
Expression of large tumour suppressor (LATS) kinases modulates chemotherapy response in advanced non-small cell lung cancer
Introduction
Lung cancer ranks the most common cancer and is also the leading cause of cancer mortality in most parts of the world (1). Non-small cell lung cancer (NSCLC) accounts for more than 80% of pulmonary tumours, in which adenocarcinoma (AD) is the predominant subtype (2). Lung cancer has been reported to be heterogeneous in terms of histological, biological characteristics, and response to chemotherapy, targeted therapy, and checkpoint inhibitor blockade (CIB) immunotherapy (3). Heterogeneity of responses to targeted therapy is largely explained by the discovery of “druggable” driver mutations for targeted and personalized treatments, such as epidermal growth factor receptor tyrosine kinase inhibitors (EGFR-TKIs) (e.g., erlotinib, gefitinib, and afatinib) targeting sensitizing mutations in the EGFR gene, and anaplastic lymphoma kinase (ALK) inhibitors targeting EML4-ALK gene rearrangements (e.g., crizotinib and ceritinib and alectinib) (4). Lung cancers that showed good response to CIB immunotherapy have several molecular biomarker predictors including expression of the target PD-L1, total tumor mutation burden (TMB) leading to high neo-antigen expression and a high degree of mutation clonality (5).
Despite the significant breakthrough in molecular targeted treatment and immunotherapy, platinum-based chemotherapy is still one of the first-line treatments for advanced stage lung cancer and also remains the mainstay of care for patients developing resistance to targeted agents (6,7). The most commonly used platinum-based regimens are cisplatin plus gemcitabine, pemetrexed, taxanes or vinorelbine (“platinum-doublet” chemotherapy) (8). The combination of cisplatin and pemetrexed is considered as a standard of care treatment option for patients with non-squamous NSCLC (AD and large-cell carcinoma) (9). However, only less than half of lung cancer patients demonstrated good response to platinum-doublet chemotherapy. Thus, a major issue in the treatment of advanced stage NSCLC is to identify biomarkers that could predict therapeutic response to platinum-doublet chemotherapy.
The human large tumour suppressor (LATS) proteins, consisting of LATS1 and LATS2, were identified as core components of the Hippo signalling pathway (10). The major function of the Hippo pathway is the regulation of organ size by coordinating cell proliferation, cell death and cell differentiation (11). De-regulation of this pathway has been shown to induce tissue over-growth (11) and this occurs in some types of human carcinomas, including lung, colorectal, breast and liver cancers (12). The upstream regulation of LATS1/2 kinases is complex and is not fully understood. In the canonical Hippo pathway, activated MST1/2 (mammalian sterile 20-like kinases 1 and 2) is associated with SAV1 phosphorylation of LATS1/2 and MOB1, leading to the formation of LATS-MOB1 complex. Within this complex, LATS1/2 kinases are fully activated by phosphorylation on both T-loop and hydrophobic sites. The resulting activated LATS kinases interact with and phosphorylate YAP (yes-associated protein)/TAZ (transcriptional co-activator with PDZ-binding motif), rendering cytoplasmic sequestering and subsequent degradation of these oncogenic transcriptional co-factors (11,13).
As homologs, LATS1 and LATS2 share some conserved features including the common composition of a C-terminal kinase domain, one protein-binding domain, two LATS conserved domains, an ubiquitin-associated domain and at least one PPxY motif which can interact with proteins possessing WW domain (10). However, each type of LATS kinase exhibits unique domains which may contribute to their distinct functions: LATS1 has a proline-rich P-stretch (14), while LATS2 shows repeats of alternating proline-alanine residues (PAPA repeat) (15). The down-regulation of LATS1 or LATS2 has been found in breast cancer (16), prostate cancer (17), colorectal cancer (18), gastric cancer (19), hepatic carcinoma (20) and certain subtypes of ovarian cancer (21). In NSCLC, decreased expression of LATS1 (22) or LATS2 (23) has been reported to correlate with poor prognosis in terms of shorter overall survival.
Not much research has investigated into the effects of LATS kinases on chemo-sensitivity in NSCLC. Furthermore, since LATS1 and LATS2 share high similarity in protein structure and exhibit redundant roles in the Hippo pathway, studies dissecting the interaction or regulation between these two homologs are needed. The hypothesis of this study was that changes in the relative expression of LATS1 and LATS2 could affect the chemotherapeutic response of lung cancer cells. Thus, we set out to explore if manipulation of the relative expression of LATS kinases would modulate cisplatin chemotherapy response in advanced stage lung AD.
Methods
Human lung AD cell lines
Ten AD cell lines were cultured in RPMI 1640 (Gibco, USA) supplemented with 1% Penicillin-Streptomycin (Gibco, USA) and 2.5% or 10% fetal bovine serum (Gibco, US). The ten lung AD cell lines used in this study were HKULC-2 (24), FA31 and FA98 established from pleural fluids of lung AD subjects, developed by the Lam lab; H2023, H1975 and H1650 from JD Minna M.D., University of Texas Southwestern Medical Center at Dallas, USA. CL1-0, CL83, H3255 and PC9 were gifts from PC Yang, M.D., National Taiwan University.
CL1-0, CL83, H2023, FA98 and HKULC-2 were EGFR wild-type cell lines; while the remaining five cell lines harboured mutations in EGFR gene (H3255 and FA31 with L858R point mutation, H1650 and PC9 carried deletions at exon 19 and H1975 has both EGFR L858R and T790M mutations). All cell lines were maintained in a humidified incubator at 37% with 5% CO2.
Plasmids and transfection
The pCMV6 full-length human LATS2 expression vector tagged with MYC/DDK was obtained from OriGene, USA. The LATS2 sequences were confirmed by DNA sequencing. Transfection of cells with this LATS2 plasmid vector was performed via Lipofectamine LTX&PLUS system (Invitrogen, USA). Briefly, cells were plated on 24-well plates on the day before transfection. For each well, 500 ng plasmid DNA in complex with Lipofectamine reagents was added for 24 hours. Culture media containing DNA-Lipofectamine complexes were replaced with G418-contained media in order to select cells that have LATS2 plasmid stably integrated into cell genome. The expression efficiency of this LATS2 plasmid was determined by protein expression assays. Cells transfected with empty vector was used as control.
Short hairpin RNA (shRNA) and infection
The lentiviral transduction particles which contained control shRNA or LATS2-targeted shRNAs cloned into the pLKO.1-puro vector (Sigma-Aldrich, USA) were utilized to establish stable knockdown of LATS2 in lung AD cells. The infection protocol was provided by the manufacturer. For each cell line, the optimal multiplicity of infection was tested empirically, and the transduced cells were selected through puromycin treatment. The surviving cell colonies were harvested to test for knockdown efficiency by means of western blotting.
Cisplatin treatment and cell viability assay
Cisplatin was purchased from Sigma-Aldrich, and was dissolved in 0.9% NaCl and kept as a stock solution (10 mM) at −80 °C. Cells were seeded in 96-well plates at appropriate density, and then exposed to different concentrations of cisplatin solution (0, 1, 10, 20, 50, 100 µM) for 72 hours. 3-(4,5-dimethyl-thiazoyl-2-yl)-2,5-diphenyl-tetrazolium bromide (MTT) solution was added to each well for testing cell viability based on manufacturer instructions (Sigma-Aldrich, USA).
Immunofluorescence staining
In brief, cells were plated on coverslips and, after proper treatment, cells were fixed with 4% paraformaldehyde in PBS for 20 min at room temperature (RT). After permeabilization with 0.1% Triton X-100 in PBS and blocking, cells were incubated with rabbit anti-human LATS2 (Cell Signaling, USA) and mouse anti-human LATS1 (Santa Cruz, USA) antibodies, followed by incubation with AlexaFluor 488- and 594-conjugated anti-rabbit and anti-mouse IgG (Molecular Probes, USA), respectively. DNA was counter-stained with DAPI (Invitrogen, USA). Images were obtained using fluorescence microscopy (Nikon Eclipse Ni-U, Japan).
Western blotting and antibodies
Immunoblotting experiments were conducted as described before (23). Primary antibodies to LATS1, LATS2, cleaved caspase-3, caspase-3 and p53 were obtained from Cell Signaling, USA. β-actin (Sigma-Aldrich, USA) was used as a loading control. The band intensity of each target was quantified with Image J software.
Statistical analysis
Graphics were analyzed with Image J software and statistical analysis was carried out using GraphPad Prism software. Data were presented as mean ± SEM, unless stated otherwise. Differences between groups were estimated using two-tailed unpaired Student’s t test. A probability level of 0.05 was used to determine statistical significance.
Results
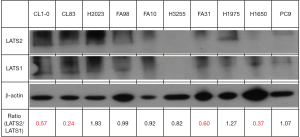
The expression of LATS2 and LATS1 at baseline in lung AD cell lines
The basal expression levels of LATS1 and LATS2 were assayed in the ten lung AD cell lines. As shown in Figure 1, these cell lines expressed LATS1 protein at a relatively consistent level except for H3255, whereas the expression levels of total LATS2 kinase were quite diverse in different cell lines. In some cell lines, such as CL1-0, CL83, FA31 and H1650, LATS2 expression levels were much lower than the levels of LATS1. We defined this group of cell lines as the “low” LATS2-to-LATS1 group. On the other hand, H2023 and H1975 cell lines were categorised as the “high” LATS2-to-LATS1 group, since these cells displayed more LATS2 kinase than LATS1. For remaining cell lines (FA98, HKULC-2, H3255 and PC9), these two LATS kinases were expressed at comparable levels.
The effect of alterations in LATS2-to-LATS1 expression ratios on cell viability after cisplatin treatment
Low LATS2-to-LATS1 cell lines CL1-0 and CL83, were transfected with LATS2-over-expression plasmid vectors for ectopic expression of LATS2 in these cells (Figure 2A,B). After selecting and expanding successfully transfected cell clones, different concentrations of cisplatin solution (0–100 μM) were added to both the parental and LATS2-transfected cells. LATS2 over-expression rendered both cell lines more resistant to cisplatin treatment, as indicated by more viable cells after cisplatin addition and the higher IC50 values in LATS2-transfected cells with larger LATS2-to-LATS1 ratios (Figure 2A,B).
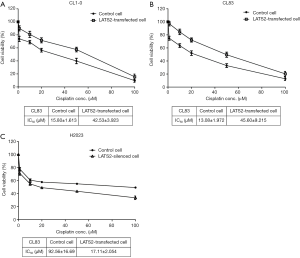
At the same time, we silenced LATS2 expression by infecting high LATS2-to-LATS1 H2023 cells with viral particles-containing control or LATS2-specific shRNA (Figure 2C). LATS2 knockdown resulted in a decrease in the LATS2-to-LATS1 ratio and sensitized H2023 cells to cisplatin. The IC50 concentration of cisplatin dropped dramatically in LATS2-silenced H2023 cells compared with H2023 cells with control shRNA (Figure 2C).
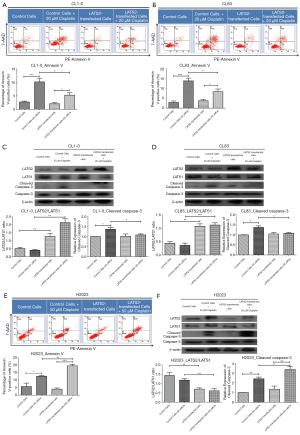
Changes in LATS2-to-LATS1 ratios modulated cisplatin-induced apoptosis in lung AD
Apart from cell viability, apoptosis in response to cisplatin treatment was altered after manipulation of LATS2-to-LATS1 ratios. For low LATS2-to-LATS1 cell lines CL1-0 (Figure 3A) and CL83 (Figure 3B), the increase of LATS2-to-LATS1 ratios by over-expressed LATS2 quantitatively inhibited cisplatin-induced apoptosis, since the percentage of apoptotic cells as well as the cleavage of caspase-3 (Figures 3C,D) were both significantly reduced in LATS2-overexpressed CL1-0 and CL83 cells after cisplatin addition. On the contrary, silencing of LATS2 in high LATS2-to-LATS1 H2023 cells promoted apoptosis triggered by cisplatin (Figure 3E), accompanied by the enhancement of caspase-3 activation (Figure 3F).
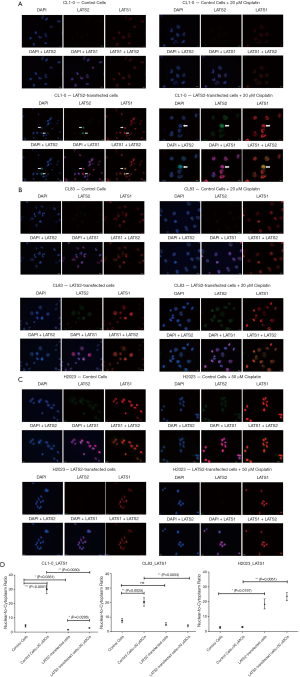
The influence of LATS2 on LATS1 subcellular location after cisplatin treatment
We studied subcellular location of LATS kinases and found that LATS2 kinase was ubiquitously present in both cytoplasm and nucleus, whereas the majority of LATS1 kinase was present in the nucleus with sparse staining in the cytoplasm (Figure 4). In two low LATS2-to-LATS1 cell lines CL1-0 and CL83, addition of cisplatin enhanced 2 to 3-folds of LATS1 translocation from cytoplasm to nucleus (Figures 4A,B upper panels; Figure 4D left and middle panels). However, this phenomenon was absent in high LATS2-to-LATS1 cell lines H2023 (Figure 4C upper panel, Figure 4D right panel). It was notable that H2023 was more resistant to cisplatin than CL1-0 and CL83 (Figure 2A,B). Furthermore, LATS2 over-expression in low LATS2-to-LATS1 cells (CL1-0 and CL83) mitigated cisplatin-induced nuclear translocation of LATS1 kinase (Figure 4A,B lower panels; Figure 4D left and middle panels). For LATS2-silenced H2023 cells which were more sensitive to cisplatin compared with control H2023 cells (Figure 2C), more LATS1 was able to translocate into nucleus (Figure 4C lower panel; Figure 4D right panel). These findings implied that LATS2 kinase could regulate the cellular localization of its homolog LATS1 and nuclear translocation of LATS1 kinase might be related to cisplatin sensitivity.
Discussion
This study was the first demonstration that manipulation of expression of LATS kinases was able to regulate sensitivity to cisplatin in lung AD cell lines. This is important because cisplatin-based doublet chemotherapy is a commonly used first-line treatment for advanced NSCLC patients. The well-known mechanism underlying the anti-neoplastic effects of cisplatin is to generate excess and irreparable DNA lesions, thus committing cells to cellular senescence or mitochondrial pathway of apoptosis (25). There is recent evidence that, in addition to its genotoxic activity, cytoplasmic cisplatin also exerts certain cytotoxic functions through accumulation of reactive oxygen species and establishment of endoplasmic reticulum stress (26,27).
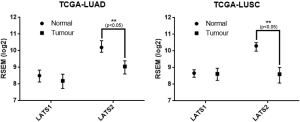
Although platinum-based chemotherapy could be effective in the initial control of advanced stage lung cancer disease, platinum drug administration is dosage limited and could not be used for maintenance chemotherapy due to potential cumulative toxicity. Some NSCLC also become resistant to chemotherapy. If there is any gene expression characteristics that predict platinum chemotherapeutic response or resistance or if that gene expression could be modulated, this may help in modifying the platinum dosage schedule and hence could reduce its potential cumulative adverse effects. Cisplatin resistance is usually multi-factorial, with potential involvement from either or both tumour cell-intrinsic and tumour cell-extrinsic (stem cell, stromal cell, or immune cell-related) pathways (25,28). In this study, we have shown that the high LATS2-to-LATS1 ratio was associated with cisplatin resistance in lung AD cells. High LATS2-to-LATS1 H2023 cells were more resistant to cisplatin than low LATS2-to-LATS1 cells (CL1-0 and CL83) (Figure 2). Furthermore, the increase in the LATS2-to-LATS1 ratio by LATS2 over-expression in CL1-0 and CL83 rendered cancer cells more insensitive to cisplatin treatment, whereas LATS2 knockdown in H2023 alleviated the LATS2-to-LATS1 ratio and actually sensitized cancer cells to cisplatin administration. Besides, according to latest publicly available TCGA data on the LATS1 and LATS2 expressions in lung cancer, LATS2 alone is significantly down-regulated in lung tumour tissue when compared with adjacent normal tissue, in both lung AD and SCC (Figure S1).
The results from this study also supported a potential role of LATS2 kinase in the regulation of LATS1 localization. In low LATS2-to-LATS1 cells, the addition of cisplatin promoted nuclear translocation of LATS1. After cisplatin treatment, however, the cellular distribution of LATS1 kinase did not change in high LATS2-to-LATS1 cells. In addition, over-expression of LATS2 in low LATS2-to-LATS1 cells considerably inhibited the nuclear entry of LATS1 kinase and moderated pro-apoptotic effects of cisplatin. The situation was reversed when LATS2 was silenced in high LATS2-to-LATS1 cells. The reduction in the LATS2-to-LATS1 ratio not only enabled more LATS1 to enter into the nucleus but also encouraged anti-neoplastic effects to take place in cancer cells. Little is known regarding the mechanisms that mediate the cellular distribution of LATS kinases. Previous studies found that LATS kinases together with other hippo pathway members were localized to centrosomes to modulate the various steps of cell cycle including centrosome numbering, spindle formation, chromosome segregation and cytokinesis (29-33), although the differential aptitudes of LATS kinases to become localized and activated in centrosomes are not fully understood. Differential functions of LATS1 and LATS2 expression have been proposed in studies by Furth et al. which suggested that the reduced expression of either LATS1 or LATS2 may re-wire breast cancer signalling networks (34). Lower LATS1 but not LATS2 mRNA expression has been found to correlate with poor survival in NSCLC (35).
The strength of our study is that manipulation of LATS2/LATS1 was shown to modulate sensitivity towards platinum chemotherapy and was associated with survival in NSCLC, with mechanistic implication of the translocation of LATS kinases in different subcellular locations. There were, however, limitations. First, this study was based on lung cancer cell lines. Direct manipulation of LATS2/LATS1 may not be possible in lung cancer patients but at least the assays of LATS2/LATS1 ratio in lung tissues or metastatic specimens such as pleural fluid, could be informative as to the potential sensitivity of the disease towards platinum-based chemotherapy. Moreover, this study focused on LATS2/LATS1 expression and yet there could be collateral or downstream signalling pathways involved in chemotherapeutic sensitivity. Further research is required to clarify the detailed mechanisms concerning the regulation of subcellular localization of LATS kinases as well as their interactions.
Conclusions
In summary, our data provided evidence that the relative expression of LATS kinases played a role in the modulation of cisplatin sensitivity in advanced lung AD. This modulation might be ascribed to the differential subcellular distribution of LATS1 which was influenced by LATS2 expression. The findings of this study warrant further translational investigation on the use of LATS2-to-LATS1 ratio as a potential biomarker or a therapeutic target.
Acknowledgments
Funding: The research project in this manuscript was partly supported by the YC Chan Scientist Award, Department of Medicine, University of Hong Kong, and the Hui Hoy & Chow Sin Lan Charity Fund and the Family of Mr Hui Ming, Department of Medicine, University of Hong Kong.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tlcr.2020.03.26). JDM received royalties for licensing human tumor and related lines from the National Institutes of Health (NIH) and University of Texas Southwestern Medical Center. JDM’s lab at UTSW receives research support from licensing fees for the distribution of the human lines. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was approved by the Institutional Review Board/Ethics Committee of the Hong Kong University/Hong Kong Hospital Authority Hong Kong West Cluster (HKU/HA HKWC IRB/EC UW 16-104).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Hong Kong Cancer Registry HA. Leading Cancer Sites in Hong Kong. 2016.
- Travis WD, Brambilla E, Noguchi M, et al. Diagnosis of lung adenocarcinoma in resected specimens: implications of the 2011 International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society classification. Arch Pathol Lab Med 2013;137:685-705. [Crossref] [PubMed]
- Abdel Karim N, Kelly K. Role of Targeted Therapy and Immune Checkpoint Blockers in Advanced Non-Small Cell Lung Cancer: A Review. Oncologist 2019;24:1270-84. [Crossref] [PubMed]
- Luo SY, Lam DC. Oncogenic driver mutations in lung cancer. Transl Respir Med 2013;1:6. [Crossref] [PubMed]
- Negrao MV, Lam VK, Reuben A, et al. PD-L1 expression, tumor mutational burden and cancer gene mutations are stronger predictors of benefit from immune checkpoint blockade than HLA class I genotype in non-small cell lung cancer. J Thorac Oncol 2019;14:1021-31. [Crossref] [PubMed]
- Yin JY, Li X, Zhou HH, et al. Pharmacogenomics of platinum-based chemotherapy sensitivity in NSCLC: toward precision medicine. Pharmacogenomics 2016;17:1365-78. [Crossref] [PubMed]
- Hellmann MD, Li BT, Chaft JE, et al. Chemotherapy remains an essential element of personalized care for persons with lung cancers. Ann Oncol 2016;27:1829-35. [Crossref] [PubMed]
- Fennell DA, Summers Y, Cadranel J, et al. Cisplatin in the modern era: The backbone of first-line chemotherapy for non-small cell lung cancer. Cancer Treat Rev 2016;44:42-50. [Crossref] [PubMed]
- Reck M, Popat S, Reinmuth N, et al. Metastatic non-small-cell lung cancer (NSCLC): ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 2014;25 Suppl 3:iii27-39. [Crossref] [PubMed]
- Visser S, Yang X. LATS tumor suppressor: a new governor of cellular homeostasis. Cell Cycle 2010;9:3892-903. [Crossref] [PubMed]
- Yu FX, Guan KL. The Hippo pathway: regulators and regulations. Genes Dev 2013;27:355-71. [Crossref] [PubMed]
- Harvey KF, Zhang X, Thomas DM. The Hippo pathway and human cancer. Nat Rev Cancer 2013;13:246-57. [Crossref] [PubMed]
- Hoa L, Kulaberoglu Y, Gundogdu R, et al. The characterisation of LATS2 kinase regulation in Hippo-YAP signalling. Cell Signal 2016;28:488-97. [Crossref] [PubMed]
- Nishiyama Y, Hirota T, Morisaki T, et al. A human homolog of Drosophila warts tumor suppressor, h-warts, localized to mitotic apparatus and specifically phosphorylated during mitosis. FEBS Lett 1999;459:159-65. [Crossref] [PubMed]
- Yabuta N, Fujii T, Copeland NG, et al. Structure, expression, and chromosome mapping of LATS2, a mammalian homologue of the Drosophila tumor suppressor gene lats/warts. Genomics 2000;63:263-70. [Crossref] [PubMed]
- Takahashi Y, Miyoshi Y, Takahata C, et al. Down-regulation of LATS1 and LATS2 mRNA expression by promoter hypermethylation and its association with biologically aggressive phenotype in human breast cancers. Clin Cancer Res 2005;11:1380-5. [Crossref] [PubMed]
- Powzaniuk M, McElwee-Witmer S, Vogel RL, et al. The LATS2/KPM tumor suppressor is a negative regulator of the androgen receptor. Mol Endocrinol 2004;18:2011-23. [Crossref] [PubMed]
- Li J, Chen X, Ding X, et al. LATS2 suppresses oncogenic Wnt signaling by disrupting beta-catenin/BCL9 interaction. Cell Rep 2013;5:1650-63. [Crossref] [PubMed]
- Zhou GX, Li XY, Zhang Q, et al. Effects of the hippo signaling pathway in human gastric cancer. Asian Pac J Cancer Prev 2013;14:5199-205. [Crossref] [PubMed]
- Li H, Wolfe A, Septer S, et al. Deregulation of Hippo kinase signalling in human hepatic malignancies. Liver Int 2012;32:38-47. [Crossref] [PubMed]
- Xu B, Sun D, Wang Z, et al. Expression of LATS family proteins in ovarian tumors and its significance. Hum Pathol 2015;46:858-67. [Crossref] [PubMed]
- Lin XY, Zhang XP, Wu JH, et al. Expression of LATS1 contributes to good prognosis and can negatively regulate YAP oncoprotein in non-small-cell lung cancer. Tumour Biol 2014;35:6435-43. [Crossref] [PubMed]
- Luo SY, Sit KY, Sihoe AD, et al. Aberrant large tumor suppressor 2 (LATS2) gene expression correlates with EGFR mutation and survival in lung adenocarcinomas. Lung cancer (Amsterdam, Netherlands) 2014;85:282-92. [Crossref] [PubMed]
- Lam DC, Girard L, Suen WS, et al. Establishment and expression profiling of new lung cancer cell lines from Chinese smokers and lifetime never-smokers. J Thorac Oncol 2006;1:932-42. [Crossref] [PubMed]
- Galluzzi L, Vitale I, Michels J, et al. Systems biology of cisplatin resistance: past, present and future. Cell Death Dis 2014;5:e1257. [Crossref] [PubMed]
- Yu F, Megyesi J, Price PM. Cytoplasmic initiation of cisplatin cytotoxicity. Am J Physiol Renal Physiol 2008;295:F44-52. [Crossref] [PubMed]
- Sancho-Martínez SM, Prieto-Garcia L, Prieto M, et al. Subcellular targets of cisplatin cytotoxicity: an integrated view. Pharmacol Ther 2012;136:35-55. [Crossref] [PubMed]
- Galluzzi L, Senovilla L, Vitale I, et al. Molecular mechanisms of cisplatin resistance. Oncogene 2012;31:1869-83. [Crossref] [PubMed]
- Mukai S, Yabuta N, Yoshida K, et al. Lats1 suppresses centrosome overduplication by modulating the stability of Cdc25B. Sci Rep 2015;5:16173. [Crossref] [PubMed]
- Bothos J, Tuttle RL, Ottey M, et al. Human LATS1 is a mitotic exit network kinase. Cancer Res 2005;65:6568-75. [Crossref] [PubMed]
- Yabuta N, Mukai S, Okada N, et al. The tumor suppressor Lats2 is pivotal in Aurora A and Aurora B signaling during mitosis. Cell Cycle 2011;10:2724-36. [Crossref] [PubMed]
- Yabuta N, Okada N, Ito A, et al. Lats2 is an essential mitotic regulator required for the coordination of cell division. J Biol Chem 2007;282:19259-71. [Crossref] [PubMed]
- Bolgioni AF, Ganem NJ. The interplay between centrosomes and the Hippo tumor suppressor pathway. Chromosome Res 2016;24:93-104. [Crossref] [PubMed]
- Furth N, Pateras IS, Rotkopf R, et al. LATS1 and LATS2 suppress breast cancer progression by maintaining cell identity and metabolic state. Life Science Alliance 2018;1:e201800171. [Crossref] [PubMed]
- Malik SA, Khan MS, Dar M, et al. Molecular Alterations and Expression Dynamics of LATS1 and LATS2 Genes in Non-Small-Cell Lung Carcinoma. Pathol Oncol Res 2018;24:207-14. [Crossref] [PubMed]


