to EGFR-targeted agents: a systematic review and meta-analysis of studies in advanced non-small-cell lung cancer and metastatic colorectal cancer. Lancet Oncol 2008;9:962-72. [Crossref] [PubMed]
- D’Incecco A, Andreozzi M, Ludovini V, et al. PD-1 and PD-L1 expression in molecularly selected non-small-cell lung cancer patients. Br J Cancer 2015;112:95-102. [Crossref] [PubMed]
- Chen N, Fang W, Lin Z, et al. KRAS mutation-induced upregulation of PD-L1 mediates immune escape in human lung adenocarcinoma. Cancer Immunol Immunother 2017;66:1175-87. [Crossref] [PubMed]
- Ji M, Liu Y, Li Q, et al. PD-1/PD-L1 expression in non-small-cell lung cancer and its correlation with EGFR/KRAS mutations. Cancer Biol Ther 2016;17:407-13. [Crossref] [PubMed]
- Scheel AH, Ansen S, Schultheis AM, et al. PD-L1 expression in non-small cell lung cancer: correlations with genetic alterations. Oncoimmunology 2016;5:e1131379. [Crossref] [PubMed]
- Gerlinger M, Rowan AJ, Horswell S, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med 2012;366:883-92. [Crossref] [PubMed]
- Murtaza M, Dawson SJ, Tsui DW, et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature 2013;497:108-12. [Crossref] [PubMed]
- Diaz LA Jr, Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol 2014;32:579-86. [Crossref] [PubMed]
- Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer 2011;11:426-37. [Crossref] [PubMed]
- Jahr S, Hentze H, Englisch S, et al. DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res 2001;61:1659-65. [PubMed]
- Nagata S. Apoptotic DNA fragmentation. Exp Cell Res 2000;256:12-8. [Crossref] [PubMed]
- Jiang P, Chan CW, Chan KC, et al. Lengthening and shortening of plasma DNA in hepatocellular carcinoma patients. Proc Natl Acad Sci U S A 2015;112:E1317-25. [Crossref] [PubMed]
- Mouliere F, Chandrananda D, Piskorz AM, et al. Enhanced detection of circulating tumor DNA by fragment size analysis. Sci Transl Med 2018. [Crossref] [PubMed]
- Underhill HR, Kitzman JO, Hellwig S, et al. Fragment length of circulating tumor DNA. PLoS Genet 2016;12:e1006162. [Crossref] [PubMed]
- Yamamoto Y, Uemura M, Fujita M, et al. Clinical significance of the mutational landscape and fragmentation of circulating tumor DNA in renal cell carcinoma. Cancer Sci 2019;110:617-28. [Crossref] [PubMed]
- Mouliere F, El Messaoudi S, Gongora C, et al. Circulating cell-free DNA from colorectal cancer patients may reveal high KRAS or BRAF mutation load. Transl Oncol 2013;6:319-28. [Crossref] [PubMed]
- Jiang P, Sun K, Tong YK, et al. Preferred end coordinates and somatic variants as signatures of circulating tumor DNA associated with hepatocellular carcinoma. Proc Natl Acad Sci U S A 2018;115:E10925-33. [Crossref] [PubMed]
- Bartels S, Persing S, Hasemeier B, et al. Molecular analysis of circulating cell-free DNA from lung cancer patients in routine laboratory practice: a cross-platform comparison of three different molecular methods for mutation detection. J Mol Diagn 2017;19:722-32. [Crossref] [PubMed]
- Chabon JJ, Simmons AD, Lovejoy AF, et al. Circulating tumour DNA profiling reveals heterogeneity of EGFR inhibitor resistance mechanisms in lung cancer patients. Nat Commun 2016;7:11815. [Crossref] [PubMed]
- Sher YP, Shih JY, Yang PC, et al. Prognosis of non-small cell lung cancer patients by detecting circulating cancer cells in the peripheral blood with multiple marker genes. Clin Cancer Res 2005;11:173-9. [PubMed]
- Hayes DC, Secrist H, Bangur CS, et al. Multigene real-time PCR detection of circulating tumor cells in peripheral blood of lung cancer patients. Anticancer Res 2006;26:1567-75. [PubMed]
- Paci M, Maramotti S, Bellesia E, et al. Circulating plasma DNA as diagnostic biomarker in non-small cell lung cancer. Lung Cancer 2009;64:92-7. [Crossref] [PubMed]
- Paez JG, Janne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 2004;304:1497-500. [Crossref] [PubMed]
- Zhang YL, Yuan JQ, Wang KF, et al. The prevalence of EGFR mutation in patients with non-small cell lung cancer: a systematic review and meta-analysis. Oncotarget 2016;7:78985-93. [PubMed]
- Xu T, Kang X, You X, et al. Cross-platform comparison of four leading technologies for detecting EGFR mutations in circulating tumor DNA from non-small cell lung carcinoma patient plasma. Theranostics 2017;7:1437-46. [Crossref] [PubMed]
- Taniguchi K, Uchida J, Nishino K, et al. Quantitative detection of EGFR mutations in circulating tumor DNA derived from lung adenocarcinomas. Clin Cancer Res 2011;17:7808-15. [Crossref] [PubMed]
- Forshew T, Murtaza M, Parkinson C, et al. Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Sci Transl Med 2012;4:136ra68. [Crossref] [PubMed]
- Liu Y, Liu B, Li XY, et al. A comparison of ARMS and direct sequencing for EGFR mutation analysis and tyrosine kinase inhibitors treatment prediction in body fluid samples of non-small-cell lung cancer patients. J Exp Clin Cancer Res 2011;30:111. [Crossref] [PubMed]
- Wang W, Song Z, Zhang Y. A comparison of ddPCR and ARMS for detecting EGFR T790M status in ctDNA from advanced NSCLC patients with acquired EGFR-TKI resistance. Cancer Med 2017;6:154-62. [Crossref] [PubMed]
- Yung TK, Chan KC, Mok TS, et al. Single-molecule detection of epidermal growth factor receptor mutations in plasma by microfluidics digital PCR in non-small cell lung cancer patients. Clin Cancer Res 2009;15:2076-84. [Crossref] [PubMed]
- Ma M, Shi C, Qian J, et al. Comparison of plasma and tissue samples in epidermal growth factor receptor mutation by ARMS in advanced non-small cell lung cancer. Gene 2016;591:58-64. [Crossref] [PubMed]
- Zhu G, Ye X, Dong Z, et al. Highly sensitive droplet digital pcr method for detection of EGFR-activating mutations in plasma cell-free DNA from patients with advanced non-small cell lung cancer. J Mol Diagn 2015;17:265-72. [Crossref] [PubMed]
- Watanabe M, Kawaguchi T, Isa SI, et al. Ultra-sensitive detection of the pretreatment EGFR T790M mutation in non-small cell lung cancer patients with an EGFR-activating mutation using droplet digital PCR. Clin Cancer Res 2015;21:3552-60. [Crossref] [PubMed]
- Vendrell JA, Mazieres J, Senal R, et al. Ultra-sensitive EGFR (T790M) detection as an independent prognostic marker for lung cancer patients harboring EGFR (del19) mutations and treated with first-generation TKIs. Clin Cancer Res 2019;25:4280-9. [Crossref] [PubMed]
- Ding PN, Becker T, Bray V, et al. Plasma next generation sequencing and droplet digital PCR-based detection of epidermal growth factor receptor (EGFR) mutations in patients with advanced lung cancer treated with subsequent-line osimertinib. Thorac Cancer 2019;10:1879-84. [Crossref] [PubMed]
- Li BT, Janku F, Jung B, et al. Ultra-deep next-generation sequencing of plasma cell-free DNA in patients with advanced lung cancers: results from the Actionable Genome Consortium. Ann Oncol 2019;30:597-603. [Crossref] [PubMed]
- Iwama E, Sakai K, Azuma K, et al. Monitoring of somatic mutations in circulating cell-free DNA by digital PCR and next-generation sequencing during afatinib treatment in patients with lung adenocarcinoma positive for EGFR activating mutations. Ann Oncol 2017;28:136-41. [Crossref] [PubMed]
- Guibert N, Hu Y, Feeney N, et al. Amplicon-based next-generation sequencing of plasma cell-free DNA for detection of driver and resistance mutations in advanced non-small cell lung cancer. Ann Oncol 2018;29:1049-55. [Crossref] [PubMed]
- Steendam CMJ, Atmodimedjo P, de Jonge E, et al. Plasma cell-free DNA testing of patients with EGFR mutant non-small-cell lung cancer: droplet digital PCR versus next-generation sequencing compared with tissue-based results. JCO Precis Oncol 2019;3:1-9.
- Vendrell JA, Taviaux S, Beganton B, et al. Detection of known and novel ALK fusion transcripts in lung cancer patients using next-generation sequencing approaches. Sci Rep 2017;7:12510. [Crossref] [PubMed]
- Lettig L, Sahnane N, Pepe F, et al. EGFR T790M detection rate in lung adenocarcinomas at baseline using droplet digital PCR and validation by ultra-deep next generation sequencing. Transl Lung Cancer Res 2019;8:584-92. [Crossref] [PubMed]
- Chang F, Li MM. Clinical application of amplicon-based next-generation sequencing in cancer. Cancer Genet 2013;206:413-9. [Crossref] [PubMed]
- Lynch TJ, Bell DW, Raffaella S, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med 2004;350:2129-39. [Crossref] [PubMed]
- Imamura F, Uchida J, Kukita Y, et al. Early responses of EGFR circulating tumor DNA to EGFR tyrosine kinase inhibitors in lung cancer treatment. Oncotarget 2016;7:71782-9. [Crossref] [PubMed]
- Husain H, Melnikova VO, Kosco K, et al. Monitoring daily dynamics of early tumor response to targeted therapy by detecting circulating tumor DNA in urine. Clin Cancer Res 2017;23:4716-23. [Crossref] [PubMed]
- Riediger AL, Dietz S, Schirmer U, et al. Mutation analysis of circulating plasma DNA to determine response to EGFR tyrosine kinase inhibitor therapy of lung adenocarcinoma patients. Sci Rep 2016;6:33505. [Crossref] [PubMed]
- Sequist LV, Waltman BA, Dias-Santagata D, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med 2011;3:75ra26. [Crossref] [PubMed]
- He J, Tan W, Tang X, et al. Variations in EGFR ctDNA correlates to the clinical efficacy of afatinib in non small cell lung cancer with acquired resistance. Pathol Oncol Res 2017;23:307-15. [Crossref] [PubMed]
- Imamura F, Uchida J, Kukita Y, et al. Monitoring of treatment responses and clonal evolution of tumor cells by circulating tumor DNA of heterogeneous mutant EGFR genes in lung cancer. Lung Cancer 2016;94:68-73. [Crossref] [PubMed]
- Hata A, Katakami N, Yoshioka H, et al. Spatiotemporal T790M heterogeneity in individual patients with EGFR-mutant non-small-cell lung cancer after acquired resistance to EGFR-TKI. J Thorac Oncol 2015;10:1553-9. [Crossref] [PubMed]
- Chen S, Zhao J, Cui L, et al. Urinary circulating DNA detection for dynamic tracking of EGFR mutations for NSCLC patients treated with EGFR-TKIs. Clin Transl Oncol 2017;19:332-40. [Crossref] [PubMed]
- Hussmann D, Madsen AT, Jakobsen KR, et al. IGF1R depletion facilitatesMET-amplification as mechanism of acquired resistance to erlotinib in HCC827 NSCLC cells. Oncotarget 2017;8:33300-15. [Crossref] [PubMed]
- Zhou J, Wang J, Zeng Y, et al. Implication of epithelial-mesenchymal transition in IGF1R-induced resistance to EGFR-TKIs in advanced non-small cell lung cancer. Oncotarget 2015;6:44332-45. [Crossref] [PubMed]
- Park JH, Choi YJ, Kim SY, et al. Activation of the IGF1R pathway potentially mediates acquired resistance to mutant-selective 3rd-generation EGF receptor tyrosine kinase inhibitors in advanced non-small cell lung cancer. Oncotarget 2016;7:22005-15. [PubMed]
- Tartarone A, Lerose R. Clinical approaches to treat patients with non-small cell lung cancer and epidermal growth factor receptor tyrosine kinase inhibitor acquired resistance. Ther Adv Respir Dis 2015;9:242-50. [Crossref] [PubMed]
- Juchum M, Gunther M, Laufer SA. Fighting cancer drug resistance: Opportunities and challenges for mutation-specific EGFR inhibitors. Drug Resist Updat 2015;20:12-28. [Crossref] [PubMed]
- Yu HA, Arcila ME, Rekhtman N, et al. Analysis of tumor specimens at the time of acquired resistance to EGFR-TKI therapy in 155 patients with EGFR-mutant lung cancers. Clin Cancer Res 2013;19:2240-7. [Crossref] [PubMed]
- Chic N, Mayo-de-Las-Casas C, Reguart N. Successful treatment with gefitinib in advanced non-small cell lung cancer after acquired resistance to osimertinib. J Thorac Oncol 2017;12:e78-80. [Crossref] [PubMed]
- Kobayashi Y, Azuma K, Nagai H, et al. Characterization of EGFR T790M, L792F, and C797S mutations as mechanisms of acquired resistance to afatinib in lung cancer. Mol Cancer Ther 2017;16:357-64. [Crossref] [PubMed]
- Niederst MJ, Hu H, Mulvey HE, et al. The allelic context of the C797S mutation acquired upon treatment with third-generation EGFR inhibitors impacts sensitivity to subsequent treatment strategies. Clin Cancer Res 2015;21:3924-33. [Crossref] [PubMed]
- Ercan D, Choi HG, Yun CH, et al. EGFR mutations and resistance to irreversible pyrimidine-based EGFR inhibitors. Clin Cancer Res 2015;21:3913-23. [Crossref] [PubMed]
- Morgillo F, Woo JK, Kim ES, et al. Heterodimerization of insulin-like growth factor receptor/epidermal growth factor receptor and induction of survivin expression counteract the antitumor action of erlotinib. Cancer Res 2006;66:10100-11. [Crossref] [PubMed]
- Yang JC, Sequist LV, Geater SL, et al. Clinical activity of afatinib in patients with advanced non-small-cell lung cancer harbouring uncommon EGFR mutations: a combined post-hoc analysis of LUX-Lung 2, LUX-Lung 3, and LUX-Lung 6. Lancet Oncol 2015;16:830-8. [Crossref] [PubMed]
- Yang JC, Shih JY, Su WC, et al. Afatinib for patients with lung adenocarcinoma and epidermal growth factor receptor mutations (LUX-Lung 2): a phase 2 trial. Lancet Oncol 2012;13:539-48. [Crossref] [PubMed]
- Ramalingam SS, O’Byrne K, Boyer M, et al. Dacomitinib versus erlotinib in patients with EGFR-mutated advanced nonsmall-cell lung cancer (NSCLC): pooled subset analyses from two randomized trials. Ann Oncol 2016;27:423-9. [Crossref] [PubMed]
- Wu YL, Cheng Y, Zhou X, et al. Dacomitinib versus gefitinib as first-line treatment for patients with EGFR-mutation-positive non-small-cell lung cancer (ARCHER 1050): a randomised, open-label, phase 3 trial. Lancet Oncol 2017;18:1454-66. [Crossref] [PubMed]
- Le X, Puri S, Negrao MV, et al. Landscape of EGFR-dependent and -independent resistance mechanisms to osimertinib and continuation therapy beyond progression in EGFR-mutant NSCLC. Clin Cancer Res 2018;24:6195-203. [Crossref] [PubMed]
- Oxnard GR, Hu Y, Mileham KF, et al. Assessment of resistance mechanisms and clinical implications in patients with EGFR T790M-positive lung cancer and acquired resistance to osimertinib. JAMA Oncol 2018;4:1527-34. [Crossref] [PubMed]
- Thress KS, Paweletz CP, Felip E, et al. Acquired EGFR C797S mutation mediates resistance to AZD9291 in non-small cell lung cancer harboring EGFR T790M. Nat Med 2015;21:560-2. [Crossref] [PubMed]
- Zhang Q, Zhang XC, Yang JJ, et al. EGFR L792H and G796R: two novel mutations mediating resistance to the third-generation EGFR tyrosine kinase inhibitor osimertinib. J Thorac Oncol 2018;13:1415-21. [Crossref] [PubMed]
- Sequist LV, Soria JC, Goldman JW, et al. Rociletinib in EGFR-mutated non-small-cell lung cancer. N Engl J Med 2015;372:1700-9. [Crossref] [PubMed]
- Patel H, Pawara R, Surana S. In-silico evidences for binding of glucokinase activators to EGFR C797S to overcome EGFR resistance obstacle with mutant-selective allosteric inhibition. Comput Biol Chem 2018;74:167-89. [Crossref] [PubMed]
- Wang S, Song Y, Liu D. EAI045: the fourth-generation EGFR inhibitor overcoming T790M and C797S resistance. Cancer Lett 2017;385:51-4. [Crossref] [PubMed]
- Jia Y, Yun CH, Park E, et al. Overcoming EGFR(T790M) and EGFR(C797S) resistance with mutant-selective allosteric inhibitors. Nature 2016;534:129-32. [Crossref] [PubMed]
- Godin-Heymann N, Ulkus L, Brannigan BW, et al. The T790M "gatekeeper" mutation in EGFR mediates resistance to low concentrations of an irreversible EGFR inhibitor. Mol Cancer Ther 2008;7:874-9. [Crossref] [PubMed]
- Wang Z, Yang JJ, Huang J, et al. Lung adenocarcinoma harboring EGFR T790M and in trans C797S responds to combination therapy of first- and third-generation EGFR TKIs and shifts allelic configuration at resistance. J Thorac Oncol 2017;12:1723-7. [Crossref] [PubMed]
- Suda K, Murakami I, Sakai K, et al. Heterogeneity in resistance mechanisms causes shorter duration of epidermal growth factor receptor kinase inhibitor treatment in lung cancer. Lung Cancer 2016;91:36-40. [Crossref] [PubMed]
- Hata AN, Niederst MJ, Archibald HL, et al. Tumor cells can follow distinct evolutionary paths to become resistant to epidermal growth factor receptor inhibition. Nat Med 2016;22:262-9. [Crossref] [PubMed]
- Graziano P, de Marinis F, Gori B, et al. EGFR-driven behavior and intrapatient T790M mutation heterogeneity of non-small-cell carcinoma with squamous histology. J Clin Oncol 2015;33:e115-8. [Crossref] [PubMed]
- Kuiper JL, Heideman DAM, Thunnissen E, et al. Incidence of T790M mutation in (sequential) rebiopsies in EGFR-mutated NSCLC-patients. Lung Cancer 2014;85:19-24. [Crossref] [PubMed]
- Oxnard GR, Arcila ME, Chmielecki J, et al. New strategies in overcoming acquired resistance to epidermal growth factor receptor tyrosine kinase inhibitors in lung cancer. Clin Cancer Res 2011;17:5530-7. [Crossref] [PubMed]
- Kim TM, Song A, Kim DW, et al. Mechanisms of acquired resistance to AZD9291: a mutation-selective, irreversible EGFR inhibitor. J Thorac Oncol 2015;10:1736-44. [Crossref] [PubMed]
- Zhao S, Li X, Zhao C, et al. Loss of T790M mutation is associated with early progression to osimertinib in Chinese patients with advanced NSCLC who are harboring EGFR T790M. Lung Cancer 2019;128:33-9. [Crossref] [PubMed]
- Soda M, Choi YL, Enomoto M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature 2007;448:561-6. [Crossref] [PubMed]
- Mitsudomi T, Morita S, Yatabe Y, et al. Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): an open label, randomised phase 3 trial. Lancet Oncol 2010;11:121-8. [Crossref] [PubMed]
- Maemondo M, Inoue A, Kobayashi K, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med 2010;362:2380-8. [Crossref] [PubMed]
- Park K, Yu CJ, Kim SW, et al. First-line erlotinib therapy until and beyond response evaluation criteria in solid tumors progression in Asian patients with epidermal growth factor receptor mutation-positive non-small-cell lung cancer: the ASPIRATION study. JAMA Oncol 2016;2:305-12. [Crossref] [PubMed]
- Goss G, Tsai CM, Shepherd FA, et al. Osimertinib for pretreated EGFR Thr790Met-positive advanced non-small-cell lung cancer (AURA2): a multicentre, open-label, single-arm, phase 2 study. Lancet Oncol 2016;17:1643-52. [Crossref] [PubMed]
- Mok TS, Wu YL, Ahn MJ, et al. Osimertinib or platinum-pemetrexed in EGFR T790M-positive lung cancer. N Engl J Med 2017;376:629-40. [Crossref] [PubMed]
- Soria JC, Ohe Y, Vansteenkiste J, et al. Osimertinib in untreated EGFR-mutated advanced non-small-cell lung cancer. N Engl J Med 2018;378:113-25. [Crossref] [PubMed]
- Shaw AT, Kim DW, Nakagawa K, et al. Crizotinib versus chemotherapy in advanced ALK-positive lung cancer. N Engl J Med 2013;368:2385-94. [Crossref] [PubMed]
- Crinò L, Ahn MJ, De Marinis F, et al. Multicenter phase II study of whole-body and intracranial activity with ceritinib in patients with ALK-rearranged non-small-cell lung cancer previously treated with chemotherapy and crizotinib: results from ASCEND-2. J Clin Oncol 2016;34:2866-73. [Crossref] [PubMed]
- Soria JC, Tan DSW, Chiari R, et al. First-line ceritinib versus platinum-based chemotherapy in advanced ALK-rearranged non-small-cell lung cancer (ASCEND-4): a randomised, open-label, phase 3 study. Lancet 2017;389:917-29. [Crossref] [PubMed]
- Shaw AT, Gandhi L, Gadgeel S, et al. Alectinib in ALK-positive, crizotinib-resistant, non-small-cell lung cancer: a single-group, multicentre, phase 2 trial. Lancet Oncol 2016;17:234-42. [Crossref] [PubMed]
- Peters S, Camidge DR, Shaw AT, et al. Alectinib versus crizotinib in untreated ALK-positive non-small-cell lung cancer. N Engl J Med 2017;377:829-38. [Crossref] [PubMed]
- Gettinger SN, Bazhenova LA, Langer CJ, et al. Activity and safety of brigatinib in ALK-rearranged non-small-cell lung cancer and other malignancies: a single-arm, open-label, phase 1/2 trial. Lancet Oncol 2016;17:1683-96. [Crossref] [PubMed]
- Solomon BJ, Besse B, Bauer TM, et al. Lorlatinib in patients with ALK-positive non-small-cell lung cancer: results from a global phase 2 study. Lancet Oncol 2018;19:1654-67. [Crossref] [PubMed]
- Kwak EL, Bang YJ, Camidge DR, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med 2010;363:1693-703. [Crossref] [PubMed]
- Solomon B, Soria JC. The continuum of care for ALK-positive NSCLC: from diagnosis to new treatment options - an overview. Ann Oncol 2016;27 Suppl 3:iii1-3. [Crossref] [PubMed]
- Solomon BJ, Mok T, Kim DW, et al. First-line crizotinib versus chemotherapy in ALK-positive lung cancer. N Engl J Med 2014;371:2167-77. [Crossref] [PubMed]
- Katayama R, Shaw AT, Khan TM, et al. Mechanisms of acquired crizotinib resistance in ALK-rearranged lung Cancers. Sci Transl Med 2012;4:120ra17. [Crossref] [PubMed]
- Dagogo-Jack I, Shaw AT. Crizotinib resistance: implications for therapeutic strategies. Ann Oncol 2016;27 Suppl 3:iii42-50. [Crossref] [PubMed]
- Lovly CM, McDonald NT, Chen H, et al. Rationale for co-targeting IGF-1R and ALK in ALK fusion-positive lung cancer. Nat Med 2014;20:1027-34. [Crossref] [PubMed]
- Choi YL, Soda M, Yamashita Y, et al. EML4-ALK mutations in lung cancer that confer resistance to ALK inhibitors. N Engl J Med 2010;363:1734-9. [Crossref] [PubMed]
- Doebele RC, Pilling AB, Aisner DL, et al. Mechanisms of resistance to crizotinib in patients with ALK gene rearranged non-small cell lung cancer. Clin Cancer Res 2012;18:1472-82. [Crossref] [PubMed]
- Toyokawa G, Hirai F, Inamasu E, et al. Secondary mutations at I1171 in the ALK gene confer resistance to both crizotinib and alectinib. J Thorac Oncol 2014;9:e86-7. [Crossref] [PubMed]
- Gainor JF, Dardaei L, Yoda S, et al. Molecular mechanisms of resistance to first- and second-generation ALK inhibitors in ALK-rearranged lung cancer. Cancer Discov 2016;6:1118-33. [Crossref] [PubMed]
- Gainor JF, Shaw AT. Emerging paradigms in the development of resistance to tyrosine kinase inhibitors in lung cancer. J Clin Oncol 2013;31:3987-96. [Crossref] [PubMed]
- Shaw AT, Kim DW, Mehra R, et al. Ceritinib in ALK-rearranged non-small-cell lung cancer. N Engl J Med 2014;370:1189-97. [Crossref] [PubMed]
- Kim DW, Tiseo M, Ahn MJ, et al. Brigatinib in patients with crizotinib-refractory anaplastic lymphoma kinase-positive non-small-cell lung cancer: a randomized, multicenter phase II trial. J Clin Oncol 2017;35:2490-8. [Crossref] [PubMed]
- Ou SH, Ahn JS, De Petris L, et al. Alectinib in crizotinib-refractory ALK-rearranged non-small-cell lung cancer: a phase II global study. J Clin Oncol 2016;34:661-8. [Crossref] [PubMed]
- Ceccon M, Mologni L, Bisson W, et al. Crizotinib-resistant NPM-ALK mutants confer differential sensitivity to unrelated ALK inhibitors. Mol Cancer Res 2013;11:122-32. [Crossref] [PubMed]
- Katayama R, Friboulet L, Koike S, et al. Two novel ALK mutations mediate acquired resistance to the next-generation ALK inhibitor alectinib. Clin Cancer Res 2014;20:5686-96. [Crossref] [PubMed]
- Friboulet L, Li N, Katayama R, et al. The ALK inhibitor ceritinib overcomes crizotinib resistance in non-small cell lung cancer. Cancer Discov 2014;4:662-73. [Crossref] [PubMed]
- Zou HY, Friboulet L, Kodack DP, et al. PF-06463922, an ALK/ROS1 inhibitor, overcomes resistance to first and second generation ALK inhibitors in preclinical models. Cancer Cell 2015;28:70-81. [Crossref] [PubMed]
- Zou HY, Li Q, Engstrom LD, et al. PF-06463922 is a potent and selective next-generation ROS1/ALK inhibitor capable of blocking crizotinib-resistant ROS1 mutations. Proc Natl Acad Sci U S A 2015;112:3493-8. [Crossref] [PubMed]
- Shaw AT, Felip E, Bauer TM, et al. Lorlatinib in non-small-cell lung cancer with ALK or ROS1 rearrangement: an international, multicentre, open-label, single-arm first-in-man phase 1 trial. Lancet Oncol 2017;18:1590-9. [Crossref] [PubMed]
- Suda K, Murakami I, Katayama T, et al. Reciprocal and complementary role of MET amplification and EGFR T790M mutation in acquired resistance to kinase inhibitors in lung cancer. Clin Cancer Res 2010;16:5489-98. [Crossref] [PubMed]
- Xu W, Tang W, Li T, et al. Overcoming resistance to AC0010, a third generation of EGFR inhibitor, by targeting c-MET and BCL-2. Neoplasia 2019;21:41-51. [Crossref] [PubMed]
- Ortiz-Cuaran S, Scheffler M, Plenker D, et al. Heterogeneous mechanisms of primary and acquired resistance to third-generation EGFR inhibitors. Clin Cancer Res 2016;22:4837-47. [Crossref] [PubMed]
- Martinez-Marti A, Felip E, Matito J, et al. Dual MET and ERBB inhibition overcomes intratumor plasticity in osimertinib-resistant-advanced non-small-cell lung cancer (NSCLC). Ann Oncol 2017;28:2451-7. [Crossref] [PubMed]
- Rosich L, Saborit-Villarroya I, López-Guerra M, et al. The phosphatidylinositol-3-kinase inhibitor NVP-BKM120 overcomes resistance signals derived from microenvironment by regulating the Akt/FoxO3a/Bim axis in chronic lymphocytic leukemia cells. Haematologica 2013;98:1739-47. [Crossref] [PubMed]
- Pan YH, Jiao L, Lin CY, et al. Combined treatment with metformin and gefitinib overcomes primary resistance to EGFR-TKIs with EGFR mutation via targeting IGF-1R signaling pathway. Biologics 2018;12:75-86. [PubMed]
- Guix M, Faber AC, Wang SE, et al. Acquired resistance to EGFR tyrosine kinase inhibitors in cancer cells is mediated by loss of IGF-binding proteins. J Clin Invest 2008;118:2609-19. [PubMed]
- Yamaoka T, Ohmori T, Ohba M, et al. Acquired resistance mechanisms to combination Met-TKI/EGFR-TKI exposure in met-amplified EGFR-TKI-resistant lung adenocarcinoma harboring an activating EGFR mutation. Mol Cancer Ther 2016;15:3040-54. [Crossref] [PubMed]
- Soucheray M, Capelletti M, Pulido I, et al. Intratumoral heterogeneity in EGFR-mutant NSCLC results in divergent resistance mechanisms in response to EGFR tyrosine kinase inhibition. Cancer Res 2015;75:4372-83. [Crossref] [PubMed]
- Shi S, Zhao J, Wang J, et al. HPIP silencing inhibits TGF-beta1-induced EMT in lung cancer cells. Int J Mol Med 2017;39:479-83. [Crossref] [PubMed]
- Zhang JX, Zhai JF, Yang XT, et al. MicroRNA-132 inhibits migration, invasion and epithelial-mesenchymal transition by regulating TGFβ1/Smad2 in human non-small cell lung cancer. Eur Rev Med Pharmacol Sci 2016;20:3793-801. [PubMed]
- Zhang F, Li T, Han L, et al. TGFβ1-induced down-regulation of microRNA-138 contributes to epithelial-mesenchymal transition in primary lung cancer cells. Biochem Biophys Res Commun 2018;496:1169-75. [Crossref] [PubMed]
- Tapon N, Harvey KF, Bell DW, et al. salvador Promotes both cell cycle exit and apoptosis in Drosophila and is mutated in human cancer cell lines. Cell 2002;110:467-78. [Crossref] [PubMed]
- Huang J, Wu S, Barrera J, et al. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell 2005;122:421-34. [Crossref] [PubMed]
- Harvey KF, Zhang X, Thomas DM. The Hippo pathway and human cancer. Nat Rev Cancer 2013;13:246-57. [Crossref] [PubMed]
- Zanconato F, Forcato M, Battilana G, et al. Genome-wide association between YAP/TAZ/TEAD and AP-1 at enhancers drives oncogenic growth. Nat Cell Biol 2015;17:1218-27. [Crossref] [PubMed]
- Lee BS, Park DI, Lee DH, et al. Hippo effector YAP directly regulates the expression of PD-L1 transcripts in EGFR-TKI-resistant lung adenocarcinoma. Biochem Biophys Res Commun 2017;491:493-9. [Crossref] [PubMed]
- McGowan M, Kleinberg L, Halvorsen AR, et al. NSCLC depend upon YAP expression and nuclear localization after acquiring resistance to EGFR inhibitors. Genes Cancer 2017;8:497-504. [PubMed]
- Hsu PC, You B, Yang YL, et al. YAP promotes erlotinib resistance in human non-small cell lung cancer cells. Oncotarget 2016;7:51922-33. [Crossref] [PubMed]
- Lee JE, Park HS, Lee D, et al. Hippo pathway effector YAP inhibition restores the sensitivity of EGFR-TKI in lung adenocarcinoma having primary or acquired EGFR-TKI resistance. Biochem Biophys Res Commun 2016;474:154-60. [Crossref] [PubMed]
- Liu XL, Zuo R, Ou WB. The hippo pathway provides novel insights into lung cancer and mesothelioma treatment. J Cancer Res Clin Oncol 2018;144:2097-106. [Crossref] [PubMed]
- Bazzani L, Donnini S, Finetti F, et al. PGE2/EP3/SRC signaling induces EGFR nuclear translocation and growth through EGFR ligands release in lung adenocarcinoma cells. Oncotarget 2017;8:31270-87. [Crossref] [PubMed]
- Brand TM, Iida M, Corrigan KL, et al. The receptor tyrosine kinase AXL mediates nuclear translocation of the epidermal growth factor receptor. Sci Signal 2017. [Crossref] [PubMed]
- Miyawaki M, Yasuda H, Tani T, et al. Overcoming EGFR bypass signal-induced acquired resistance to ALK tyrosine kinase inhibitors in ALK-translocated lung cancer. Mol Cancer Res 2017;15:106-14. [Crossref] [PubMed]
- Tanimoto A, Yamada T, Nanjo S, et al. Receptor ligand-triggered resistance to alectinib and its circumvention by Hsp90 inhibition in EML4-ALK lung cancer cells. Oncotarget 2014;5:4920-8. [Crossref] [PubMed]
- Tani T, Yasuda H, Hamamoto J, et al. Activation of EGFR bypass signaling by TGFalpha overexpression induces acquired resistance to alectinib in ALK-translocated lung cancer cells. Mol Cancer Ther 2016;15:162-71. [Crossref] [PubMed]
- Tanizaki J, Okamoto I, Okabe T, et al. Activation of HER family signaling as a mechanism of acquired resistance to ALK inhibitors in EML4-ALK-positive non-small cell lung cancer. Clin Cancer Res 2012;18:6219-26. [Crossref] [PubMed]
- Sasaki T, Koivunen J, Ogino A, et al. A novel ALK secondary mutation and EGFR signaling cause resistance to ALK kinase inhibitors. Cancer Res 2011;71:6051-60. [Crossref] [PubMed]
- Yamaguchi N, Lucena-Araujo AR, Nakayama S, et al. Dual ALK and EGFR inhibition targets a mechanism of acquired resistance to the tyrosine kinase inhibitor crizotinib in ALK rearranged lung cancer. Lung Cancer 2014;83:37-43. [Crossref] [PubMed]
- Yamada T, Takeuchi S, Nakade J, et al. Paracrine receptor activation by microenvironment triggers bypass survival signals and ALK inhibitor resistance in EML4-ALK lung cancer cells. Clin Cancer Res 2012;18:3592-602. [Crossref] [PubMed]
- Kogita A, Togashi Y, Hayashi H, et al. Activated MET acts as a salvage signal after treatment with alectinib, a selective ALK inhibitor, in ALK-positive non-small cell lung cancer. Int J Oncol 2015;46:1025-30. [Crossref] [PubMed]
- Feng Y, Minca EC, Lanigan C, et al. High MET receptor expression but not gene amplification in ALK 2p23 rearrangement positive non-small-cell lung cancer. J Thorac Oncol 2014;9:646-53. [Crossref] [PubMed]
- Berger LA, Janning M, Velthaus JL, et al. Identification of a high-level MET amplification in CTCs and cfTNA of an ALK-positive NSCLC patient developing evasive resistance to crizotinib. J Thorac Oncol 2018;13:e243-6. [Crossref] [PubMed]
- Yang H, Wang F, Deng Q, et al. Predictive and prognostic value of phosphorylated c-KIT and PDGFRA in advanced non-small cell lung cancer harboring ALK fusion. Oncol Lett 2019;17:3071-6. [PubMed]
- Dziadziuszko R, Le AT, Wrona A, et al. An activating KIT mutation induces crizotinib resistance in ROS1-positive lung cancer. J Thorac Oncol 2016;11:1273-81. [Crossref] [PubMed]
- Wilson C, Nimick M, Nehoff H, et al. ALK and IGF-1R as independent targets in crizotinib resistant lung cancer. Sci Rep 2017;7:13955. [Crossref] [PubMed]
- Galkin AV, Melnick JS, Kim S, et al. Identification of NVP-TAE684, a potent, selective, and efficacious inhibitor of NPM-ALK. Proc Natl Acad Sci U S A 2007;104:270-5. [Crossref] [PubMed]
- Estrach S, Lee SA, Boulter E, et al. CD98hc (SLC3A2) loss protects against ras-driven tumorigenesis by modulating integrin-mediated mechanotransduction. Cancer Res 2014;74:6878-89. [Crossref] [PubMed]
- Zhu B, Cheng D, Hou L, et al. SLC3A2 is upregulated in human osteosarcoma and promotes tumor growth through the PI3K/Akt signaling pathway. Oncol Rep 2017;37:2575-82. [Crossref] [PubMed]
- Gollamudi M, Nethery D, Liu J, et al. Autocrine activation of ErbB2/ErbB3 receptor complex by NRG-1 in non-small cell lung cancer cell lines. Lung Cancer 2004;43:135-43. [Crossref] [PubMed]
- Shin DH, Lee D, Hong DW, et al. Oncogenic function and clinical implications of SLC3A2-NRG1 fusion in invasive mucinous adenocarcinoma of the lung. Oncotarget 2016;7:69450-65. [Crossref] [PubMed]
- Shin DH, Jo JY, Han JY. Dual targeting of ERBB2/ERBB3 for the treatment of SLC3A2-NRG1-mediated lung cancer. Mol Cancer Ther 2018;17:2024-33. [Crossref] [PubMed]
- Fernandez-Cuesta L, Plenker D, Osada H, et al. CD74-NRG1 fusions in lung adenocarcinoma. Cancer Discov 2014;4:415-22. [Crossref] [PubMed]
- Kimura M, Endo H, Inoue T, et al. Analysis of ERBB ligand-induced resistance mechanism to crizotinib by primary culture of lung adenocarcinoma with EML4-ALK fusion gene. J Thorac Oncol 2015;10:527-30. [Crossref] [PubMed]
- Dong X, Fernandez-Salas E, Li E, et al. Elucidation of resistance mechanisms to second-generation ALK inhibitors alectinib and ceritinib in non-small cell lung cancer cells. Neoplasia 2016;18:162-71. [Crossref] [PubMed]
- Chen H, Zhang Q, Zhang Y, et al. Afatinib reverses ceritinib resistance (CR) in ALK/ROS1-positive non-small-cell lung cancer cell (NSCLC) via suppression of NRG1 pathway. Onco Targets Ther 2018;11:8201-9. [Crossref] [PubMed]
- Murayama T, Nakaoku T, Enari M, et al. Oncogenic fusion gene CD74-NRG1 confers cancer stem cell-like properties in lung cancer through a IGF2 autocrine/paracrine circuit. Cancer Res 2016;76:974-83. [Crossref] [PubMed]
- Jones MR, Lim H, Shen Y, et al. Successful targeting of the NRG1 pathway indicates novel treatment strategy for metastatic cancer. Ann Oncol 2017;28:3092-7. [Crossref] [PubMed]
- Muscarella LA, Trombetta D, Fabrizio FP, et al. ALK and NRG1 fusions coexist in a patient with signet ring cell lung adenocarcinoma. J Thorac Oncol 2017;12:e161-3. [Crossref] [PubMed]
- Nakaoku T, Tsuta K, Ichikawa H, et al. Druggable oncogene fusions in invasive mucinous lung adenocarcinoma. Clin Cancer Res 2014;20:3087-93. [Crossref] [PubMed]
- Meetze K, Vincent S, Tyler S, et al. Neuregulin 1 expression is a predictive biomarker for response to AV-203, an ERBB3 inhibitory antibody, in human tumor models. Clin Cancer Res 2015;21:1106-14. [Crossref] [PubMed]
- Xu X, Mao L, Xu W, et al. AC0010, an irreversible EGFR inhibitor selectively targeting mutated EGFR and overcoming T790M-induced resistance in animal models and lung cancer patients. Mol Cancer Ther 2016;15:2586-97. [Crossref] [PubMed]
- Ma Y, Zheng X, Zhao H, et al. First-in-human phase I study of AC0010, a mutant-selective EGFR inhibitor in non-small cell lung cancer: safety, efficacy, and potential mechanism of resistance. J Thorac Oncol 2018;13:968-77. [Crossref] [PubMed]
- Fan W, Tang Z, Yin L, et al. MET-independent lung cancer cells evading EGFR kinase inhibitors are therapeutically susceptible to BH3 mimetic agents. Cancer Res 2011;71:4494-505. [Crossref] [PubMed]
- Iwai Y, Ishida M, Tanaka Y, et al. Involvement of PD-L1 on tumor cells in the escape from host immune system and tumor immunotherapy by PD-L1 blockade. Proc Natl Acad Sci U S A 2002;99:12293-7. [Crossref] [PubMed]
- Ota K, Azuma K, Kawahara A, et al. Induction of PD-L1 expression by the EML4-ALK oncoprotein and downstream signaling pathways in non-small cell lung cancer. Clin Cancer Res 2015;21:4014-21. [Crossref] [PubMed]
- Azuma K, Ota K, Kawahara A, et al. Association of PD-L1 overexpression with activating EGFR mutations in surgically resected nonsmall-cell lung cancer. Ann Oncol 2014;25:1935-40. [Crossref] [PubMed]
- Chen N, Fang W, Zhan J, et al. Upregulation of PD-L1 by EGFR activation mediates the immune escape in EGFR-driven NSCLC: implication for optional immune targeted therapy for NSCLC patients with EGFR mutation. J Thorac Oncol 2015;10:910-23. [Crossref] [PubMed]
- Borghaei H, Paz-Ares L, Horn L, et al. Nivolumab versus docetaxel in advanced nonsquamous non-small-cell lung cancer. N Engl J Med 2015;373:1627-39. [Crossref] [PubMed]
- Vokes EE, Ready N, Felip E, et al. Nivolumab versus docetaxel in previously treated advanced non-small-cell lung cancer (CheckMate 017 and CheckMate 057): 3-year update and outcomes in patients with liver metastases. Ann Oncol 2018;29:959-65. [Crossref] [PubMed]
- Reck M, Rodriguez-Abreu D, Robinson AG, et al. Pembrolizumab versus chemotherapy for PD-L1-positive non-small-cell lung cancer. N Engl J Med 2016;375:1823-33. [Crossref] [PubMed]
- Fehrenbacher L, Spira A, Ballinger M, et al. Atezolizumab versus docetaxel for patients with previously treated non-small-cell lung cancer (POPLAR): a multicentre, open-label, phase 2 randomised controlled trial. Lancet 2016;387:1837-46. [Crossref] [PubMed]
- Rittmeyer A, Barlesi F, Waterkamp D, et al. Atezolizumab versus docetaxel in patients with previously treated non-small-cell lung cancer (OAK): a phase 3, open-label, multicentre randomised controlled trial. Lancet 2017;389:255-65. [Crossref] [PubMed]
- Gandhi L, Rodriguez-Abreu D, Gadgeel S, et al. Pembrolizumab plus chemotherapy in metastatic non-small-cell lung cancer. N Engl J Med 2018;378:2078-92. [Crossref] [PubMed]
- Antonia SJ, Villegas A, Daniel D, et al. Durvalumab after chemoradiotherapy in stage III non-small-cell lung cancer. N Engl J Med 2017;377:1919-29. [Crossref] [PubMed]
- Antonia SJ, Villegas A, Daniel D, et al. Overall survival with durvalumab after chemoradiotherapy in stage III NSCLC. N Engl J Med 2018;379:2342-50. [Crossref] [PubMed]
- Barlesi F, Vansteenkiste J, Spigel D, et al. Avelumab versus docetaxel in patients with platinum-treated advanced non-small-cell lung cancer (JAVELIN Lung 200): an open-label, randomised, phase 3 study. Lancet Oncol 2018;19:1468-79. [Crossref] [PubMed]
- Schneider H, Downey J, Smith A, et al. Reversal of the TCR stop signal by CTLA-4. Science 2006;313:1972-5. [Crossref] [PubMed]
- Huerter MM, Ganti AK. Predictive biomarkers for immune checkpoint inhibitor therapy: we need to keep searching. J Thorac Dis 2018;10:S2195-7. [Crossref] [PubMed]
- Hellmann MD, Rizvi NA, Goldman JW, et al. Nivolumab plus ipilimumab as first-line treatment for advanced non-small-cell lung cancer (CheckMate 012): results of an open-label, phase 1, multicohort study. Lancet Oncol 2017;18:31-41. [Crossref] [PubMed]
- Gubens MA, Sequist LV, Stevenson JP, et al. Pembrolizumab in combination with ipilimumab as second-line or later therapy for advanced non-small-cell lung cancer: KEYNOTE-021 cohorts D and H. Lung Cancer 2019;130:59-66. [Crossref] [PubMed]
- Antonia S, Goldberg SB, Balmanoukian A, et al. Safety and antitumour activity of durvalumab plus tremelimumab in non-small cell lung cancer: a multicentre, phase 1b study. Lancet Oncol 2016;17:299-308. [Crossref] [PubMed]
- Brahmer J, Reckamp KL, Baas P, et al. Nivolumab versus docetaxel in advanced squamous-cell non-small-cell lung cancer. N Engl J Med 2015;373:123-35. [Crossref] [PubMed]
- Herbst RS, Baas P, Kim DW, et al. Pembrolizumab versus docetaxel for previously treated, PD-L1-positive, advanced non-small-cell lung cancer (KEYNOTE-010): a randomised controlled trial. Lancet 2016;387:1540-50. [Crossref] [PubMed]
- Mok TSK, Wu YL, Kudaba I, et al. Pembrolizumab versus chemotherapy for previously untreated, PD-L1-expressing, locally advanced or metastatic non-small-cell lung cancer (KEYNOTE-042): a randomised, open-label, controlled, phase 3 trial. Lancet 2019;393:1819-30. [Crossref] [PubMed]
- Camidge DR, Doebele RC, Kerr KM. Comparing and contrasting predictive biomarkers for immunotherapy and targeted therapy of NSCLC. Nat Rev Clin Oncol 2019;16:341-55. [Crossref] [PubMed]
- Topalian SL, Hodi FS, Brahmer JR, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med 2012;366:2443-54. [Crossref] [PubMed]
- Herbst RS, Soria JC, Kowanetz M, et al. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature 2014;515:563-7. [Crossref] [PubMed]
- Boutros C, Tarhini A, Routier E, et al. Safety profiles of anti-CTLA-4 and anti-PD-1 antibodies alone and in combination. Nat Rev Clin Oncol 2016;13:473-86. [Crossref] [PubMed]
- Singal G, Miller PG, Agarwala V, et al. Association of patient characteristics and tumor genomics with clinical outcomes among patients with non-small cell lung cancer using a clinicogenomic database. JAMA 2019;321:1391-9. [Crossref] [PubMed]
- Le DT, Durham JN, Smith KN, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 2017;357:409-13. [Crossref] [PubMed]
- Hellmann MD, Nathanson T, Rizvi H, et al. Genomic features of response to combination immunotherapy in patients with advanced non-small-cell lung cancer. Cancer Cell 2018;33:843-52.e4. [Crossref] [PubMed]
- Goodman AM, Kato S, Bazhenova L, et al. Tumor mutational burden as an independent predictor of response to immunotherapy in diverse cancers. Mol Cancer Ther 2017;16:2598-608. [Crossref] [PubMed]
- Peters S, Gettinger S, Johnson ML, et al. Phase II trial of atezolizumab as first-line or subsequent therapy for patients with programmed death-ligand 1-selected advanced non-small-cell lung cancer (BIRCH). J Clin Oncol 2017;35:2781-9. [Crossref] [PubMed]
- Rizvi NA, Hellmann MD, Snyder A, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 2015;348:124-8. [Crossref] [PubMed]
- Schuler M, Yang JC, Park K, et al. Afatinib beyond progression in patients with non-small-cell lung cancer following chemotherapy, erlotinib/gefitinib and afatinib: phase III randomized LUX-Lung 5 trial. Ann Oncol 2016;27:417-23. [Crossref] [PubMed]
- Chaft JE, Oxnard GR, Sima CS, et al. Disease flare after tyrosine kinase inhibitor discontinuation in patients with EGFR-mutant lung cancer and acquired resistance to erlotinib or gefitinib: implications for clinical trial design. Clin Cancer Res 2011;17:6298-303. [Crossref] [PubMed]
- Martínez Vila C, Garcia Garcia Y, Carcereny Costa E. Prolonged partial response to osimertinib after disease progression to rociletinib in metastasic EGFR T790M-positive non-small cell lung cancer. J Thorac Oncol 2018;13:e77-9. [Crossref] [PubMed]
- Sequist LV, Piotrowska Z, Niederst MJ, et al. Osimertinib responses after disease progression in patients who had been receiving rociletinib. JAMA Oncol 2016;2:541-3. [Crossref] [PubMed]
- Ramalingam SS, Yang JC, Lee CK, et al. Osimertinib as first-line treatment of EGFR mutation-positive advanced non-small-cell lung cancer. J Clin Oncol 2018;36:841-9. [Crossref] [PubMed]
- Lipson EJ, Velculescu VE, Pritchard TS, et al. Circulating tumor DNA analysis as a real-time method for monitoring tumor burden in melanoma patients undergoing treatment with immune checkpoint blockade. J Immunother Cancer 2014;2:42. [Crossref] [PubMed]
- Ashida A, Sakaizawa K, Uhara H, et al. Circulating tumour DNA for monitoring treatment response to anti-PD-1 immunotherapy in melanoma patients. Acta Derm Venereol 2017;97:1212-8. [Crossref] [PubMed]
- Gray ES, Rizos H, Reid AL, et al. Circulating tumor DNA to monitor treatment response and detect acquired resistance in patients with metastatic melanoma. Oncotarget 2015;6:42008-18. [Crossref] [PubMed]
- Xi L, Pham TH, Payabyab EC, et al. Circulating tumor DNA as an early indicator of response to T-cell transfer immunotherapy in metastatic melanoma. Clin Cancer Res 2016;22:5480-6. [Crossref] [PubMed]
- Guibert N, Mazieres J, Delaunay M, et al. Monitoring of KRAS-mutated ctDNA to discriminate pseudo-progression from true progression during anti-PD-1 treatment of lung adenocarcinoma. Oncotarget 2017;8:38056-60. [Crossref] [PubMed]
- Lee JH, Long GV, Menzies AM, et al. Association between circulating tumor DNA and pseudoprogression in patients with metastatic melanoma treated with anti-programmed cell death 1 antibodies. JAMA Oncol 2018;4:717-21. [Crossref] [PubMed]
- Cabel L, Riva F, Servois V, et al. Circulating tumor DNA changes for early monitoring of anti-PD1 immunotherapy: a proof-of-concept study. Ann Oncol 2017;28:1996-2001. [Crossref] [PubMed]
- Goldberg SB, Narayan A, Kole AJ, et al. Early assessment of lung cancer immunotherapy response via circulating tumor DNA. Clin Cancer Res 2018;24:1872-80. [Crossref] [PubMed]
- Vanderwalde A, Spetzler D, Xiao N, et al. Microsatellite instability status determined by next-generation sequencing and compared with PD-L1 and tumor mutational burden in 11,348 patients. Cancer Med 2018;7:746-56. [Crossref] [PubMed]
- Weiss GJ, Beck J, Braun DP, et al. Tumor cell-free DNA copy number instability predicts therapeutic response to immunotherapy. Clin Cancer Res 2017;23:5074-81. [Crossref] [PubMed]
- Johnson DB, Frampton GM, Rioth MJ, et al. Targeted next generation sequencing identifies markers of response to PD-1 blockade. Cancer Immunol Res 2016;4:959-67. [Crossref] [PubMed]
- Rizvi H, Sanchez-Vega F, La K, et al. Molecular determinants of response to anti-programmed cell death (PD)-1 and anti-programmed death-ligand 1 (PD-L1) blockade in patients with non-small-cell lung cancer profiled with targeted next-generation sequencing. J Clin Oncol 2018;36:633-41. [Crossref] [PubMed]
- Wang Z, Duan J, Cai S, et al. Assessment of blood tumor mutational burden as a potential biomarker for immunotherapy in patients with non-small cell lung cancer with use of a next-generation sequencing cancer gene panel. JAMA Oncol 2019;5:696-702. [Crossref] [PubMed]
- Gandara DR, Paul SM, Kowanetz M, et al. Blood-based tumor mutational burden as a predictor of clinical benefit in non-small-cell lung cancer patients treated with atezolizumab. Nat Med 2018;24:1441-8. [Crossref] [PubMed]
- Forschner A, Battke F, Hadaschik D, et al. Tumor mutation burden and circulating tumor DNA in combined CTLA-4 and PD-1 antibody therapy in metastatic melanoma - results of a prospective biomarker study. J Immunother Cancer 2019;7:180. [Crossref] [PubMed]
- Gainor JF, Shaw AT, Sequist LV, et al. EGFR mutations and ALK rearrangements are associated with low response rates to PD-1 pathway blockade in non-small cell lung cancer: a retrospective analysis. Clin Cancer Res 2016;22:4585-93. [Crossref] [PubMed]
- Peng S, Wang R, Zhang X, et al. EGFR-TKI resistance promotes immune escape in lung cancer via increased PD-L1 expression. Mol Cancer 2019;18:165. [Crossref] [PubMed]
- Haratani K, Hayashi H, Tanaka T, et al. Tumor immune microenvironment and nivolumab efficacy in EGFR mutation-positive non-small-cell lung cancer based on T790M status after disease progression during EGFR-TKI treatment. Ann Oncol 2017;28:1532-9. [Crossref] [PubMed]
- Yang JC, Gadgeel SM, Sequist LV, et al. Pembrolizumab in combination with erlotinib or gefitinib as first-line therapy for advanced NSCLC with sensitizing EGFR mutation. J Thorac Oncol 2019;14:553-9. [Crossref] [PubMed]
- Ma BBY, Rudin CM, Cervantes A, et al. 441O Preliminary safety and clinical activity of erlotinib plus atezolizumab from a Phase Ib study in advanced NSCLC. Ann Oncol 2016;27:mdw594.005.
- Gibbons DL, Chow LQ, Kim DW, et al. 57O Efficacy, safety and tolerability of MEDI4736 (durvalumab [D]), a human IgG1 anti-programmed cell death-ligand-1 (PD-L1) antibody, combined with gefitinib (G): a phase I expansion in TKI-naïve patients (pts) with EGFR mutant NSCLC. J Thorac Oncol 2016;11:S79. [Crossref]


