
Increased number of subclones in lung squamous cell carcinoma elicits overexpression of immune related genes
Introduction
Lung cancer is the most common cancer. In 2018, there were an estimated 2.09 million cases of lung cancer and 1.76 million deaths due to lung cancer worldwide (1). Squamous cell lung cancer (SqCC) is a distinct histological subtype of lung cancer that accounts for approximately 25–30% of all lung cancers, following adenocarcinomas which account for approximately 40% of all lung cancers. SqCC is more commonly located in the central lung and frequently invades the proximal bronchus and large blood vessels (2). Compared with the stage of nonsquamous non-small cell lung cancers, SqCCs are often at an advanced stage at the time of diagnosis and this is frequently accompanied by comorbidities such as chronic obstructive lung disease and heart disease, which makes SqCC challenging to treat (3-6). Moreover, activating mutations such as the epidermal growth factor receptor and anaplastic lymphoma kinase fusion, which lead to remarkable changes in the treatment of lung adenocarcinoma, are typically not present in SqCC and targeted agents that are used with adenocarcinoma are largely not effective with SqCC (7-10).
Through tumor genome profiling, via the development of next-generation sequencing, detailed information on carcinogenesis, including tumor development, progression, therapeutic response, and drug resistance, has been obtained. The discoveries of multiple studies, based on tumor sequencing, suggest that “intratumor heterogeneity”, which describes the uneven distribution of genetically diverse tumor subpopulations within a tumor, plays a key role in treatment failure and drug resistance (11-14).
The cause of intratumor heterogeneity can be explained by genomic instability that results from exposure to exogenous mutagens (such as UV radiation and exposure to cigarette smoke) and aberrations in endogenous processes (such as DNA replication, error repair, and oxidative stress), which are maintained by selective processes (13,15).
Methods for estimating intratumoral heterogeneity are: extensive tumor dissection (16-18), ultradeep sequencing of mutations (19), and single-nucleus sequencing by isolating individual nuclei (20,21). Most of these methods are not only technically difficult to perform but are also practically difficult to carry out in clinical settings. In this study, we adopted a method to infer the number of subclones from variant allele frequency (VAF), which could be more feasible to apply clinically than the methods mentioned.
VAF is the percentage of sequence reads that match a specific DNA variant, divided by the overall coverage at that locus (22). VAF represents the percentage of tumor cells that harbor a specific mutation, assuming a relatively pure tumor sample (23,24). By clustering VAF, the number of subclones in the tumor can be inferred and heterogeneity can be estimated. Mutant-allele tumor heterogeneity (MATH) score, which is calculated from the ratio of the width to the center of the distribution of the VAF in the tumor-specific mutated loci, could provide a straightforward measure of one type of intratumor heterogeneity (25).
In this study, we investigated whether the number of subclones that represent intratumor heterogeneity is related to genomic mutations and cancer stage. We also investigated the genes that are related to intratumor heterogeneity.
Methods
Data acquisition
The results shown here are based upon data generated by The Cancer Genome Atlas (TCGA) Research Network: https://www.cancer.gov/tcga. The following data were downloaded from 504 SqCC cases shared in the TCGA project: (I) mutation annotation format (MAF) files for single nucleotide variants (SNV) analyzed with VarScan2 variant Aggregation and Masking workflow; (II) Masked Copy Number Segment analyzed with Affymetrix SNP 6.0; (III) RNA sequencing analyzed with HTSeq; (IV) Clinical information. Of the 504 The Cancer Genome Atlas-Lung Squamous Cell Carcinoma (TCGA-LUSC) cases, 488 had SNV, copy number segment, RNA sequencing, and clinical information, which made them eligible for study analyses. The Tumor Sample Barcodes of those cases were confirmed and analyzed based on the data obtained from the primary solid tumor. Among them, three cases (TCGA-18-3409, TCGA-90-A4ED, and TCGA-21-1079), which had exceptionally high numbers of mutations and four cases (TCGA-34-2604, TCGA-34-2605, TCGA-34-2609, and TCGA-77-8146) in which the SNVs were 0, were excluded. Finally, 481 cases were recruited for this analysis (Table S1). To improve the positive predictive value of the low allele frequency, SNVs with a total read depth of less than 40 and SNV data that did not meet the limit of detection as suggested by Shin et al. were excluded (24).

Full table
Calculations of subclone numbers and differentially expressed gene (DEG) analysis
To estimate the number of subclones SciClone (http://github.com/genome/sciclone) was used, which estimates the number of subclones by clustering a variant with similar allele frequencies (26). Computational efficiency is achieved by clustering VAFs using a variational Bayesian mixture model (27). To identify genes related to intratumor heterogeneity that were represented by the number of subclones, DEG analysis between high clone and low clone groups was performed. Patients who had five subclones were paired with patients who had one subclone. Potential confounding variables such as age, sex, stage, and smoking status were adjusted by the propensity matching method while pairing the two groups using the “MatchIt” package in R. Analyzing the DEG between high clone and low clone groups was demonstrated using the “DEseq2” package in R. When the ratio of gene expression in the experimental groups to the control group was more than 2 or less than 1/2 and the P-adjusted value was less than 0.05, it was considered a significant DEG and further analysis was performed. The ontology of the DEG was confirmed using ToppGene (https://toppgene.cchmc.org/).
Statistical analysis
The distribution of variables was examined using the Shapiro-Wilk test. Continuous variables of three or more groups were analyzed using the Kruskal-Wallis test. Categorical variables were analyzed using the chi-squared distribution and Fisher’s exact test. In all cases, P-values <0.05 were considered statistically significant. Statistical analyses were performed using R statistical software, version 3.6.0 (the R Foundation for Statistical Computing, Vienna, Austria).
Results
Study cohort
Data from 481 LUSCs in stages I–IV that had not experienced any treatment for lung cancer were collected from the TCGA. The demographic characteristics are described in Table 1. The median age of the study cohort was 68.6 (62.3–73.9) years and 356 (74.0%) were male. The study population was comprised of 232 cases (48.2%) in stage I, 157 (32.6%) in stage II, 81 (16.8%) in stage III, and 7 (1.4%) in stage IV. Regarding smoking status, 453 cases (94.2%) were ever-smokers, 18 (3.7%) were never smoked, and the smoking status of 10 (2.1%) were unavailable. The data of smoking amount (packs-years) were available in 407 cases among ever smokers and the median was 54.0 (21.0–70.0) pack-years.

Full table
Number of subclones constituting the primary tumor was related to the number of variants and tumor stage
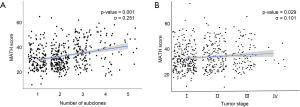
Whole exome sequencing of 481 LUSCs identified a total of 117,869 variants with a median of 209.0 (128.0–319.0) variants per tumor (Figure 1A). SNVs occupied most of these variations, followed by deletions and insertions. When the number of subclones that comprised the single primary tumor was inferred by SciClone (Representative figures are shown in Figure 1B,C), the median number of subclones that comprised a tumor was 2 (1-3). The total variation in a tumor showed a significant positive correlation with the number of subclones in a tumor (σ=0.388, P-value <0.001, Figure 1D) and when these variations were classified into SNVs and indels, they showed significant positive correlations with the number of subclones (σ=0.389, P-value <0.001; σ=0.241, P-value <0.001, respectively, Table 2). According to the variant effect predictor (VEP) as presented in the TCGA, the variants were further classified into high, moderate, low impact variants, and modifier and analyzed according to the number of subclones. The number of high and moderate impact variants increased as the number of subclones increased (σ=0.357, P-value <0.001; σ=0.383, P-value <0.001, respectively; Figure 1E, Table 2). Because the staging system is still the most useful parameter for predicting the clinical outcome in patients with lung cancer, we compared the number of subclones with the stage of lung cancer and discovered that there was a significant positive correlation between the number of subclones and stage (σ=0.099, P-value =0.030, Figure 1F). These results suggested that the number of subclones that comprised a primary tumor was positively related to the number of variants and stage of the corresponding tumor, which suggested that the number of subclones reflected the biological aspect of a tumor.
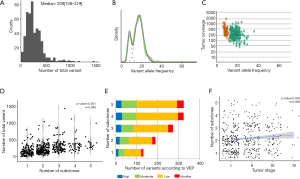

Full table
Gender was related to the number of subclones
To determine the factors associated with the increased number of subclones, we examined the relationship between the number of subclones and clinical parameters that were known to be associated with SqCCs. Because smoking is one of the main causes of lung cancer by causing C > A transversions of DNA, we divided the patients into non-smokers and ever-smokers and compared the number of subclones between the two groups. Tumors of ever-smokers consisted of a significantly larger number of subclones than those of never-smokers (2.2 vs. 1.7, P-value =0.043, Kruskal-Wallis rank-sum test, Figure 2A, http://cdn.amegroups.cn/static/application/b0dcd306dde9c330e80f00e4985ce55f/tlcr-19-589-1.pdf). Therefore, we further investigated whether the expansion of clone number was dependent on the amount of smoking and discovered that there was no significant relationship between pack-years and the number of subclones (Figure 2B). As age-related mutations were observed in most malignancies, including lung cancer, and there was a significant positive correlation between lung cancer incidence and age (28), the relationship between age at diagnosis of lung cancer and the number of subclones was investigated. Age at diagnosis and number of subclones did not show a significant correlation (Figure 2C). These results suggested that smoking and aging had no major effect on the propagation of subclones.
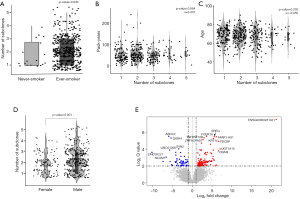
Finally, the influence of gender on the increase in subclone number was investigated. The number of subclones that constituted a tumor was significantly higher in patients who were male than those who were female (P=0.001, Wilcoxon rank-sum test, Figure 2D). To verify whether gender was a truly significant factor that influenced the number of subclones, 105 female TCGA-LUSC cases, which had identifiable smoking status, were 1:1 matched with male cases considering age, pack-years, and stage using the “MatchIt” R package (Figure S1). In this analysis, male tumors consisted of significantly more subclones than female LUSCs, which indicated that gender is the sole parameter related to the number of subclones (2.25 vs. 1.86, male vs. female: P-value =0.002, Welch Two Sample t-test).
Inferring clonal expansion via DEG
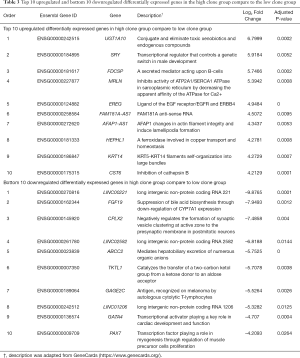
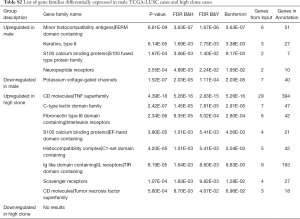
To uncover the possible etiology that increased the number of subclones that constituted a tumor, an additional analysis was performed using RNA sequencing data. Using the propensity score matching of the “MatchIt” R package, the LUSC cases in which tumors were composed of five subclones, which is the largest number of subclones among the study cases, were matched with those in which the tumor was composed of a single subclone by age, sex, pack-years, and stage (Figure 1B, http://cdn.amegroups.cn/static/application/92369042f5e4ab643e2c8b83670a2049/tlcr-19-589-2.pdf). After performing three independent matching and DESeq2 analyses, we obtained a set of intersections from the results of each analysis and performed gene ontology analysis using ToppGene (https://toppgene.cchmc.org/). DEG analysis revealed that 291 genes were upregulated and 102 genes were downregulated in the tumors composed of 5 subclones compared to single subclone tumors (http://cdn.amegroups.cn/static/application/ff5c281257c4bebadda9eaff792d56a5/tlcr-19-589-3.pdf). Among these, the most upregulated genes were UGT1A10, SRY, FDCSP, MRLN, and EREG in order of magnitude of overexpression (Table 3,Figure 2E) and the overexpressed genes were significantly enriched in the (I) tumor necrosis factor superfamily cluster of differentiation molecules, (II) C-type lectin domain family, (II) fibronectin type III domain that contained interleukin receptors, (IV) EF-hand domain that contained S100 calcium-binding proteins (Table S2). The biological function of these upregulated genes was strongly enriched for the positive regulation of immune processes and inflammatory responses. The downregulated genes were LINC00221, FGF19, CPLX2, LINC02582, and ABCC2 in order of magnitude of downregulation; however, these downregulated genes were not significantly enriched for specific gene families. Because a relationship was found between sex and the increase in subclone number, the DEG between the two genders was obtained by matching pack-year, age at diagnosis, and tumor stage after propensity score matching (http://cdn.amegroups.cn/static/application/96e916da4746e0ecf63d3d9d39787278/tlcr-19-589-4.pdf, http://cdn.amegroups.cn/static/application/9d1e5289244e24d7e455384d8c5dc314/tlcr-19-589-5.pdf). Then, the intersection of DEGs upregulated in the male LUSCs and those in which the tumor was comprised of five subclones were obtained. Six DEGs were observed, five of which were upregulated and one of which was downregulated in both groups (Figure 2E,Table S3). The biological function of upregulated genes LRRC38, FDCSP, SRY, FAM181A-AS1, and PICSAR was closely related to the positive regulation of potassium ion transmembrane transport and male gender differentiation. However, the limited number of common genes observed in DEG analysis in male to female LUSC tumors and DEG analysis according to the number of clones suggested that masculinity did not play a critical role in clonal expansion.
Full table
Full table
Full table
Overall, positive regulator genes involved in inflammatory and immune responses were significantly overexpressed in the LUSC tumors with high clone numbers.
Discussion
In this study, we discovered that intratumor heterogeneity inferred from the number of subclones that constituted a tumor positively correlated with the number of somatic variants and cancer stage. The number of subclones was significantly higher in males than that in females, whereas smoking status and age did not show a significant relationship with the number of subclones. This suggested that masculinity was a factor that affected clonal expansion. In DEG analysis, which was performed to identify genes related to clonal expansion, genes, whose function is related to the positive regulation of immune processes and inflammatory responses, were enriched in the LUSCs that were composed of high subclone numbers. However, it was assumed that masculinity did not play a critical role in clonal expansion in DEG analysis.
There are several statistical models for the inference of clonal population structure in a tumor, including SciClone’s (26) variational Bayesian mixture model, MAFtools’ (29) Gaussian mixture, and PyClone’s (30) beta-binomial emission densities. Among these statistical models, we calculated the number of subclones with Sciclone. The MATH scores, calculated from the ratio of the width to the center of the distribution of VAF to represent intratumor heterogeneity, were evaluated in the study population. There was a significant correlation between the number of subclones inferred from either SciClone or MAFtools and MATH score (Figure S2). The MATH score in this study was positively correlated with the number of somatic variants and cancer stage and the median was significantly higher in males than that in females, whereas smoking and age did not show a significant correlation with the MATH score. These were the same results as those evaluated by the number of subclones calculated by SciClone (data not shown), which suggested there is no significant difference among the results obtained by different methods of inferring intratumoral clonality.
Despite interest that heterogeneity could be the main cause of treatment failure, clinical indicators that properly reflect heterogeneity are not widely used in the medical field. In this study, by analyzing genome and transcriptome data of 481 TCGA-LUSC cases, we revealed that masculinity could affect clonal expansion, whereas smoking status and age have no major effect on the propagation of heterogeneity. To verify these results, we analyzed the mutational signature suggested by Alexandrov et al. (28). In 481 TCGA-LUSC patients with high clonality, whose tumors consisted of five clones, only Signature 4 (smoking) and Signature 2 (APOBEC) were extracted in mutational signature analysis, which is no different to the signatures observed in the TCGA-LUSC cases (Figure S3), which suggests that the effect of specific mutational signatures on clonal expansion are insignificant.
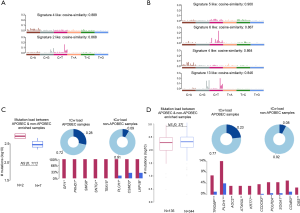
Representative genes that were overexpressed in tumors that consisted of a high number of subclones compared to single clone tumors are UGT1A10, SRY, FDCSP, MRLN, and EREG. The most significant difference between the tumors is in regard to UGT1A10. UDP-glucuronosyltransferases (UGTs), estimated as a gene involved in clonal expansion by DEG analysis, catalyzes the conjugation of glucuronic acid with the polar groups (e.g., hydroxyl, thiol, carboxyl, and amines) from xenobiotic substances to facilitate their elimination as well as the elimination of endogenous molecules, such as bile acids and hormones (31). To date, many studies were performed to reveal the links between UGTs and cancer risk, based on the idea that the alterations of UGT function may significantly influence the clearance of carcinogens and sex hormones. UGT1A1, 1A6, 1A7, and 1A8 are involved in the metabolism of dietary carcinogens and genotypes with low functional properties would highly increase the risk of colorectal, esophageal, and proximal digestive tract cancers (31-33). Estrogen-metabolism related UGT1A1, 1A6, and 2B4 and androgen-metabolism related 2B15 and 2B17 genes are closely related to breast, endometrial, and prostate cancers (34-37). UGT1A6, 1A7, 2B7, and 2B17 are involved in the metabolism of tobacco carcinogens and are linked to lung, oropharyngeal, and bladder cancers (38-40). Although the majority of studies are related to the low functional activities of UGTs and increased cancer risk, there are also several studies that reported higher UGT activities with higher cancer risk and decreased UGT activities with lower cancer risk (41,42). UGTs are the key metabolic enzymes involved in not only the detoxification of many carcinogens but also the elimination of antioxidant, anti-proliferative substances, and therapeutic drugs. An imbalance in UGT activity, both upregulated and downregulated, can be interpreted in association with carcinogenesis based on previous studies. In the TCGA-LUSC cases matched by sex, age, and stage, UGT was highly upregulated in the high clonality group and it was inferred that the high functional activity of UGT was related to the clonal expansion of SqCC. This requires further research to determine whether increased activity of UGT derives or assists clonal expansion.
This study has a few limitations. Firstly, none of the methods for the prediction of heterogeneity is complete. Single-cell analysis may be ideal for evaluating intratumoral heterogeneity, but it has many challenges, including difficulty in the selection of target lesion and analyzing only 5,000 to 10,000 cells per selected area, which suggest that this method also has the same common limitations. Secondly, due to the lack of data, we could not demonstrate the association between clonality and outcome (disease recurrence, survival, and treatment responses). However, by the result that patients with higher numbers of subclones showed more advanced cancer stages, we could infer poor prognosis with high clonality indirectly.
Conclusions
Intratumor heterogeneity represented by the number of subclones was positively correlated with somatic mutation burden and cancer stage. Considering that the tumor mutation burden predicts the response of the immune checkpoint inhibitors, it could be inferred that there is a relationship between the number of subclones and the immune response. The number of subclones was higher in males than in females, whereas smoking status and age were not correlated with clonality. In DEG analysis, a significant enrichment in genes whose function is related to the positive regulation of immune processes and inflammatory responses in high clone tumors compared to that in single clone tumors. In-depth studies are required to determine whether the overexpression of the positive regulation of immune process and inflammatory response genes in high clone number tumors is a driver of clonal expansion.
Acknowledgments
Funding: This work was supported by the National Research Foundation of Korea (grant number NRF-2020R1A2B5B01001883).
Footnote
Data Sharing Statement: available at http://dx.doi.org/10.21037/tlcr-19-589.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tlcr-19-589). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- American Cancer Society, Cancer Statistics Center. (accessed 13 March 2019). Available online: https://cancerstatisticscenter.cancer.org/#!/cancer-site/Lung%20and%20bronchus
- Rosado-de-Christenson ML, Templeton PA, Moran CA. Bronchogenic carcinoma: radiologic-pathologic correlation. RadioGraphics 1994;14:429-46. [Crossref] [PubMed]
- Janssen-Heijnen ML, Schipper RM, Razenberg PP, et al. Prevalence of co-morbidity in lung cancer patients and its relationship with treatment: a population-based study. Lung Cancer 1998;21:105-13. [Crossref] [PubMed]
- Subramanian J, Morgensztern D, Goodgame B, et al. Distinctive Characteristics of Non-small Cell Lung Cancer (NSCLC) in the Young: A Surveillance, Epidemiology, and End Results (SEER) Analysis. J Thorac Oncol 2010;5:23-8. [Crossref] [PubMed]
- Socinski MA, Obasaju C, Gandara D, et al. Current and Emergent Therapy Options for Advanced Squamous Cell Lung Cancer. J Thorac Oncol 2018;13:165-83. [Crossref] [PubMed]
- Papi A, Casoni G, Caramori G, et al. COPD increases the risk of squamous histological subtype in smokers who develop non-small cell lung carcinoma. Thorax 2004;59:679. [Crossref] [PubMed]
- The Cancer Genome Atlas Research Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature 2012;491:288. [Crossref] [PubMed]
- Zhao W, Choi YL, Song JY, et al. ALK, ROS1 and RET rearrangements in lung squamous cell carcinoma are very rare. Lung Cancer 2016;94:22-7. [Crossref] [PubMed]
- Miyamae Y, Shimizu K, Hirato J, et al. Significance of epidermal growth factor receptor gene mutations in squamous cell lung carcinoma. Oncology Reports 2011;25:921-8. [PubMed]
- Rekhtman N, Paik PK, Arcila ME, et al. Clarifying the spectrum of driver oncogene mutations in biomarker-verified squamous carcinoma of lung: lack of EGFR/KRAS and presence of PIK3CA/AKT1 mutations. Clin Cancer Res 2012;18:1167-76. [Crossref] [PubMed]
- McGranahan N, Swanton C. Clonal Heterogeneity and Tumor Evolution: Past, Present, and the Future. Cell 2017;168:613-28. [Crossref] [PubMed]
- Greaves M. Evolutionary determinants of cancer. Cancer Discov 2015;5:806-20. [Crossref] [PubMed]
- Marusyk A, Almendro V, Polyak K. Intra-tumour heterogeneity: a looking glass for cancer? Nat Rev Cancer 2012;12:323-34. [Crossref] [PubMed]
- Garraway LA, Lander ES. Lessons from the cancer genome. Cell 2013;153:17-37. [Crossref] [PubMed]
- Dagogo-Jack I, Shaw AT. Tumour heterogeneity and resistance to cancer therapies. Nat Rev Clin Oncol 2018;15:81-94. [Crossref] [PubMed]
- Maley CC, Galipeau PC, Finley JC, et al. Genetic clonal diversity predicts progression to esophageal adenocarcinoma. Nat Genet 2006;38:468-73. [Crossref] [PubMed]
- Jovanovic L, Delahunt B, McIver B, et al. Most multifocal papillary thyroid carcinomas acquire genetic and morphotype diversity through subclonal evolution following the intra-glandular spread of the initial neoplastic clone. J Pathol 2008;215:145-54. [Crossref] [PubMed]
- Yachida S, Jones S, Bozic I, et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature 2010;467:1114-7. [Crossref] [PubMed]
- Shah SP, Roth A, Goya R, et al. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature 2012;486:395-9. [Crossref] [PubMed]
- Navin N, Kendall J, Troge J, et al. Tumour evolution inferred by single-cell sequencing. Nature 2011;472:90-4. [Crossref] [PubMed]
- Park SY, Gonen M, Kim HJ, et al. Cellular and genetic diversity in the progression of in situ human breast carcinomas to an invasive phenotype. J Clin Invest 2010;120:636-44. [Crossref] [PubMed]
- Strom SP. Current practices and guidelines for clinical next-generation sequencing oncology testing. Cancer Biol Med 2016;13:3-11. [Crossref] [PubMed]
- Sallman DA, Padron E. Integrating mutation variant allele frequency into clinical practice in myeloid malignancies. Hematol Oncol Stem Cell Ther 2016;9:89-95. [Crossref] [PubMed]
- Shin HT, Choi YL, Yun JW, et al. Prevalence and detection of low-allele-fraction variants in clinical cancer samples. Nat Commun 2017;8:1377. [Crossref] [PubMed]
- Mroz EA, Rocco JW. MATH, a novel measure of intratumor genetic heterogeneity, is high in poor-outcome classes of head and neck squamous cell carcinoma. Oral Oncol 2013;49:211-5. [Crossref] [PubMed]
- Miller CA, White BS, Dees ND, et al. SciClone: inferring clonal architecture and tracking the spatial and temporal patterns of tumor evolution. PLoS Comput Biol 2014;10:e1003665. [Crossref] [PubMed]
- Bishop CM. Pattern recognition and machine learning. Information science and statistics, 2006:738.
- Alexandrov LB, Nik-Zainal S, Wedge DC, et al. Signatures of mutational processes in human cancer Nature 2013;500:415-21. [Crossref] [PubMed]
- Mayakonda A, Lin DC, Assenov Y, et al. Maftools: efficient and comprehensive analysis of somatic variants in cancer. Genome Res 2018;28:1747-56. [Crossref] [PubMed]
- Roth A, Khattra J, Yap D, et al. PyClone: statistical inference of clonal population structure in cancer. Nat Methods 2014;11:396-8. [Crossref] [PubMed]
- Kiang TK, Ensom MH, Chang TK. UDP-glucuronosyltransferases and clinical drug-drug interactions. Pharmacol Ther 2005;106:97-132. [Crossref] [PubMed]
- van der Logt EM, Bergevoet SM, Roelofs HM, et al. Genetic polymorphisms in UDP-glucuronosyltransferases and glutathione S-transferases and colorectal cancer risk. Carcinogenesis 2004;25:2407-15. [Crossref] [PubMed]
- Tang KS, Chiu HF, Chen HH, et al. Link between colorectal cancer and polymorphisms in the uridine-diphosphoglucuronosyltransferase 1A7 and 1A1 genes. World J Gastroenterol 2005;11:3250-4. [Crossref] [PubMed]
- Sparks R, Ulrich CM, Bigler J, et al. UDP-glucuronosyltransferase and sulfotransferase polymorphisms, sex hormone concentrations, and tumor receptor status in breast cancer patients. Breast Cancer Res 2004;6:R488-98. [Crossref] [PubMed]
- Justenhoven C, Winter S, Dunnebier T, et al. Combined UGT1A1 and UGT1A6 genotypes together with a stressful life event increase breast cancer risk. Breast Cancer Res Treat 2010;124:289-92. [Crossref] [PubMed]
- Sun C, Huo D, Southard C, et al. A signature of balancing selection in the region upstream to the human UGT2B4 gene and implications for breast cancer risk. Hum Genet 2011;130:767-75. [Crossref] [PubMed]
- Bélanger A, Pelletier G, Labrie F, Barbier O, Chouinard S. Inactivation of androgens by UDP-glucuronosyltransferase enzymes in humans. Trends Endocrinol Metab 2003;14:473-9. [Crossref] [PubMed]
- Strassburg CP, Strassburg A, Nguyen N, Li Q, et al. Regulation and function of family 1 and family 2 UDP-glucuronosyltransferase genes (UGT1A, UGT2B) in human oesophagus. Biochem J 1999;338:489-98. [Crossref] [PubMed]
- Araki J, Kobayashi Y, Iwasa M, et al. Polymorphism of UDP-glucuronosyltransferase 1A7 gene: a possible new risk factor for lung cancer. Eur J Cancer 2005;41:2360-5. [Crossref] [PubMed]
- Kua LF, Ross S, Lee SC, et al. UGT1A6 polymorphisms modulated lung cancer risk in a Chinese population. PloS One 2012;7:e42873. [Crossref] [PubMed]
- Dura P, Salomon J, Te Morsche RH, et al. High enzyme activity UGT1A1 or low activity UGT1A8 and UGT2B4 genotypes increase esophageal cancer risk. Int J Oncol 2012;40:1789-96. [PubMed]
- Duguay Y, McGrath M, Lepine J, et al. The functional UGT1A1 promoter polymorphism decreases endometrial cancer risk. Cancer Res 2004;64:1202-7. [Crossref] [PubMed]


