
Mechanisms of drugs-resistance in small cell lung cancer: DNA-related, RNA-related, apoptosis-related, drug accumulation and metabolism procedure
Introduction
Comparing with other types of cancer, lung cancer has the highest morbidity and is the major reason of malignant neoplasms related-death around the world, while small-cell lung cancer (SCLC) accounting for 10–15% (1,2). SCLC, as the highest malignant cancer amongst different types of lung cancer, has the feature of lower differentiation, rapid progression and poor survival rate. Although SCLC patients are dramatically sensitive to various types of treatment methods at the beginning, including chemotherapy, radiotherapy and immunotherapy, the emergence of drug resistance is still a grandly clinical challenge (3,4). Taking all these reasons into account, the 5-year survival rate of SCLC is lower than 7% (5).
In terms of classification, SCLC differentiates itself from other types of cancers which traditionally use the international TNM staging. At present, to particular clinical condition, SCLC is capable of being classified on the basis of the extension of the disease. The International Association for the Study of Lung Cancer (IASLC) regarded that limited disease (LD) includes all patients without distant metastasis, while extensive disease (ED) comprises all patients with sites of disease beyond the definition of LD (6).
As of yet, surgery is still one of the principally clinic choices for treating a large proportion of malignant tumors. Unfortunately, it is rarely applied to the SCLC. And specifically, merely LD-SCLC patients (2–5%) who do not have mediastinal lymph node metastasis get a chance to receive surgical treatment. Over the past several decades, a great number of oncologists devoted themselves to developing valid therapy for SCLC patients and has achieved a great deal. Meaningfully, cisplatin combination etoposide (EP) regimen has been firstly proven to be effective by Evans et al. in 1985 (7). From then on, platinum-based combination regimen, such as a platinum compounds (cisplatin or carboplatin) in combination with the EP or irinotecan (IP), which are both parts of topoisomerase-II inhibitor, has been recommended as the first-line therapy for SCLC, with the chance to accompany radiotherapy (8-11). According to the data, ED-SCLC patients who are treated with EP has an objective response rate (ORR) surpass 60–70% (12). On the other side, comparing with EP strategy, there is not any glaring improvement in the ORR and overall survival (OS) of the ED-SCLC patients who are treated with IP (11,13,14). The finite differences of treatment outcomes of IP strategy is due to the national differences (15-17).
In recent years, thanks to the great efforts made by oncologists worldwide, novel therapeutic strategies for SCLC, such as targeted therapy and immunotherapy are round the corner (3,18-22). Nowadays, multiple clinical trials, including larger phase II/III studies, are conducted in the matter of untreated, refractory, and relapsed SCLC, and we are looking forward to the meaningful outcomes. In spite of various kinds of SCLC therapeutic strategies are emerging and developing promptly, over 90% of patients ultimately came up primary or secondary resistance in diverse drugs and had poor prognosis (3,23). This review is to summarize the complex mechanisms of drug resistance during SCLC therapy from four aspects, including DNA-related process, RNA-related process, apoptosis-related mechanism, as well as the procedure of drug accumulation and metabolism, thus having a more comprehensive and systematic understanding of the mechanisms that still not be fully elucidated. Moreover, the discussion of promising strategies to tackle drugs-resistance and enhance curative effect in SCLC was presented.
DNA-related process
A large amount of anti-tumor drugs which are applied in SCLC patients treatment act effectively by means of DNA damage, including injuring the structure and function of DNA, thus causing tremendous impact on the survival of rapid proliferating tumor cells. To date, however, a crowd of studies have revealed that one of the major reasons of the occurrence of drug resistance is DNA-related process, including via promoting the repairing of cellular DNA damage, preventing anti-tumor drugs from interaction with DNA and aberrant DNA methylation.
The repairing of DNA damage
The repairing of DNA damage was composed of DNA double-strand break repair, base excision repair, nucleotide excision repair (NER), and mismatch repair (MMR), etc. which had significant influence in the drug response (24). Glutathione (GSH), one kind of thiol-containing molecules, is available to augment the repair of DNA damage and prevent apoptosis so as to further mediate resistance to cisplatin (25). The increasing degree of the repairing of cisplatin-induced DNA damage in the human drugs-resistant cells also resulted from the higher activity of NER (26). Heyza et al. revealed that p53 was able to regulate the interstrand crosslinks to make up for the loss of ERCC1, so it was considered as a potential confounding variable (27). The decreasing of MMR could straightly give rise to the lower probability-of-detection DNA damage, inhibition of apoptosis and higher mutation rate, leading the drugs-resistance (28). MMR-deficient cells owned higher spontaneous mutation rates, including the point mutation of TOPIIA (29).
Preventing anti-tumor drugs from interaction with DNA
Nowadays, a mass of researches have demonstrated the mechanisms of resistance to anti-tumor drugs in the matter of preventing the interaction between drugs and DNA. GSH also participated in the process of drug resistance as it could fortify inactivation of drugs that resulting in the reduction of interaction between cisplatin and DNA (25). Furthermore, the γ-glutamyl cysteine synthetase (γ-GCS) gene was deemed to be one kind of resistance mechanisms because it could boost the expression of GSH in cells (30). The increasing number of resistant tumor cells also caused by the function of GSH-S-transferase π (GSTπ), which could catalyze the coalition between active GSH and cisplatin (31,32). Adequate evidences of these resistant mechanisms were reflected in various researches. In 1990, Meijer et al. used multiple methods, such as D,L-buthionine-S, R-sulphoximine (BSO), to draw a conclusion that GSH mainly through restraining the repairing of DNA damage to cause the resistance of in the SCLC cell line (33). Besides, according to the research of Kurokawa et al., the human γ-GCS gene was transfected into the SCLC cell line, SBC-3, for the sake of producing SBC-3/GCS (34). The experimental data illustrated that, in comparison with the control group, SBC-3/GCS group doubled in the level of intracellular GSH and had higher resistance (relative resistance value of 7.4) to cisplatin. The higher levels of GSH in resistant cell lines or xenografts were investigated by other researches as well (35). Nevertheless, the meaningful overexpression of GSH in drug-resistance cells was not being able to be confirmed by all studies. Campling et al. illustrated that GSH had no statistical correlation with the resistance of chemotherapeutic agents via measuring the levels of GSH in a panel of 20 SCLC cell lines (36). In the field of resistance in SCLC, many published researches paid attention to the function of GSH. More in depth and width clinical studies on the interaction between anti-cancer drugs and cancer-related DNA in refractory SCLC is worth expecting.
Aberrant DNA methylations
As one of epigenetic mechanisms, the abnormal methylation level of DNA was indicated as a kind of mechanisms of drugs-resistance in cancer cells by recent researches (37). Gardner et al. found that histone H3 lysine 27 trimethylation (H3K27me3) that regulated by zeste homologue 2 (EZH2) resulted in the development of resistance in SCLC cells (3). By means of multiple experimental tactics, including immunohistochemical (IHC), western blotting, flow cytometry and bisulfite sequencing PCR, Fang et al. found that H3K27me3 could modulate the expression of HOX transcript antisense RNA (HOTAIR) to have an impact on the methylation process of HOXA1 DNA, which eventually led to the drugs-resistance in SCLC cells (38,39). As a long non-coding RNA (lncRNA), HOTAIR also played a major role in cancer invasiveness and metastasis through influencing H3K27me3 and EZH2 (40). Similarly, Tsai et al. demonstrated that lncRNA was a kind of regulatory factors to chromatin states and the model of histone modifications (41). However, seldom researches were launched to estimate the epigenetic interaction between DNA and RNA in resistant SCLC. More basic and clinical studies are necessary to further explore and validate the resistance-associated epigenetic mechanisms in SCLC.
RNA-related process
MicroRNAs (miRNAs), as endogenous RNAs of approximately 22 nucleotides in length, play vital regulatory roles in cancer (42,43). MicroRNAs were previously found to be closely correlated with proliferation, invasion and drugs-resistance in lung cancer cells. Several researches found the differential expression of miRNAs between drugs-resistance malignant cells and normal cells (44,45). However, at the very start, oncologists mainly payed their attention to the function of human miRNAs in non-small cell lung cancer (NSCLC). Hence, this review will principally summarize the published RNA-related mechanisms of drugs-resistance in SCLC patients.
MiR-7 is not only recognized as a human tumor suppressor gene but also as an oncogene in malignant neoplasm. Mounting evidences revealed that MiR-7 is potential to be employed as a promising therapeutic target for SCLC to help patients who suffer from drugs-resistance. According to the data from the research of Lai et al., in comparison with parallel control group (H69 and H446 cells), the expression level of miR-7-5p was remarkably declined about quadrupled (P<0.05) in doxorubicin-resistant SCLC group (H69AR and H446AR cells) (46). Guo and colleagues demonstrated that miR-7-5p and the Ras/MAPK pathway could modulate the expression of inwardly rectifying potassium channel Kir2.1 (KCNJ2), which finally led to the multiple drug resistance (MDR) in SCLC cells. KCNJ2/Kir2.1 would take effect in drugs-resistance through regulating multidrug resistance-associated protein 1 (MRP1/ABCC1) expression (47). By means of a bioinformatic assay, IHC analysis and so on, Liu et al. once again illustrated that miR-7 could exert its suppressive effects on MRP1/ABCC1 gene, which was an atypical ABC transporter, resulting in the drugs-resistance in SCLC cells (48). In addition, the expression of MRP1/ABCC1 gene was also directly mediated by certain miRNAs, consisting of negative regulator miR-134 and positive control factors like miR-326, miR-199a, miR-199b and miR-296 (49). What’s more, a credibly experimental evidence showed that miR-7-5p could directly downregulate the express of poli [ADP-ribose] polymerase-1 (PARP1), which was a nuclear enzyme that catalyzes the processes of DNA repair and gene transcription (50,51). Taken all these into consideration, one kind of the pathway of drugs-resistance in SCLC cells was mediated by miR-7-5p, which was able to decrease the expression of BRCA1 and Rad51, and hold back the homologous recombination (HR) repair through regulating PARP1. It was indicated the clinical value of miR-7-5p and the re-expression of miR-7-5p might guide to a novel strategy for overcoming drugs-resistance in SCLC.
Aberrant expression of miR-339-5p has been reported to play a crucial role in a variety of malignant neoplasm, including lung carcinoma (52-54). Li et al. demonstrated that miR-339-5p acted as a cancer suppressor in the migration and invasion of tumor cells and was linked with the tumor-node-metastasis (TNM) staging of NSCLC patients (54). MiR-339-5p was likely to be a valid agent for hampering and reversing MDR in tumor cells. In 2017, according to the experimental data of Gan et al., the downregulation of miR-339-5p was distinctly observed in the Taxol-resistant A549 subclone compared with that in normal A549 cells group (55). Furthermore, it was indicated that miR-339-5p could affect drugs-resistance and apoptosis of SCLC through regulating a1,2-fucosyltransferase 1 (FUT1) and the downstream protein Lewis y (LeY). FUT1, as a fucosylation synthase, primarily control the synthesis of LeY and could inhibit the proliferation and metastasis of cancer (56,57).
In previous studies, mounting evidences were obtained for the function of MiR-335 in the process of proliferation, invasion, metastatic and drug-activity in a variety of cancers (58). Gong et al. revealed that the loss of miR-335 could downregulate the expression of insulin-like growth factor-I receptor (IGF-IR) and affect the RANKL pathway, and gave rise to the SCLC metastatic skeletal lesions (59). Tang and the colleagues investigated that drug sensitivity in SCLC patients were induced by miR-335, which downregulated the WW domain binding protein 5 (WBP5) protein level by the Hippo pathway (60). Rather, the overexpression of WBP5 in SCLC prompted MDR and suppressed apoptosis (60,61). Collectively these results indicated that WBP5 was potential to be serviced as an effective target for attenuating and interfering with drugs-resistance in SCLC.
MiR-200 family, which includes miR-200a, miR-200b, miR-200c, miR-141 and miR-429, is supposed to be a family of tumor suppressor miRNAs. In recent studies, miR-200 was closely related to the process of resistance, cell division, apoptosis and so on (62). For cancer treatments, the dysfunction of miR-200 promoted the MDR in terms of various tumors, such as lung cancer, breast cancer and gastric carcinoma (63-65). A research that chiefly explored the role of miR-200 in drugs-resistance of SCLC was conducted by Guo et al. (66). By means of silico analysis and luciferase reporter gene assay, the research showed that ZincfingerE-box-bindinghomeobox2 (ZEB2) was the target gene of miR-200b and its expression could decline accompany with the upregulation of miR-200b, thus eventually caused drugs-resistance in SCLC. Similarly, miR-200 also modulated the resistance of targeted therapy and chemotherapy in bladder and ovarian cancer by target ZEB2 (67,68).
The miR-134 cluster locates on chromosome 14q32.31 and consists of a huge cluster of 13 miRNA genes within 10 kb (69,70). MiR-134 took part in the biological process of human malignant neoplasm, such as cell proliferation, apoptosis, drug resistance, as well as malignant tumor treatment and prognosis (71). However, because of the functions of miR-134 are extremely sophisticated and interactional, the mechanisms in the drug-resistance in SCLC are still not clear yet. Guo et al. found that miR-134 downregulated the expression of MRP1/ABCC1 to mediate drug-resistance of SCLC (72). In addition, the research also revealed that the cell survival in drug-resistant SCLC (H69AR cells) was increased as the result of G1 arrest that induced by miR-134.
Next to the RNAs mentioned above, an increasing number RNAs are also recognized as affecting factors in the field of drug-resistance in SCLC. By means of miRNA and cDNA microarrays, Guo et al. found that 61 miRNAs and 2,383 genes were differentially expressed between SCLC resistant cells and parental cells (H69) (72). In summary, although the understanding of the RNA-related molecular mechanisms of MDR in SCLC had made great progress, there is a long way to go as the insufficient and ambiguous mechanisms.
Apoptosis-related mechanism
Apoptosis, also called programmed cell death (PCD), is a biological process of energy-dependence, cellular autonomy and orderliness. There are two major pathways of apoptosis; one of them is the extrinsic pathway which is mainly induced by caspase, while the other is the intrinsic pathway which is also referred to as the mitochondrial pathway. The caspases, as a family of intracellular cysteine aspartic acid specific proteases, are the key enzymes of the process of the apoptotic cascade. On one hand, the abnormal process of apoptosis pathways account for the high probability of gene mutations and the immune escape of immortalized cells, which finally lead to the occurrence of cancer (73,74). On the other hand, previous studies had supported that the drugs-resistance in a variety of cancers was the consequence of the disability of inducing apoptosis.
In the situation of the resistance to platinum in SCLC, the aberrations of the apoptotic cascade have been reported to be closely related. The early release of mitochondrial cytochrome c, activation of Apaf-1and caspase 9, cleavage of downstream, or the self-amplifying cascade of specific caspases are the characters of the cascade of apoptosis. By IHC, Stefanaki et al. examined the expression of B cell lymphoma-2 (Bcl-2), P53 and the wild-type (WT) p53-induced proteins mdm2 and p21/waf1 in 31 cases of SCLC patients (75). The data showed that Bcl was extremely overexpressed in 77% of SCLC specimens, while p53 protein accounted for 42%. In ovarian carcinoma cell systems, the mutations of TP53 gene in exons 4 to 9 played a part in the development of cisplatin-resistance because of the inability of TP53 gene to transactivate bax, a gene of enhancing apoptosis (76). Besides the aforementioned Bcl-2, p53 and bax, XIAP and surviving were detected to be widely overexpressed in platinum-resistant cells (77). XIAP and surviving belong to the members of the anti-apoptotic molecules family which could prevent the activation of caspases. When it comes to the apoptosis-related mechanism of resistance to EP, a topoisomerase inhibitor, a study conducted by Kaplan et al. revealed that the lower expression levels of MLH1 and MSH2 in resistant group and the level of EP-resistance were highly interrelated (78). MLH1, a component of the conserved DNA MMR complex, was involved in the process of apoptosis through its function as a proapoptotic factor in cytoplasm (79). However, to date, the apoptosis-related mechanism of resistance to topoisomerase inhibitors in SCLC patients still has not been clearly and directly pointed out.
Several genes induced the MDR in SCLC by the apoptosis-related mechanisms, including EPHA3, SPHK1, RLIP76, WIG-1, and fibroblast growth factor-inducible 14 (Fn14). EPHA3, a member of the Eph receptor family, could suppress the activation of AKT to increased apoptosis (80). Ross et al. performed that the rate of actionable alteration of EPHA3 genes is 3% via comprehensive genomic profiling of 98 SCLC patterns (81). In 2016, Peng et al. had investigated the role of EPHA3 genes in drugs-resistance by loss- and gain-of-function approaches in different SCLC cell lines, such as H69, H69AR, H446, H146, and H1688 (82). The results demonstrated that EPHA3 could attenuate drugs-resistance by means of promoting apoptosis and G0/G1 arrest, along with suppressing the phosphorylation of PI3K/BMX/STAT3 signaling pathway. However, the concrete cellular and molecular mechanisms of the role of EPHA3 genes in SCLC chemoresistance have rarely been fully detected.
SPHK1 is the rate-limiting enzyme of producing S1P, which plays an important role in metabolism of lipid, affecting apoptosis, and stimulating cell proliferation (83). In previous study, the up-regulation of SPHK1 alleviated the drug-resistance in prostate cancer PC3 cells (84). In terms of SCLC, Yang et al. found that the expression of SPHK1 in H69AR cells group was distinctly higher that in H69 cells groups (P<0.01) (85). Furthermore, similar to the function of EPHA3, the overexpression of SPHK1 was not only able to suppress the apoptosis of cancer cells, but also able to G0/G1 arrest.
RLIP76, a GSH-conjugate transporter, is also known as Ralbp1 that belongs to the Ras superfamily. According to the research of Haixia et al., the expression of RLIP76 was significant difference in human drug-resistant SCLC cells line (H69AR) and drug-sensitive SCLC cells line (H69) (86). RLIP76 was closely related to the multi-drug resistance in SCLC (P<0.05). It was not only RLIP76 could regulate the cellular internalization and membrane plasticity so as to control the accumulation of proapoptotic peroxidation liposomes, but also it could combine with various biological factors, such as Ral, Hsf-1, cyclin-dependent kinase 1 (CDK1), and so on, thus activating different signal pathways.
WIG-1 is the target gene of p53 which could regulate the expression of WIG-1 and cellular apoptosis via binding to the promoter region of WIG-1 (87,88). By means of real-time PCR and other experimental methods, Luo et al. found that the expression of WIG-1 that regulated by p53 was significantly different between H69AR and H69 (89). In addition, after suppressing the expression of WIG-1, the difference of apoptotic rate between H69AR-Si-WIG-1 negative control and blank group was statistically significant, which indicated the participation of WIG-1 in MDR in SCLC.
Fn14 gene, known as an oncogene, is a member of the tumor necrosis factor (TNF) superfamily of receptors (90). A certain amount of studies had been demonstrated that the expression of Fn14 were dramatically increased and played a significant role in cellular processes of various malignant neoplasm, including NSCLC and SCLC (91-96). More than half of SCLC patients (50.98%) were detected the expression of Fn14 via immunohistochemistry (96). Further, promoting apoptosis and inducing cell cycle arrest were the mechanisms of the Fn14 to take effect in the cell growth and MDR of SCLC. At the molecular level, similar to other type of cancers (97,98), Fn14 is also capable of upregulating the expression of Bcl-xl by means of the nuclear factor-κB (NF-κB) pathway, thus resulting the drugs-resistance of SCLC (96).
On one hand, mitochondrial dysfunction was reported to be of benefit to the growth of tumor cells (99). On the other hand, the mutation of mitochondrial DNA (mtDNA) has been illustrated to be distinctly correlated with the process of apoptosis and the MDR in various kinds of cancers, except for SCLC (100-102). It was not until the year of 2015 that a study was carried by Ma et al. which distinguished whether mitochondrial dysfunction or mtDNA mutation took effect on the development of drugs-resistance in SCLC (103). The results of the study revealed that the sequence mutation of mtDNA had little to do with the drugs-resistance in SCLC cell groups, while mitochondrial dysfunction clearly involved in the development of MDR via affecting the reactive oxygen species (ROS)-mediated mitochondrial apoptotic pathway. Mitochondrion, as an intracellular semi-autonomous organelle, could generate energy and ROS which lead to the poor condition of the mitochondrion, such as the situation of swelling, apoptotic death and so on (104,105).
Drugs accumulation and metabolism procedure
Drugs accumulation procedure
The use of platinum drugs, including cisplatin and carboplatin, has carried giant implications on the treatment of SCLC, while the decline of drug accumulation in cells was proved to be one of its major mechanisms of resistance. Preventing intake or accelerating excretion, or both these two factors, may lead to the failure of maintaining a sufficient intracellular anti-tumor drugs concentration (26). The overexpression of the human MRP2, an ABC-family membrane protein, was capable of regulating the drugs efflux in drug-resistant tumor cells, whereas there was not more researches to enhance the persuasion of this discovery (106). In addition, the copper-transporting ATPase proteins, including ATP7A and ATP7B, mediated drugs efflux and overexpressed in resistant cells of ovarian cancer cells as well (107). On the other hand, in terms of the mechanisms of the platinum-resistance, copper transporter 1 (CTR1), normally served as the major copper influx transporter, could also take clear effect in increasing the intracellular drugs accumulation via influence the process of drugs uptake (108,109).
Topoisomerase inhibitors are commonly applied in combination with platinum drugs for SCLC treatment because of their synergistic effect. However, it is inevitable that there would be the occurrence of drugs-resistance during the SCLC therapy. TOPIIA, the gene that encoding the topoisomerase II alpha, is the direct target of topoisomerase inhibitors. In breast cancer, according to the PCR analysis, the dramatically lower expression of TOPIIA in EP-resistant sublines MCF-7/1E and MCF-7/4E was regarded as a general cellular mechanism of EP-resistance, while the activity of TOPIIA almost does not matter (110). In addition, on the basis of mounting evidences, the higher expression of MRP1/ABCC1 had obvious and powerful impact on the drug accumulation as well as drug resistance in variety cancer, including SCLC. In EP/IP-resistant SCLC cell lines, the MRP1/ABCC1 promoted efflux of EP from the cells (110-115).
Drugs metabolism procedure
There are two factors that could affect the drug sensitivity to IP, including the higher activity of CYP3A4/CYP3A5 and the lower activity of CES1/2. As mentioned above, these abnormal conditions of activity could mainly lead to the aberrant IP metabolism in cells, such as mounting inactivation and declining drug activation. Moreover, in cancer cells, the glucuronidation of SN-38 by uridine diphosphate glucuronosyltransferase 1As (UGT1As) is vital to IP metabolism as well. At this point, the efficacy and sensitivity of IP treatment are related to the polymorphisms in UGT1A1 (116). In colorectal cancer and gynecologic cancers, the significance of UGT1A1 in the procedure of drugs metabolism was reported (117-119). With respect to the mechanisms of drugs metabolism in drugs-resistance, at present, there are limited studies that are conducted by oncologists to investigate the molecular pathway in SCLC.
Novel strategies to tackle drug-resistance in SCLC
Immune checkpoints inhibitors
Immune checkpoints inhibitors, including the programmed death ligands-1 (PD-L1) inhibitors, the programmed death-1 (PD-1) inhibitors, and the cytotoxic T-lymphocyte-associated protein 4 (CTLA-4) inhibitors, were considered as effective drugs among various solid tumors, including chemotherapy-resistant SCLC. We summarized the completed and ongoing clinical trials which involved immune checkpoints inhibitors in Table 1.
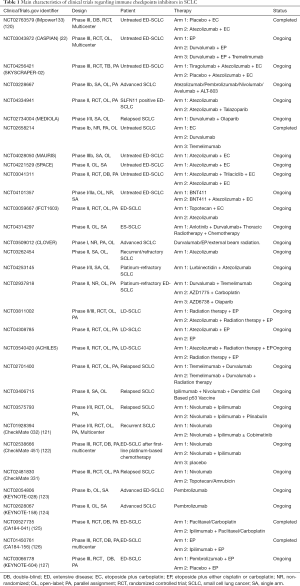
Full table
In regard to immunotherapy, the landmark event is that atezolizumab, the PD-L1 inhibitor, which was the first drug to make obvious achievement in OS advantage and lower mortality risk in ED-SCLC. In comparison with the group of using chemotherapy solely, IMpower133 (NCT02763579), a phase III clinical trial, showed that the another group which atezolizumab plus chemotherapy are enrolled, achieved 2 months OS advantage (P=0.007), nearly a year median progression free survival (mPFS) improvements (P=0.02), and 30% reduction in mortality risk (20). As another PD-L1 inhibitor, durvalumab also showed its clinical benefits in SCLC. A randomized phase III clinical trial, CASPIAN (NCT03043872), demonstrated the prognostic improvements of first-line durvalumab plus chemotherapy regimen in ED-SCLC compared with EP regimen (OS 13.0 vs. 10.3 months, P=0.0047) (22). The ORR of durvalumab group higher than it of chemotherapy group (67.9% vs. 57.6%), which highlighted the potential application of durvalumab for refractory SCLC patients. Recently, growing number of clinical trials are in progress so as to further explore the promising PD-L1 inhibitor-included combination therapy for untreated ED-SCLC patients, such as atezolizumab-EP-tiragolumab (TIGIT inhibitors) regimen (SKYSCRAPER-02, NCT04256421), atezolizumab-ALT-803 (novel IL-15 complex) regimen (NCT03228667), and PD-L1 inhibitor plus PARP inhibitors regimen (NCT04334941 and MEDIOLA, NCT02734004), and durvalumab monotherapy (NCT02658214), etc. For SCLC patients who were resistant to platinum drugs, several phase I/II clinical trials will apply atezolizumab monotherapy (NCT03262454), atezolizumab plus lurbinectedin treatment (NCT04253145), and durvalumab plus the CTLA-4 inhibitor tremelimumab (NCT02937818), so as to assess the clinical feasibility and safety of different treatment. In addition to ED-SCLC, atezolizumab is also being studied as the potential drugs for LD-SCLC patients in some researches (NCT03811002, NCT04308785, and NCT03540420).
Furthermore, the efficacy of PD-1 inhibitors, including nivolumab and pembrolizumab, was tested in the second- or third-line treatment of SCLC. In the light of the data of Checkmate 032 (NCT01928394), nivolumab in combination with ipilimumab (anti-CTLA-4) demonstrated clinically promising activity in recurrent SCLC, with 19% ORR (120). Nevertheless, two phase III clinical trials, CheckMate-451 (NCT02538666) (121) and CheckMate-331 (NCT02481830, Bristol-Myers Squibb, 2020, unpublished data), then demonstrated the failure of nivolumab in SCLC second-line therapy. For pembrolizumab monotherapy, the ORR of phase II clinical trial KEYNOTE-028 (NCT02054806) and phase Ib clinical trial KEYNOTE-158 (NCT02628067) was 33.3% and 18.7%, respectively (122,123). In spite of the relatively lower ORR, both nivolumab and pembrolizumab demonstrated persistent clinical response, thus being granted by FDA for SCLC third-line treatment. However, the efficacy of PD-L1 inhibitors and CTLA-4 inhibitors were not performed well in the SCLC first-line treatment. Although the ipilimumab plus EP group obtained better immune-related PFS in phase II clinical trial CA184-041 (NCT00527735) (124), the OS and mPFS improvement in ED-SCLC patients who received ipilimumab plus EP therapy were not statistically significant in the phase III CA184-156 (NCT01450761) (125). A phase III randomized clinical trial KEYNOTE604 (NCT03066778), which compared pembrolizumab plus EP regimen with EP alone, was recently announced to fail to meet the primary endpoint (OS: HR 0.80; 95% CI, 0.64–0.98) (126). The detailed data of KEYNOTE604 have not yet been reported.
Compared with the success of PD-L1 inhibitors in SCLC, the unsatisfying results of PD-1 inhibitors illustrated the difference between these immune checkpoints inhibitors, which should arouse more attention in clinical use. Considering the complicated immune signaling pathways and the limited prognostic improvements of immunotherapy, the seeking of biomarkers which could help identify SCLC patients who may benefit from the immunotherapy is necessary. The challenges of drug-resistance in SCLC call for better combination therapy and we look forward to the results from other ongoing clinical trials.
Novel targeted drugs
An admirable revolution of novel targeted drugs in oncotherapy is taking place. On account of increasing scientific researches in SCLC treatment, oncologists have a far better understanding of SCLC biology, peculiar deviation in different pathways and potential target. More and more targeted drugs, such as PARP inhibitors, Bcl-2 inhibitors, delta-like canonical Notch ligand 3 (DLL3) monoclonal antibody, and EZH2 inhibitor, are going through clinical studies at present and revealed unlikeliness anti-tumor activity as well (3,18-20).
PARP participated in the process of drug-resistance in SCLC, such as the DNA-related and RNA-related procedure, indicating that target PARP may effectively reverse drug-resistance and improve clinical prognosis. The efficacy of PARP inhibitors in SCLC, such as olaparib, talazoparib, rucaparib, niraparib, and veliparib, were evaluated in some clinical trials (Table 2). A phase I clinical trial (NCT01286987) which applied talazoparib monotherapy to relapsed SCLC patients reported the promising anti-cancer activity and the tolerable side effects of talazoparib (127). In terms of PARP inhibitor in combination with EP, Florence Atrafi et al. promoted the safety of veliparib-EP regimen for newly diagnosed ED-SCLC patients by the phase I clinical trial (NCT02289690) (128). A phase II ECOG-ACRIN 2511 (NCT01642251) which aimed at comparing the performance of veliparib-EP regimen with EP alone for first-line treatment showed the statistical mPFS improvement in the veliparib-EP group (6.1 vs. 5.5 months, P=0.01), whereas no significance was found in the OS between two arms (10.3 vs. 8.9 months, P=0.17) (129). Another phase II study discovered the better ORR in the veliparib plus temozolomide group than it of placebo plus temozolomide group (39% vs. 14%, P=0.016), and put forward a hypothesis that SLFN11 may be served as the potential biomarker for PARP inhibitors (NCT01638546) (130). For SLFN11 positive ED-SCLC patients, there is an ongoing phase II clinical trial (NCT04334941) which is carried out for exploring the efficacy of combining atezolizumab with talazoparib and atezolizumab monotherapy. Several clinical trials in SCLC will further investigate the efficacy of PARP inhibitor monotherapy or PARP inhibitor plus other drugs, such as immune checkpoints inhibitors, chemotherapeutic drugs, or WEE1 inhibitors (Table 2).
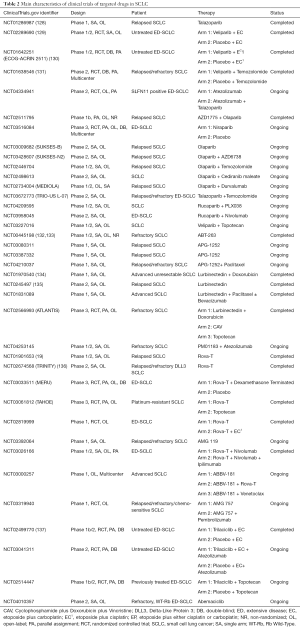
Full table
As Bcl-2 protein family played a major role in the drug-resistance of SCLC by apoptosis-related mechanisms and RNA-related mechanisms as well as the common overexpression of Bcl-2 in drug-resistant SCLC cancer cells, many clinical studies of refractory or relapsed SCLC patients attempted to assess the safety and efficacy of Bcl-2 inhibitors, including navitoclax (ABT-263) and APG-1252 (Table 2). Navitoclax and APG-1252, both selective targeted drugs that inhibit Bcl-2, Bcl-XL, and Bcl-W, kills tumor cells by restoring the mechanism of PCD. The phase I/IIa study of (NCT00445198) reported the clinical performance of navitoclax in refractory SCLC patients, with 25.6% ORR, 1.5 months mPFS, and 3.2 months OS (131,132). Three single-arm clinical trials currently study the therapeutic prospect of APG-1252 monotherapy (NCT03080311 and NCT03387332) or APG-1252 plus paclitaxel (NCT04210037) in SCLC. However, the modest clinical response of Bcl-2 inhibitors monotherapy urgently called for the better combination regimens to enhance the curative effect and reduce drugs toxicities. Although some potential biomarkers for Bcl-2 inhibitors treatment had been investigated, such as pro-gastrin releasing peptide (pro-GRP) and cytokeratin 19 fragment antigen 21-1 (131,132), more clinical studies are needed for further evaluation and application.
Lurbinectedin (PM01183), as the RNA polymerase II inhibitor, performed encouraging activity in the SCLC second-line treatment, thus granted as the orphan drug by FDA. According to the published data of a phase I study (NCT01970540), the ORR of all relapsed SCLC patients who received the therapy of lurbinectedin plus doxorubicin was 58% (133). Specially, the percentage of enrolled drug-resistant participants occurred partial response (PR) was 33.3% in second-line treatment, and 20.0% in third-line treatment (133). Another phase II study emphasized the certain safety and efficacy of lurbinectedin monotherapy in the second-line therapy of both drug-sensitive and drug-resistant SCLC (134). The detailed information of two completed clinical trials, including phase I study (NCT01831089) which employed lurbinectedin plus paclitaxel, with or without bevacizumab in advanced SCLC, and phase III ATLANTIS (NCT02566993) which compared lurbinectedin plus doxorubicin arm with cyclophosphamide plus doxorubicin and vincristine arm, and topotecan single agent arm in refractory SCLC is not available. A phase I/II study of lurbinectedin plus atezolizumab therapy (NCT04253145) is ongoing. As the sample size of currently published researches is too small to fully realize the efficacy of lurbinectedin. Future researches of larger sample size and more reasonable combination therapy in resistant SCLC patients are worthy of expectation.
DLL3 that frequently overexpressed in SCLC tumor tissues and cells is considered as the effective target in drug-resistant SCLC (135,138). The novel DLL3 inhibitors, including rovalpituzumab tesirine (Rova-T), AMG757, and AMG119. Two completed clinical trials, a phase I study (NCT01901653) and TRINITY (NCT02674568) which was a single-arm phase II study, respectively adapted Rova-T monotherapy as the second- or third-line treatment in SCLC patients (19,139). The former study reported the controllable toxicities and 18% ORR of Rova-T monotherapy (19), while the later one with 339 participants presented the high incidence of adverse effects that over grade 3 (63%) and the limited clinical benefit (ORR 12.4% and mOS 5.6 months) (139). Moreover, the negatively prognostic results of two phase III clinical trials, MERU (NCT03033511) which compared Rova-T plus dexamethasone with placebo, and TAHOE (NCT03061812) which compared Rova-T with topotecan, indicating the restrictions to the application of Rova-T monotherapy and Rova-T plus dexamethasone in SCLC. Some clinical studies of ED-SCLC, including phase I randomized study which employed Rova-T and Rova-T plus EP in the first-line treatment (NCT02819999), phase I study of AMG 119 monotherapy in SCLC patients who resistant to chemotherapy (NCT03392064), a phase I/II studies that combined Rova-T with immunotherapy (NCT03026166 and NCT03000257), and phase I study of AMG 757 in combination of immunotherapy (NCT03319940), is not disclosed the research data (Table 2). It is necessary to synthesize the results of these clinical trials to make a comprehensive evaluation of DLL3 inhibitors in untreated and resistant SCLC.
Trilaciclib, the CDK4/6 inhibitor, showed promising anti-cancer activity in SCLC. When compared to the placebo-EP regimen, a phase Ib/II trial (NCT02499770) of untreated ED-SCLC patients demonstrated that combing Trilaciclib with EP contributed to higher ORR (66.7% vs. 56.8%) and security enhancement, indicating that EP plus CDK4/6 inhibitors therapy may help decline the ratio of primary resistance as well as therapeutic side effects in SCLC (136). More clinical trials will continue to further explore the efficacy of CDK4/6 inhibitors and other targeted drugs in SCLC (Table 2).
Epigenetic inhibitors
Epigenetic mechanisms which contain the methylation of cancer-related DNA, and the aberrant expression levels of lncRNAs and miRNAs, etc. could induce the process of drugs-resistance in SCLC (137,140). The main characteristics of clinical trials of epigenetic inhibitors in SCLC were summed up in Table 3. RRx-001 takes effect by regulating the epigenetic characteristics of tumor-related immunocytes, thus being identified as the epigenetic and immunity inhibitor. QUADRUPLE THREAT (NCT02489903) was a phase II trial that firstly demonstrated the manageable side effects and cheerful efficacy of RRx-001 plus EP treatment in SCLC patents who were resistant to the platinum-based chemotherapy (143). The open-label trial of 26 enrolled SCLC patients observed that the ORR, half-year OS, and 1-year OS of RRx-001 plus EP therapy was 26.9%, 66.2%, and 44.1%, respectively (143). At present, a phase III study REPLATINUM (NCT03699956) is conducted to further assess the possibility of RRx-001 in reversing the chemotherapy resistance in SCLC, and its efficacy in the third-line or above treatment in SCLC (141).
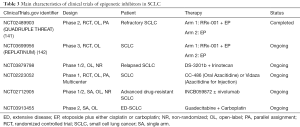
Full table
EZH2 is the histone methyltransferase gene that has broader impacts on the methylation level in SCLC (142). A preclinical research of drug-resistant mice and cell lines demonstrated that the employment of EZH2 inhibitor promoted the expression of SLFN11 and inhibited the H3K27me3 level by epigenetic regulation, thus contributed to the re-sensitivity of chemotherapy in SCLC (3). DS-3201b, also known as valemomestat, is the EZH1/2 inhibitor that have potential to re-sensitizing the tumor cells to chemotherapy by epigenetic mechanism. A newly phase I/II study will make an investigation about the efficacy of DS-3201b plus IP in relapsed SCLC (NCT03879798). Other epigenetic inhibitors, such as CC-486, Vidaza, and INCB059872, guadecitabine (SGI-110) are also subjected to clinical trials in SCLC (Table 3). The exploration of epigenetic inhibitors in refractory SCLC is still in the beginning. Revealing the specific and common epigenetic alterations in drugs-resistant SCLC by more researches is expected, which help fully discovery effectively sites to tackle drugs-resistance and improve drug efficacy.
Conclusions and prospective
At present, in the treatment of SCLC, although a large number of studies were conducted by scientists and oncologists, the development of resistance against anti-cancer drugs is still inevitable and severe. MDR, as one of the major difficulties in SCLC therapy, is hard to be broken through due to the currently conflicting evidences on it and the lack of substantial therapeutic progress made over the past few years. What’s more, it is disappointing that there are not adequate concentrated clinical studies which mainly focus on the analysis of the mechanisms of MDR in SCLC patients. Therefore, there is an urgent need to clarify the sophisticated mechanisms of MDR in SCLC so as to develop promising strategies that overcome drug resistance and confer significant survival benefits for patients. Firstly, the developing molecular biotechnology can provide new methods for the studies of drug-resistant analysis and offer more novel target sites for SCLC therapy. Moreover, future basic and clinical studies as well as updated real-world data are required for the validation of treatment efficacy, the exploration of novel combination therapy, and the search of efficient and convenient biomarkers.
Acknowledgments
Funding: This study was supported in part by a grant from National Natural Science Foundation of China (81802255), Shanghai Pujiang Program (17PJD036) and a grant from Shanghai Municipal Commission of Health and Family Planning Program (20174Y0131), national key research & development project (2016YFC0902300), major disease clinical skills enhancement program of three-year action plan for promoting clinical skills and clinical innovation in municipal hospitals, Shanghai Shen Kang Hospital Development Center Clinical Research Plan of SHDC (16CR1001A), “Dream Tutor” Outstanding Young Talents Program (fkyq1901), key disciplines of Shanghai Pulmonary Hospital (2017ZZ02012), grant of Shanghai Science and Technology Commission (16JC1405900).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tlcr-19-547). YH serves as an unpaid editorial board member of Translational Lung Cancer Research. CZ serves as an unpaid Editor in-Chief of Translational Lung Cancer Research from Mar 2012 to Mar 2022. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69:7-34. [Crossref] [PubMed]
- Tsoukalas N, Aravantinou-Fatorou E, Baxevanos P, et al. Advanced small cell lung cancer (SCLC): new challenges and new expectations. Ann Transl Med 2018;6:145. [Crossref] [PubMed]
- Gardner EE, Lok BH, Schneeberger VE, et al. Chemosensitive Relapse in Small Cell Lung Cancer Proceeds through an EZH2-SLFN11 Axis. Cancer Cell 2017;31:286-99. [Crossref] [PubMed]
- Lawson MH, Cummings NM, Rassl DM, et al. Two novel determinants of etoposide resistance in small cell lung cancer. Cancer Res 2011;71:4877-87. [Crossref] [PubMed]
- Byers LA, Rudin CM. Small cell lung cancer: where do we go from here? Cancer 2015;121:664-72. [Crossref] [PubMed]
- Micke P, Faldum A, Metz T, et al. Staging small cell lung cancer: Veterans Administration Lung Study Group versus International Association for the Study of Lung Cancer--what limits limited disease? Lung Cancer 2002;37:271-6. [Crossref] [PubMed]
- Evans WK, Shepherd FA, Feld R, et al. VP-16 and cisplatin as first-line therapy for small-cell lung cancer. J Clin Oncol 1985;3:1471-7. [Crossref] [PubMed]
- Farago AF, Keane FK. Current standards for clinical management of small cell lung cancer. Transl Lung Cancer Res 2018;7:69-79. [Crossref] [PubMed]
- Früh M, De Ruysscher D, Popat S, et al. Small-cell lung cancer (SCLC): ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 2013;24 Suppl 6:vi99-vi105. [Crossref] [PubMed]
- Stinchcombe TE, Gore EM. Limited-stage small cell lung cancer: current chemoradiotherapy treatment paradigms. Oncologist 2010;15:187-95. [Crossref] [PubMed]
- Hanna N, Bunn PA Jr, Langer C, et al. Randomized phase III trial comparing irinotecan/cisplatin with etoposide/cisplatin in patients with previously untreated extensive-stage disease small-cell lung cancer. J Clin Oncol 2006;24:2038-43. [Crossref] [PubMed]
- Kalemkerian GP, Akerley W, Bogner P, et al. Small cell lung cancer. J Natl Compr Canc Netw 2013;11:78-98. [Crossref] [PubMed]
- Lara PN Jr, Natale R, Crowley J, et al. Phase III trial of irinotecan/cisplatin compared with etoposide/cisplatin in extensive-stage small-cell lung cancer: clinical and pharmacogenomic results from SWOG S0124. J Clin Oncol 2009;27:2530-5. [Crossref] [PubMed]
- Noda K, Nishiwaki Y, Kawahara M, et al. Irinotecan plus cisplatin compared with etoposide plus cisplatin for extensive small-cell lung cancer. N Engl J Med 2002;346:85-91. [Crossref] [PubMed]
- Kondo R, Watanabe S, Shoji S, et al. A Phase II Study of Irinotecan for Patients with Previously Treated Small-Cell Lung Cancer. Oncology 2018;94:223-32. [Crossref] [PubMed]
- Pallis AG, Agelidou A, Agelaki S, et al. A multicenter randomized phase II study of the irinotecan/gemcitabine doublet versus irinotecan monotherapy in previously treated patients with extensive stage small-cell lung cancer. Lung Cancer 2009;65:187-91. [Crossref] [PubMed]
- Masuda N, Fukuoka M, Kusunoki Y, et al. CPT-11: a new derivative of camptothecin for the treatment of refractory or relapsed small-cell lung cancer. J Clin Oncol 1992;10:1225-9. [Crossref] [PubMed]
- Leonetti A, Facchinetti F, Minari R, et al. Notch pathway in small-cell lung cancer: from preclinical evidence to therapeutic challenges. Cell Oncol (Dordr) 2019;42:261-73. [Crossref] [PubMed]
- Rudin CM, Pietanza MC, Bauer TM, et al. Rovalpituzumab tesirine, a DLL3-targeted antibody-drug conjugate, in recurrent small-cell lung cancer: a first-in-human, first-in-class, open-label, phase 1 study. Lancet Oncol 2017;18:42-51. [Crossref] [PubMed]
- Satoh MS, Lindahl T. Role of poly (ADP-ribose) formation in DNA repair. Nature 1992;356:356-8. [Crossref] [PubMed]
- Horn L, Mansfield AS, Szczęsna A, et al. First-Line Atezolizumab plus Chemotherapy in Extensive-Stage Small-Cell Lung Cancer. N Engl J Med 2018;379:2220-9. [Crossref] [PubMed]
- Paz-Ares L, Dvorkin M, Chen Y, et al. Durvalumab plus platinum-etoposide versus platinum-etoposide in first-line treatment of extensive-stage small-cell lung cancer (CASPIAN): a randomised, controlled, open-label, phase 3 trial. Lancet 2019;394:1929-39. [Crossref] [PubMed]
- Jett JR, Schild SE, Kesler KA, et al. Treatment of small cell lung cancer: Diagnosis and management of lung cancer, 3rd ed: American College of Chest Physicians evidence-based clinical practice guidelines. Chest 2013;143:e400S-19S.
- Goldstein M, Kastan MB. The DNA damage response: implications for tumor responses to radiation and chemotherapy. Annu Rev Med 2015;66:129-43. [Crossref] [PubMed]
- Kelland LR. New platinum antitumor complexes. Crit Rev Oncol Hematol 1993;15:191-219. [Crossref] [PubMed]
- Siddik ZH. Cisplatin: mode of cytotoxic action and molecular basis of resistance. Oncogene 2003;22:7265-79. [Crossref] [PubMed]
- Heyza JR, Lei W, Watza D, et al. Identification and Characterization of Synthetic Viability with ERCC1 Deficiency in Response to Interstrand Crosslinks in Lung Cancer. Clin Cancer Res 2019;25:2523-36. [Crossref] [PubMed]
- Fink D, Aebi S, Howell SB. The role of DNA mismatch repair in drug resistance. Clin Cancer Res 1998;4:1-6. [PubMed]
- de las Alas MM, Aebi S, Fink D, et al. Loss of DNA mismatch repair: effects on the rate of mutation to drug resistance. J Natl Cancer Inst 1997;89:1537-41. [Crossref] [PubMed]
- Hamaguchi K, Godwin AK, Yakushiji M, et al. Cross-resistance to diverse drugs is associated with primary cisplatin resistance in ovarian cancer cell lines. Cancer Res 1993;53:5225-32. [PubMed]
- Sakamoto M, Kondo A, Kawasaki K, et al. Analysis of gene expression profiles associated with cisplatin resistance in human ovarian cancer cell lines and tissues using cDNA microarray. Hum Cell 2001;14:305-15. [PubMed]
- Goto S, Iida T, Cho S, et al. Overexpression of glutathione S-transferase pi enhances the adduct formation of cisplatin with glutathione in human cancer cells. Free Radic Res 1999;31:549-58. [Crossref] [PubMed]
- Meijer C, Mulder NH, Hospers GA, et al. The role of glutathione in resistance to cisplatin in a human small cell lung cancer cell line. Br J Cancer 1990;62:72-7. [Crossref] [PubMed]
- Kurokawa H, Ishida T, Nishio K, et al. Gamma-glutamylcysteine synthetase gene overexpression results in increased activity of the ATP-dependent glutathione S-conjugate export pump and cisplatin resistance. Biochem Biophys Res Commun 1995;216:258-64. [Crossref] [PubMed]
- Caffrey PB, Frenkel GD, McAndrew KL, et al. A Model of the Development of Cisplatin Resistance in Human Small Cell Lung Cancer Xenografts. In Vivo 2016;30:745-9. [Crossref] [PubMed]
- Campling BG, Baer K, Baker HM, et al. Do glutathione and related enzymes play a role in drug resistance in small cell lung cancer cell lines? Br J Cancer 1993;68:327-35. [Crossref] [PubMed]
- Teschendorff AE, Lee SH, Jones A, et al. HOTAIR and its surrogate DNA methylation signature indicate carboplatin resistance in ovarian cancer. Genome Med 2015;7:108. [Crossref] [PubMed]
- Fang S, Shen Y, Chen B, et al. H3K27me3 induces multidrug resistance in small cell lung cancer by affecting HOXA1 DNA methylation via regulation of the lncRNA HOTAIR. Ann Transl Med 2018;6:440. [Crossref] [PubMed]
- Fang S, Gao H, Tong Y, et al. Long noncoding RNA-HOTAIR affects chemoresistance by regulating HOXA1 methylation in small cell lung cancer cells. Lab Invest 2016;96:60-8. [Crossref] [PubMed]
- Gupta RA, Shah N, Wang KC, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010;464:1071-6. [Crossref] [PubMed]
- Tsai MC, Manor O, Wan Y, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science 2010;329:689-93. [Crossref] [PubMed]
- Ambros V. microRNAs: tiny regulators with great potential. Cell 2001;107:823-6. [Crossref] [PubMed]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004;116:281-97. [Crossref] [PubMed]
- Li J, Guan J, Long X, et al. mir-1-mediated paracrine effect of cancer-associated fibroblasts on lung cancer cell proliferation and chemoresistance. Oncol Rep 2016;35:3523-31. [Crossref] [PubMed]
- Ma W, Ma CN, Li XD, et al. Examining the effect of gene reduction in miR-95 and enhanced radiosensitivity in non-small cell lung cancer. Cancer Gene Ther 2016;23:66-71. [Crossref] [PubMed]
- Lai J, Yang H, Zhu Y, et al. MiR-7-5p-mediated downregulation of PARP1 impacts DNA homologous recombination repair and resistance to doxorubicin in small cell lung cancer. BMC Cancer 2019;19:602. [Crossref] [PubMed]
- Liu H, Huang J, Peng J, et al. Upregulation of the inwardly rectifying potassium channel Kir2.1 (KCNJ2) modulates multidrug resistance of small-cell lung cancer under the regulation of miR-7 and the Ras/MAPK pathway. Mol Cancer 2015;14:59. [Crossref] [PubMed]
- Liu H, Wu X, Huang J, et al. miR-7 modulates chemoresistance of small cell lung cancer by repressing MRP1/ABCC1. Int J Exp Pathol 2015;96:240-7. [Crossref] [PubMed]
- Liang Z, Wu H, Xia J, et al. Involvement of miR-326 in chemotherapy resistance of breast cancer through modulating expression of multidrug resistance-associated protein 1. Biochem Pharmacol 2010;79:817-24. [Crossref] [PubMed]
- Kraus WL, Lis JT. PARP goes transcription. Cell 2003;113:677-83. [Crossref] [PubMed]
- Ray Chaudhuri A, Nussenzweig A. The multifaceted roles of PARP1 in DNA repair and chromatin remodelling. Nat Rev Mol Cell Biol 2017;18:610-21. [Crossref] [PubMed]
- Wu ZS, Wu Q, Wang CQ, et al. MiR-339-5p inhibits breast cancer cell migration and invasion in vitro and may be a potential biomarker for breast cancer prognosis. BMC Cancer 2010;10:542. [Crossref] [PubMed]
- Shan W, Li J, Bai Y, et al. miR-339-5p inhibits migration and invasion in ovarian cancer cell lines by targeting NACC1 and BCL6. Tumour Biol 2016;37:5203-11. [Crossref] [PubMed]
- Li Y, Zhao W, Bao P, et al. miR-339-5p inhibits cell migration and invasion in vitro and may be associated with the tumor-node-metastasis staging and lymph node metastasis of non-small cell lung cancer. Oncol Lett 2014;8:719-25. [Crossref] [PubMed]
- Gan CZ, Li G, Luo QS, et al. miR-339-5p downregulation contributes to Taxol resistance in small-cell lung cancer by targeting α1,2-fucosyltransferase 1. IUBMB Life 2017;69:841-9. [Crossref] [PubMed]
- Ma B, Simala-Grant JL, Taylor DE. Fucosylation in prokaryotes and eukaryotes. Glycobiology 2006;16:158R-84R. [Crossref] [PubMed]
- Zhang Z, Sun P, Liu J, et al. Suppression of FUT1/FUT4 expression by siRNA inhibits tumor growth. Biochim Biophys Acta 2008;1783:287-96. [Crossref] [PubMed]
- Kim Y, Kim H, Park D, et al. miR-335 Targets SIAH2 and Confers Sensitivity to Anti-Cancer Drugs by Increasing the Expression of HDAC3. Mol Cells 2015;38:562-72. [Crossref] [PubMed]
- Gong M, Ma J, Guillemette R, et al. miR-335 inhibits small cell lung cancer bone metastases via IGF-IR and RANKL pathways. Mol Cancer Res 2014;12:101-10. [Crossref] [PubMed]
- Tang R, Lei Y, Hu B, et al. WW domain binding protein 5 induces multidrug resistance of small cell lung cancer under the regulation of miR-335 through the Hippo pathway. Br J Cancer 2016;115:243-51. [Crossref] [PubMed]
- Bedford MT, Chan DC, Leder P. FBP WW domains and the Abl SH3 domain bind to a specific class of proline-rich ligands. EMBO J 1997;16:2376-83. [Crossref] [PubMed]
- Feng X, Wang Z, Fillmore R, et al. MiR-200, a new star miRNA in human cancer. Cancer Lett 2014;344:166-73. [Crossref] [PubMed]
- Rui W, Bing F, Hai-Zhu S, et al. Identification of microRNA profiles in docetaxel-resistant human non-small cell lung carcinoma cells (SPC-A1). J Cell Mol Med 2010;14:206-14. [Crossref] [PubMed]
- Zhu W, Xu H, Zhu D, et al. miR-200bc/429 cluster modulates multidrug resistance of human cancer cell lines by targeting BCL2 and XIAP. Cancer Chemother Pharmacol 2012;69:723-31. [Crossref] [PubMed]
- Pogribny IP, Filkowski JN, Tryndyak VP, et al. Alterations of microRNAs and their targets are associated with acquired resistance of MCF-7 breast cancer cells to cisplatin. Int J Cancer 2010;127:1785-94. [Crossref] [PubMed]
- Fang S, Zeng X, Zhu W, et al. Zinc finger E-box-binding homeobox 2 (ZEB2) regulated by miR-200b contributes to multi-drug resistance of small cell lung cancer. Exp Mol Pathol 2014;96:438-44. [Crossref] [PubMed]
- Adam L, Zhong M, Choi W, et al. miR-200 expression regulates epithelial-to-mesenchymal transition in bladder cancer cells and reverses resistance to epidermal growth factor receptor therapy. Clin Cancer Res 2009;15:5060-72. [Crossref] [PubMed]
- Haslehurst AM, Koti M, Dharsee M, et al. EMT transcription factors snail and slug directly contribute to cisplatin resistance in ovarian cancer. BMC Cancer 2012;12:91. [Crossref] [PubMed]
- Seitz H, Royo H, Bortolin ML, et al. A large imprinted microRNA gene cluster at the mouse Dlk1-Gtl2 domain. Genome Res 2004;14:1741-8. [Crossref] [PubMed]
- Luk JM, Burchard J, Zhang C, et al. DLK1-DIO3 genomic imprinted microRNA cluster at 14q32.2 defines a stemlike subtype of hepatocellular carcinoma associated with poor survival. J Biol Chem 2011;286:30706-13. [Crossref] [PubMed]
- Pan JY, Zhang F, Sun CC, et al. miR-134: A Human Cancer Suppressor? Mol Ther Nucleic Acids 2017;6:140-9. [Crossref] [PubMed]
- Guo L, Liu Y, Bai Y, et al. Gene expression profiling of drug-resistant small cell lung cancer cells by combining microRNA and cDNA expression analysis. Eur J Cancer 2010;46:1692-702. [Crossref] [PubMed]
- Reed JC. Dysregulation of apoptosis in cancer. J Clin Oncol 1999;17:2941-53. [Crossref] [PubMed]
- Kerr JF, Winterford CM, Harmon BV. Apoptosis. Its significance in cancer and cancer therapy. Cancer 1994;73:2013-26. [Crossref] [PubMed]
- Stefanaki K, Rontogiannis D, Vamvouka C, et al. Immunohistochemical detection of bcl2, p53, mdm2 and p21/waf1 proteins in small-cell lung carcinomas. Anticancer Res 1998;18:1689-95. [PubMed]
- Perego P, Giarola M, Righetti SC, et al. Association between cisplatin resistance and mutation of p53 gene and reduced bax expression in ovarian carcinoma cell systems. Cancer Res 1996;56:556-62. [PubMed]
- Ikeguchi M, Liu J, Kaibara N. Expression of survivin mRNA and protein in gastric cancer cell line (MKN-45) during cisplatin treatment. Apoptosis 2002;7:23-9. [Crossref] [PubMed]
- Kaplan E, Gunduz U. Expression analysis of TOP2A, MSH2 and MLH1 genes in MCF7 cells at different levels of etoposide resistance. Biomed Pharmacother 2012;66:29-35. [Crossref] [PubMed]
- Chen F, Arseven OK, Cryns VL. Proteolysis of the mismatch repair protein MLH1 by caspase-3 promotes DNA damage-induced apoptosis. J Biol Chem 2004;279:27542-8. [Crossref] [PubMed]
- Zhuang G, Song W, Amato K, et al. Effects of cancer-associated EPHA3 mutations on lung cancer. J Natl Cancer Inst 2012;104:1182-97. [Crossref] [PubMed]
- Ross JS, Wang K, Elkadi OR, et al. Next-generation sequencing reveals frequent consistent genomic alterations in small cell undifferentiated lung cancer. J Clin Pathol 2014;67:772-6. [Crossref] [PubMed]
- Peng J, Wang Q, Liu H, et al. EPHA3 regulates the multidrug resistance of small cell lung cancer via the PI3K/BMX/STAT3 signaling pathway. Tumour Biol 2016;37:11959-71. [Crossref] [PubMed]
- Wallington-Beddoe CT, Bradstock KF, Bendall LJ. Oncogenic properties of sphingosine kinases in haematological malignancies. Br J Haematol 2013;161:623-38. [Crossref] [PubMed]
- Akao Y, Banno Y, Nakagawa Y, et al. High expression of sphingosine kinase 1 and S1P receptors in chemotherapy-resistant prostate cancer PC3 cells and their camptothecin-induced up-regulation. Biochem Biophys Res Commun 2006;342:1284-90. [Crossref] [PubMed]
- Yang L, Hu H, Deng Y, et al. Role of SPHK1 regulates multi-drug resistance of small cell lung cancer and its clinical significance. Zhongguo Fei Ai Za Zhi 2014;17:769-77. [PubMed]
- Haixia P, Yifeng B, Honglin H. Role and clinical significance of RLIP76 in regulation of multi-drug resistance of small cell lung cancer. Zhonghua Zhong Liu Za Zhi 2015;37:266-71. [PubMed]
- Vilborg A, Glahder JA, Wilhelm MT, et al. The p53 target Wig-1 regulates p53 mRNA stability through an AU-rich element. Proc Natl Acad Sci U S A 2009;106:15756-61. [Crossref] [PubMed]
- Vilborg A, Bersani C, Wickstrom M, et al. Wig-1, a novel regulator of N-Myc mRNA and N-Myc-driven tumor growth. Cell Death Dis 2012;3:e298. [Crossref] [PubMed]
- Luo S, Bai Y, Lan H. Influence of interference of WIG-1 on the multi-drug resistance in small cell lung cancer. Zhonghua Zhong Liu Za Zhi 2014;36:733-8. [PubMed]
- Meighan-Mantha RL, Hsu DK, Guo Y, et al. The mitogen-inducible Fn14 gene encodes a type I transmembrane protein that modulates fibroblast adhesion and migration. J Biol Chem 1999;274:33166-76. [Crossref] [PubMed]
- Han H, Bearss DJ, Browne LW, et al. Identification of differentially expressed genes in pancreatic cancer cells using cDNA microarray. Cancer Res 2002;62:2890-6. [PubMed]
- Tran NL, McDonough WS, Donohue PJ, et al. The human Fn14 receptor gene is up-regulated in migrating glioma cells in vitro and overexpressed in advanced glial tumors. Am J Pathol 2003;162:1313-21. [Crossref] [PubMed]
- Tran NL, McDonough WS, Savitch BA, et al. Increased fibroblast growth factor-inducible 14 expression levels promote glioma cell invasion via Rac1 and nuclear factor-kappaB and correlate with poor patient outcome. Cancer Res 2006;66:9535-42. [Crossref] [PubMed]
- Whitsett TG, Cheng E, Inge L, et al. Elevated expression of Fn14 in non-small cell lung cancer correlates with activated EGFR and promotes tumor cell migration and invasion. Am J Pathol 2012;181:111-20. [Crossref] [PubMed]
- Huang M, Narita S, Tsuchiya N, et al. Overexpression of Fn14 promotes androgen-independent prostate cancer progression through MMP-9 and correlates with poor treatment outcome. Carcinogenesis 2011;32:1589-96. [Crossref] [PubMed]
- Li X, Zhu W, Chen Z, et al. Fibroblast growth factor-inducible 14 regulates cell growth and multidrug resistance of small-cell lung cancer through the nuclear factor-kappaB pathway. Anticancer Drugs 2014;25:1152-64. [Crossref] [PubMed]
- Kwon OH, Park SJ, Kang TW, et al. Elevated fibroblast growth factor-inducible 14 expression promotes gastric cancer growth via nuclear factor-kappaB and is associated with poor patient outcome. Cancer Lett 2012;314:73-81. [Crossref] [PubMed]
- Tran NL, McDonough WS, Savitch BA, et al. The tumor necrosis factor-like weak inducer of apoptosis (TWEAK)-fibroblast growth factor-inducible 14 (Fn14) signaling system regulates glioma cell survival via NFkappaB pathway activation and BCL-XL/BCL-W expression. J Biol Chem 2005;280:3483-92. [Crossref] [PubMed]
- Compton S, Kim C, Griner NB, et al. Mitochondrial dysfunction impairs tumor suppressor p53 expression/function. J Biol Chem 2011;286:20297-312. [Crossref] [PubMed]
- Chen JZ, Gokden N, Greene GF, et al. Extensive somatic mitochondrial mutations in primary prostate cancer using laser capture microdissection. Cancer Res 2002;62:6470-4. [PubMed]
- Heddi A, Faure-Vigny H, Wallace DC, et al. Coordinate expression of nuclear and mitochondrial genes involved in energy production in carcinoma and oncocytoma. Biochim Biophys Acta 1996;1316:203-9. [Crossref] [PubMed]
- Vivekanandan P, Daniel H, Yeh MM, et al. Mitochondrial mutations in hepatocellular carcinomas and fibrolamellar carcinomas. Mod Pathol 2010;23:790-8. [Crossref] [PubMed]
- Ma L, Wang R, Duan H, et al. Mitochondrial dysfunction rather than mtDNA sequence mutation is responsible for the multi-drug resistance of small cell lung cancer. Oncol Rep 2015;34:3238-46. [Crossref] [PubMed]
- Degli Esposti M. Mitochondria in apoptosis: past, present and future. Biochem Soc Trans 2004;32:493-5. [Crossref] [PubMed]
- Salet C, Moreno G, Ricchelli F, et al. Singlet oxygen produced by photodynamic action causes inactivation of the mitochondrial permeability transition pore. J Biol Chem 1997;272:21938-43. [Crossref] [PubMed]
- Kool M, van der Linden M, de Haas M, et al. Expression of human MRP6, a homologue of the multidrug resistance protein gene MRP1, in tissues and cancer cells. Cancer Res 1999;59:175-82. [PubMed]
- Samimi G, Safaei R, Katano K, et al. Increased expression of the copper efflux transporter ATP7A mediates resistance to cisplatin, carboplatin, and oxaliplatin in ovarian cancer cells. Clin Cancer Res 2004;10:4661-9. [Crossref] [PubMed]
- Kim ES. Chemotherapy Resistance in Lung Cancer. Adv Exp Med Biol 2016;893:189-209. [Crossref] [PubMed]
- Howell SB, Safaei R, Larson CA, et al. Copper transporters and the cellular pharmacology of the platinum-containing cancer drugs. Mol Pharmacol 2010;77:887-94. [Crossref] [PubMed]
- Alpsoy A, Yasa S, Gunduz U. Etoposide resistance in MCF-7 breast cancer cell line is marked by multiple mechanisms. Biomed Pharmacother 2014;68:351-5. [Crossref] [PubMed]
- Wang YL, Yan YL, Zhou NJ, et al. Mechanism of multidrug resistance of human small cell lung cancer cell line H446/VP. Chin Med J 2010;123:3299-303. [PubMed]
- Brock I, Hipfner DR, Nielsen BS, et al. Sequential coexpression of the multidrug resistance genes MRP and mdr1 and their products in VP-16 (etoposide)-selected H69 small cell lung cancer cells. Cancer Res 1995;55:459-62. [PubMed]
- Minato K, Kanzawa F, Nishio K, et al. Characterization of an etoposide-resistant human small-cell lung cancer cell line. Cancer Chemother Pharmacol 1990;26:313-7. [Crossref] [PubMed]
- Doyle LA, Ross DD, Ordonez JV, et al. An etoposide-resistant lung cancer subline overexpresses the multidrug resistance-associated protein. Br J Cancer 1995;72:535-42. [Crossref] [PubMed]
- Beretta GL, Gatti L, Perego P, et al. Camptothecin resistance in cancer: insights into the molecular mechanisms of a DNA-damaging drug. Curr Med Chem 2013;20:1541-65. [Crossref] [PubMed]
- Takano M, Sugiyama T. UGT1A1 polymorphisms in cancer: impact on irinotecan treatment. Pharmgenomics Pers Med 2017;10:61-8. [Crossref] [PubMed]
- Etienne-Grimaldi MC, Boyer JC, Thomas F, et al. UGT1A1 genotype and irinotecan therapy: general review and implementation in routine practice. Fundam Clin Pharmacol 2015;29:219-37. [Crossref] [PubMed]
- Takano M, Kato M, Yoshikawa T, et al. Clinical significance of UDP-glucuronosyltransferase 1A1*6 for toxicities of combination chemotherapy with irinotecan and cisplatin in gynecologic cancers: a prospective multi-institutional study. Oncology 2009;76:315-21. [Crossref] [PubMed]
- Toffoli G, Cecchin E, Corona G, et al. The role of UGT1A1*28 polymorphism in the pharmacodynamics and pharmacokinetics of irinotecan in patients with metastatic colorectal cancer. J Clin Oncol 2006;24:3061-8. [Crossref] [PubMed]
- Antonia SJ, López-Martin JA, Bendell J, et al. Nivolumab alone and nivolumab plus ipilimumab in recurrent small-cell lung cancer (CheckMate 032): a multicentre, open-label, phase 1/2 trial. Lancet Oncol 2016;17:883-95. [Crossref] [PubMed]
- Owonikoko TK, Kim HR, Govindan R, et al. Nivolumab (nivo) plus ipilimumab (ipi), nivo, or placebo (pbo) as maintenance therapy in patients (pts) with extensive disease small cell lung cancer (ED-SCLC) after first-line (1L) platinum-based chemotherapy (chemo): Results from the double-blind, randomized phase III CheckMate 451 study. Ann Oncol 2019;30 Suppl 2:ii77. [Crossref]
- Ott PA, Elez E, Hiret S, et al. Pembrolizumab in patients with extensive-stage small-cell lung cancer: results from the phase Ib KEYNOTE-028 Study. J Clin Oncol 2017;35:3823-9. [Crossref] [PubMed]
- Strosberg J, Mizuno N, Doi T, et al. Efficacy and Safety of Pembrolizumab in Previously Treated Advanced Neuroendocrine Tumors: Results From the Phase II KEYNOTE-158 Study. Clin Cancer Res 2020;26:2124-30. [Crossref] [PubMed]
- Reck M, Bondarenko I, Luft A, et al. Ipilimumab in combination with paclitaxel and carboplatin as first-line therapy in extensive-disease-small-cell lung cancer: results from a randomized, double-blind, multicenter phase 2 trial. Ann Oncol 2013;24:75-83. [Crossref] [PubMed]
- Reck M, Luft A, Szczesna A, et al. Phase III Randomized Trial of Ipilimumab Plus Etoposide and Platinum Versus Placebo Plus Etoposide and Platinum in Extensive-Stage Small-Cell Lung Cancer. J Clin Oncol 2016;34:3740-8. [Crossref] [PubMed]
- Merck’s KEYTRUDA® (pembrolizumab) in Combination with Chemotherapy Significantly Improved Progression-Free Survival Compared to Chemotherapy Alone as First-Line Treatment for Extensive Stage Small Cell Lung Cancer 2020.
- de Bono J, Ramanathan RK, Mina L, et al. Phase I, Dose-Escalation, Two-Part Trial of the PARP Inhibitor Talazoparib in Patients with Advanced Germline BRCA1/2 Mutations and Selected Sporadic Cancers. Cancer Discov 2017;7:620-9. [Crossref] [PubMed]
- Atrafi F, Groen HJM, Byers LA, et al. A Phase I Dose-Escalation Study of Veliparib Combined with Carboplatin and Etoposide in Patients with Extensive-Stage Small Cell Lung Cancer and Other Solid Tumors. Clin Cancer Res 2019;25:496-505. [Crossref] [PubMed]
- Owonikoko TK, Dahlberg SE, Sica GL, et al. Randomized Phase II Trial of Cisplatin and Etoposide in Combination With Veliparib or Placebo for Extensive-Stage Small-Cell Lung Cancer: ECOG-ACRIN 2511 Study. J Clin Oncol 2019;37:222-9. [Crossref] [PubMed]
- Pietanza MC, Waqar SN, Krug LM, et al. Randomized, Double-Blind, Phase II Study of Temozolomide in Combination With Either Veliparib or Placebo in Patients With Relapsed-Sensitive or Refractory Small-Cell Lung Cancer. J Clin Oncol 2018;36:2386-94. [Crossref] [PubMed]
- Gandhi L, Camidge DR, Ribeiro de Oliveira M, et al. Phase I study of Navitoclax (ABT-263), a novel Bcl-2 family inhibitor, in patients with small-cell lung cancer and other solid tumors. J Clin Oncol 2011;29:909-16. [Crossref] [PubMed]
- Rudin CM, Hann CL, Garon EB, et al. Phase II study of single-agent navitoclax (ABT-263) and biomarker correlates in patients with relapsed small cell lung cancer. Clin Cancer Res 2012;18:3163-9. [Crossref] [PubMed]
- Calvo E, Moreno V, Flynn M, et al. Antitumor activity of lurbinectedin (PM01183) and doxorubicin in relapsed small-cell lung cancer: results from a phase I study. Ann Oncol 2017;28:2559-66. [Crossref] [PubMed]
- Trigo J, Leary A, Besse B, et al. Efficacy and safety of lurbinectedin (PM1183) in small cell lung cancer (SCLC): Results from a phase 2 study. J Thorac Oncol 2018.36.
- Tanaka K, Isse K, Fujihira T, et al. Prevalence of Delta-like protein 3 expression in patients with small cell lung cancer. Lung Cancer 2018;115:116-20. [Crossref] [PubMed]
- Weiss JM, Csoszi T, Maglakelidze M, et al. Myelopreservation with the CDK4/6 inhibitor trilaciclib in patients with small-cell lung cancer receiving first-line chemotherapy: a phase Ib/randomized phase II trial. Ann Oncol 2019;30:1613-21. [Crossref] [PubMed]
- Quintanal-Villalonga A, Molina-Pinelo S. Epigenetics of lung cancer: a translational perspective. Cell Oncol (Dordr) 2019;42:739-56. [Crossref] [PubMed]
- Saunders LR, Bankovich AJ, Anderson WC, et al. A DLL3-targeted antibody-drug conjugate eradicates high-grade pulmonary neuroendocrine tumor-initiating cells in vivo. Sci Transl Med 2015;7:302ra136. [Crossref] [PubMed]
- Morgensztern D, Besse B, Greillier L, et al. Efficacy and Safety of Rovalpituzumab Tesirine in Third-Line and Beyond Patients with DLL3-Expressing, Relapsed/Refractory Small-Cell Lung Cancer: Results From the Phase II TRINITY Study. Clin Cancer Res 2019;25:6958-66. [Crossref] [PubMed]
- Duruisseaux M, Esteller M. Lung cancer epigenetics: From knowledge to applications. Semin Cancer Biol 2018;51:116-28. [Crossref] [PubMed]
- Oronsky B, Reid TR, Larson C, et al. REPLATINUM Phase III randomized study: RRx-001 + platinum doublet versus platinum doublet in third-line small cell lung cancer. Future Oncol 2019;15:3427-33. [Crossref] [PubMed]
- Poirier JT, Gardner EE, Connis N, et al. DNA methylation in small cell lung cancer defines distinct disease subtypes and correlates with high expression of EZH2. Oncogene 2015;34:5869-78. [Crossref] [PubMed]
- Morgensztern D, Rose M, Waqar SN, et al. RRx-001 followed by platinum plus etoposide in patients with previously treated small-cell lung cancer. Br J Cancer 2019;121:211-7. [Crossref] [PubMed]


