
EGFR circulating tumour DNA testing: identification of predictors of ctDNA detection and implications for survival outcomes
Introduction
Epidermal growth factor receptor (EGFR) circulating tumour deoxyribonucleic acid (ctDNA) testing has been incorporated into routine testing for molecular characterization of non-small cell lung cancer (NSCLC). In addition to being utilized to determine of first line therapy options for patients with insufficient tissue, it is used to screen patients with advanced EGFR mutant NSCLC progressing on 1st or 2nd generation TKIs for the presence of the EGFR T790M mutation (1). ctDNA EGFR mutation detection can facilitate determination of appropriate first, second or third generation EGFR TKI therapy without the risks of a tissue biopsy. Clinically it is equally effective in patients with the mutation detected on tissue or plasma genotyping (2-5).
The relative paucity of ctDNA in the circulation mandates the use of an ultrasensitive detection method such as next generation sequencing, droplet digital polymerase chair reaction (ddPCR) or real-time PCR methods (6-9). Despite very sensitive technologies, ctDNA detection can be difficult in patients on EGFR targeted therapy due to the low burden of residual disease, genetically heterogeneous clonal populations and slow rate of disease progression on acquired resistance. Therefore, the identification of the original activating mutation in plasma may be problematic for these reasons and detection of the subpopulation of T790M clones represents an even greater challenge.
Several commercially available kits are now available for EGFR ctDNA testing. Regulatory approved EGFR ctDNA testing include the Cobas EGFR Mutations Test version 2 (Roche Molecular Diagnostics; FDA approval for EGFR del19, L858R, and T790M testing), Therascreen EGFR Rotor-Gene Q (RGQ) Plasma polymerase chain reaction (PCR) kit (Qiagen Inc.; FDA and European Union approved for EGFR del19 and EGFR L858R detection) and AmoyDx Super Amplification Refractory Mutation System (ARMS) EGFR mutation test (AmoyDx; Chinese Food and Drug Administration approval for detection of EGFR del19, 858R, and T790M) (10-12). Multi-gene next-generation sequencing liquid biopsy platforms are also readily accessible (13,14). While a multitude of commercial options are available, the sensitivity of ctDNA testing can be challenging and can delay treatment decision making if the result is inconclusive and a biopsy is required.
The goal of our study was to identify patient factors that predict for the detection of the activating and resistance EGFR mutations in the ctDNA using a laboratory developed test and to assess the value of ctDNA detection as an indicator of aggressiveness of disease and outcomes.
Methods
All patients with advanced EGFR mutant NSCLC undergoing ctDNA testing from February 2018 to March 2019 at BC Cancer were identified. Baseline characteristics, imaging data at time of ctDNA testing and follow-up data were collected retrospectively. Baseline CT scan results nearest to the date of ctDNA testing were reviewed by one clinical investigator (AP) to identify number and location of sites of progressive disease with randomized quality assurance assessments by a second clinical investigator (CH). Metastatic sites were defined as metastases noted on CT imaging. Progression sites were defined as individual lesions that increased by a minimum of 20% in the greatest dimension or new lesions that developed on CT imaging. Progression categories were selected based on local practices for stereotactic body radiation therapy for ≤5 lesions. Overall survival was calculated from the timepoint of the first EGFR ctDNA test until death.
Twenty mL of peripheral blood was collected in Streck tubes (Streck, La Vista, Nebraska) which were processed to separate the plasma within 3 days. Plasma separation was performed using a two-step centrifugation which included a 1,900 g spin for 10 minutes at 4 °C followed by a 16,000 g spin for 10 minutes at 4 °C, and stored at −80 °C in CryoVials. Extraction of cell-free DNA (cfDNA) from 10 ml plasma was performed using two separate columns of the QIAamp Circulating Nucleic Acid kit (Qiagen, Hliden, Germany). Eluted volumes were combined and quantified by a Qubit™ fluorometer (ThermoFisher Scientific, Waltham, MA, USA) with the dsDNA High Sensitivity assay kit.
Custom ddPCR assays were used to test for the appropriate EGFR activating mutation (exon 19 deletion, L858R or G719A/C/S) and the EGFR T790M resistance mutation using the Bio-Rad QX200 system (Bio-Rad Laboratories, Hercules, California). The limit of detection of the custom designed ddPCR assays was ≤0.15% variant allele fraction.
The presence or absence of the original activating mutation and EGFR T790M was reported. In our clinical testing algorithm, if the ctDNA specimen was positive for the original mutation and negative for the T790M mutation it was considered a true negative for T790M. The rationale was that if T790M could not be detected in the presence of the original activating mutation then it did not represent a clinically significant proportion of the disease burden to determine next line of therapy. If the original activation mutation and/or T790M was not detectable the result was reported as indeterminate.
The primary objective was to identify clinical predictors of a positive ctDNA test. Statistical analysis included Chi squared test, Fisher’s exact test and logistical regression. The secondary outcome measured was overall survival (OS) calculated from the date of ctDNA testing to death or last follow-up. The first ctDNA test was done at the time of clinically significant disease progression as decided by the treating physician. Repeat ctDNA tests were performed at clinician discretion. Survival outcomes were compared using the Kaplan Meier method, log rank test and Cox proportional hazards test. All statistical analysis was carried out using SPSS version 25 (Armonk, NY: IBM Corp.).
All data was collected and analyzed as per local ethical approval (H18-02964, University of British Columbia, BC Cancer Research Ethics Board). Patient consent was not obtained for this retrospective review.
Results
EGFR ctDNA tests were performed on 177 patients with EGFR mutant NSCLC (Table 1). A total of 136 patients had only one ctDNA test, 33 had two tests and 8 had three or more. At the time of first ctDNA testing, the median age was 66 years, 63% of patients were female, 61% were never smokers and 55% were of Asian ethnicity. Routine tissue genotyping at diagnosis showed that 54% patients had tumours with an EGFR exon 19 deletion, 35% had an EGFR L858R mutation, 5% had more than 1 EGFR mutation, 5% had an uncommon EGFR mutation and 1% had a de novo EGFR T790M mutation. Prior EGFR TKI treatment was with a first generation TKI in 65% and second generation TKI in 35%. The median duration of initial EGFR TKI therapy was 14.9 months (m).
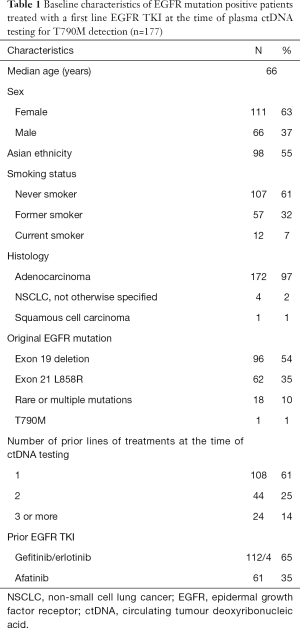
Full table
The median time from diagnosis of metastatic disease to ctDNA testing was 16.9 m. At the time of first EGFR ctDNA test, 101 (57%) had undetectable original or T790M EGFR mutation, 51 (29%) were positive for the original mutation and EGFR T790M, 23 (13%) were positive for the original EGFR mutation alone and 2 (1%) were positive for EGFR T790M alone. The sensitivity of the test was 43% for EGFR ctDNA detection.
Imaging data was available for 175 patients at the time of first EGFR ctDNA testing with the location and number of sites of progressive disease as shown in Table 2. 40% of patients had greater than 10 sites of metastatic disease. All sites of progression were recorded for each patient at the time of first ctDNA testing, the most common sites of progression were lung 65%, bone 31% and pleura/peritoneum 21%. Progression was seen in 0–2 sites in 38% of patients, 3–5 sites 31%, 6 or greater sites 31%.
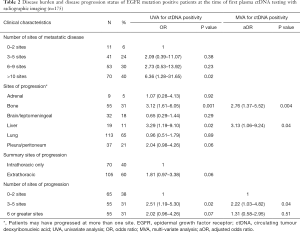
Full table
ctDNA positivity with the original EGFR mutation or original mutation and T790M was noted in 76 of the 175 patients at first ctDNA test with radiographic imaging available. On univariate analysis, predictors for ctDNA first test positivity included the presence of greater than 10 metastases, bone metastases, liver metastases and 3–5 sites of disease progression (Table 2). On multivariate analysis including bony progression, liver progression and number of progressing sites, the presence of liver or bone metastases and 3–5 progressing sites remained statistically significant predictors of ctDNA positivity at the time of first ctDNA test.
Of the 177 patients, 96 received subsequent lines of treatment; 66 (37%) received osimertinib, 30 (17%) platinum doublet chemotherapy and 81 (46%) continued the first line EGFR TKI or were deceased before initiation of next line of treatment. There were 28 (29%) patients who received 3rd line treatment; platinum doublet (36%), osimertinib (25%), first or second generation EGFR TKI (14%), single agent chemotherapy (14%) and immunotherapy (11%).
Repeated ctDNA testing was conducted in 41 patients, with up to 2 repeat ctDNA tests 12 patients were identified to have a T790M mutation. The median time from first to second ctDNA test was 1.6 m (interquartile range, 0.7–2.9 m). Forty-seven patients that had an initial ctDNA test had a subsequent tissue-based EGFR test of which 15 were T790M positive. 27 of the 101 patients with an indeterminate first ctDNA test result went on to be T790M positive on subsequent testing. In total, there were 80 patients who had a detectable T790M mutation by initial or repeat plasma or tissue-based testing, which accounts for 45% of the whole population. After repeat testing, there were a total of 64 patients who had an indeterminate result for T790M status due to undetectable EGFR ctDNA, inability to biopsy or death.
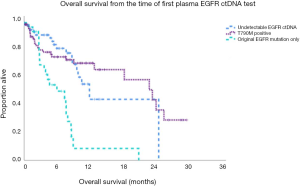
The median overall survival (OS) from time of first ctDNA testing was 13.2 m including all patients regardless of ctDNA testing results. The median OS was 8.18 m in the patients who had detectable EGFR ctDNA versus 25.3 m with undetectable EGFR ctDNA at first testing (P=0.001). The median OS from first ctDNA test was 7.6 m for original EGFR mutation only (n=33), 12.3 m for undetectable ctDNA (n=64) and 24.1 m with T790M (n=80) (Figure 1, P=0.001). In univariate analysis for OS, the presence of the original EGFR mutation only and 6 or greater sites of progression had a higher risk of death (Table 3). In multivariate analysis, the original EGFR mutation only was predictive of mortality.
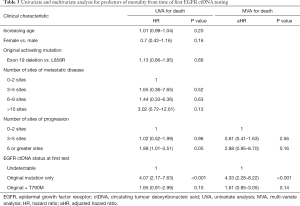
Full table
Discussion
Our study demonstrated that in patients progressing on first line EGFR TKI, the presence of bone or liver metastases and progression in 3–5 sites were predictors for EGFR ctDNA detection. Awareness of ctDNA shedding predictive factors may facilitate decision-making with respect to understanding the likelihood of an informative outcome from an EGFR ctDNA test and the need for arranging alternative tissue-based testing. Our study also noted that the detection of the original EGFR ctDNA mutation only at the time of disease progression on first line EGFR TKI is a negative prognostic indicator, potentially reflecting disease activity in the absence of a targetable treatment option.
The widespread adoption of EGFR ctDNA testing to genotype NSCLC has facilitated improved care of EGFR mutation positive NSCLC lung cancer patients. Regulatory approved liquid biopsy EGFR testing platforms have been evaluated in the real world. In the ASSESS study, newly diagnosed treatment-naive advanced NSCLC patients provided diagnostic tissue/cytologic and plasma samples. Comparison of the Cobas EGFR Mutation plasma test versus tumour tissue demonstrated that the plasma testing had a sensitivity of 46% with tissue testing as the gold standard (15). Similarly, IGNITE, a study with a similar design and the same testing platform involving over 2500 matched specimens, noted a sensitivity of 46.9% for the plasma test (16). In the post first line EGFR TKI population, a pooled analysis of the AURA extension and AURA2 phase II studies of osimertinib for T790M positive patients noted that the Cobas plasma test detected the T790M mutation in 61% of tumor tissue T790M mutation-positive patients (17). In our study, knowing that all patients had an EGFR mutation at the outset, the sensitivity of our laboratory developed EGFR ctDNA test was 43% for the original and/or T790M mutation. This result is similar to other studies noting that many other studies were done at baseline prior to EGFR TKI administration when patients may have higher levels of ctDNA due to the untreated disease status.
Predictors of ctDNA positivity can provide useful clinical guidance for patients who require timely transition of therapy. In patients with progressive disease that has an aggressive trajectory, the treating physician may opt for arranging a more invasive test if the likelihood of EGFR ctDNA detection is low. Our study identified liver and bone progression as being associated with EGFR ctDNA detection. This is in agreement with other cohorts that demonstrated that bone and higher burden of metastatic disease is more frequently associated with a positive EGFR ctDNA test (18,19). Passiglia et al. noted that it is more difficult to detect EGFR ctDNA in the first line setting when limited to intrathoracic (M1a) disease as compared with extrathoracic (M1b) disease, a trend that was also noted in our data (Table 2, P=0.06) (20). In a resource limited environment this information may also guide physicians as to when to consider testing in slowly progressive disease. The number of sites of progressive disease also predicted for ctDNA detection when there were 3–5 progressive sites but not for patients with 6–10 sites of progressive disease. This may reflect that the anatomical site of disease progression is more important than the absolute number of sites of progressive disease e.g., the influence of progression in multiple bony metastases as compared with multiple pulmonary nodules. The small sample size for this group may have also made it more difficult to detect differences. Our assay evaluates both the original EGFR mutation and EGFR T790M and so our data suggests that these features are predictive of EGFR ctDNA after first line EGFR TKI, not specifically T790M positivity in contrast to other studies.
Patients with detectable ctDNA for the original mutation only had poorer outcomes than those with detectable EGFR T790M ctDNA as demonstrated in Figure 1. This may reflect the limited therapeutic options for patients with an EGFR T790M independent mechanism of acquired resistance to EGFR TKIs in contrast to the options for patients with T790M (4). Of patients who did not have any detectable EGFR ctDNA at first testing, 28% had subsequent identification of a T790M mutation by plasma or tissue. After the multiple iterations of testing, the median OS of patients with undetectable EGFR ctDNA was 12.3 months, potentially due to a lower burden of progressing disease. The timepoint of first EGFR ctDNA test was selected to calculate the OS as it was felt to represent the timepoint at which the treating physician felt there was sufficient progression to consider a change in systemic therapy. Lee et al. found that both progression free survival (PFS) and OS were superior in patients who had undetectable EGFR ctDNA after 2 months of EGFR TKI therapy. Longitudinal follow up of these patients also noted the ability to detect progression by EGFR ctDNA levels earlier than the radiographic investigations (21). In another study, lower levels of EGFR ctDNA at baseline was associated with better PFS, however not OS (22). Detectable ctDNA is a useful tool to gauge aggressiveness of disease and the need to plan further investigations or subsequent lines of treatment for patient care.
Our study is novel and of value as it reflects real-world use of EGFR ctDNA testing in North America and examines the influence of the site and number of progressive disease sites on ctDNA detection. Furthermore, we demonstrate the prognostic implication of different patterns of EGFR ctDNA detected at this clinical timepoint. Our data validates the results of other studies that have examined the prognostic value in the post-treatment and treatment naive setting and location of metastasis in the post-first line EGFR TKI setting (18,20,23). The study has the limitation of using an in-house laboratory developed test at BC Cancer which had not been directly compared to regulatory approved EGFR ctDNA tests and we did not have a gold standard tissue T790M testing on every patient. The use of droplet digital PCR for EGFR ctDNA detection however has been widely adopted and this assay had been cross validated nationally (24,25). Several studies have already demonstrated the comparison between mutation detection in plasma and tissue and highlighted that due to tumor heterogeneity, plasma ctDNA may more accurately reflect clinically relevant clones (3,9). In addition, the centralized laboratory services used in this study provide cancer genetic analysis for a total population of 4.6 million individuals over a geographic area of 944,735 km2, allowing for a spectrum of clinical and logistical situations to be included in the analysis.
Conclusions
In conclusion, liver or bone metastases and a higher number of progressing lesions were predictors for detectable EGFR ctDNA with our laboratory developed test. The performance of our test was similar to regulatory approved EGFR ctDNA platforms examined in the real world. The presence of detectable EGFR ctDNA in the absence of a T790M mutation is a negative prognostic indicator associated with a higher burden progressing disease and poorer overall survival. EGFR ctDNA testing provides valuable information for patients progressing on first line EGFR TKIs from a treatment selection and prognostic perspective.
Acknowledgments
Funding: The BC Cancer Foundation and Roche Canada for supporting the BC Cancer Post-graduate Fellowship award for Dr. Alexandra Pender. Grants to Dr. Aly Karsan from Genome BC and Astra Zeneca for supporting ddPCR test development.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tlcr-9-581). AP reports personal fees from Bristol-Myers Squibb, Guardant Health, grants from Roche Canada during the conduct of the study. CH reports other from AstraZeneca outside the submitted work. TT reports grants from Astra Zeneca outside the submitted work. JL reports grants from Roche, AstraZeneca, personal fees from Roche, Pfizer, Takeda outside the submitted work. SY reports personal fees from Amgen, Bayer, Novartis, Roche outside the submitted work. CH reports grants from Astra Zeneca during the conduct of the study; grants and personal fees from Astra Zeneca, Eisai, personal fees and other from Boehringer Ingelheim, Roche, personal fees from BMS, Lilly, Merck, Bayer, Novartis outside the submitted work. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). All data was collected and analyzed as per local ethical approval (H18-02964, University of British Columbia, BC Cancer Research Ethics Board). Patient consent was not obtained for this retrospective review. The outcome of this study does not impact patient care.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- National Comprehensive Cancer Network. NCCN Clinical Practice Guidelines in Oncology Non-Small Cell Lung Cancer 2019; Available online: https://www.nccn.org/professionals/physician_gls/pdf/nscl.pdf. Accessed October, 2019.
- Goto K, Ichinose Y, Ohe Y, et al. Epidermal growth factor receptor mutation status in circulating free DNA in serum: from IPASS, a phase III study of gefitinib or carboplatin/paclitaxel in non-small cell lung cancer. J Thorac Oncol 2012;7:115-21. [Crossref] [PubMed]
- Douillard JY, Ostoros G, Cobo M, et al. Gefitinib treatment in EGFR mutated caucasian NSCLC: circulating-free tumor DNA as a surrogate for determination of EGFR status. J Thorac Oncol 2014;9:1345-53. [Crossref] [PubMed]
- Mok TS, Wu Y, Ahn M, et al. Osimertinib or Platinum-Pemetrexed in EGFR T790M-Positive Lung Cancer. N Engl J Med 2017;376:629-40. [Crossref] [PubMed]
- Park CK, Cho HJ, Choi YD, et al. A Phase II Trial of Osimertinib in the Second-Line Treatment of Non-small Cell Lung Cancer with the EGFR T790M Mutation, Detected from Circulating Tumor DNA: LiquidLung-O-Cohort 2. Cancer Res Treat 2019;51:777-87. [Crossref] [PubMed]
- Thress KS, Brant R, Carr TH, et al. EGFR mutation detection in ctDNA from NSCLC patient plasma: A cross-platform comparison of leading technologies to support the clinical development of AZD9291. Lung Cancer 2015;90:509-15. [Crossref] [PubMed]
- Passiglia F, Rizzo S, Di Maio M, et al. The diagnostic accuracy of circulating tumor DNA for the detection of EGFR-T790M mutation in NSCLC: a systematic review and meta-analysis. Sci Rep 2018;8:13379-018-30780-4.
- Lindeman NI, Cagle PT, Aisner DL, et al. Updated Molecular Testing Guideline for the Selection of Lung Cancer Patients for Treatment With Targeted Tyrosine Kinase Inhibitors: Guideline From the College of American Pathologists, the International Association for the Study of Lung Cancer, and the Association for Molecular Pathology. J Thorac Oncol 2018;13:323-58. [Crossref] [PubMed]
- Lettig L, Sahnane N, Pepe F, et al. EGFR T790M detection rate in lung adenocarcinomas at baseline using droplet digital PCR and validation by ultra-deep next generation sequencing. Transl Lung Cancer Res 2019;8:584-92. [Crossref] [PubMed]
- Roche Diagnostic. cobas® EGFR Mutation Test v2 For in vitro diagnostic use. 2016 (October).
- Qiagen therascreen® EGFR RGQ PCR Kit Handbook. 2019.
- Amoy Diagnostics. Super-ARMS® EGFR Mutation Detection Kit. 2018.
- Foundation Medicine. FoundationOne Liquid. 2019; Available online: https://www.foundationmedicine.com/genomic-testing/foundation-one-liquid. Accessed October, 2019.
- Guardant Health. Guardant 360 Liquid Biopsy Assay. 2019.
- Reck M, Hagiwara K, Han B, et al. ctDNA Determination of EGFR Mutation Status in European and Japanese Patients with Advanced NSCLC: The ASSESS Study. J Thorac Oncol 2016;11:1682-9. [Crossref] [PubMed]
- Han B, Tjulandin S, Hagiwara K, et al. EGFR mutation prevalence in Asia-Pacific and Russian patients with advanced NSCLC of adenocarcinoma and non-adenocarcinoma histology: The IGNITE study. Lung Cancer 2017;113:37-44. [Crossref] [PubMed]
- Jenkins S, Yang JC, Ramalingam SS, et al. Plasma ctDNA Analysis for Detection of the EGFR T790M Mutation in Patients with Advanced Non-Small Cell Lung Cancer. J Thorac Oncol 2017;12:1061-70. [Crossref] [PubMed]
- Zhu YJ, Zhang HB, Liu YH, et al. Quantitative cell-free circulating EGFR mutation concentration is correlated with tumor burden in advanced NSCLC patients. Lung Cancer 2017;109:124-7. [Crossref] [PubMed]
- Mondaca S, Offin M, Borsu L, et al. Lessons learned from routine, targeted assessment of liquid biopsies for EGFR T790M resistance mutation in patients with EGFR mutant lung cancers. Acta Oncol 2019;58:1634-9. [Crossref] [PubMed]
- Passiglia F, Rizzo S, Rolfo C, et al. Metastatic Site Location Influences the Diagnostic Accuracy of ctDNA EGFR- Mutation Testing in NSCLC Patients: a Pooled Analysis. Curr Cancer Drug Targets 2018;18:697-705. [Crossref] [PubMed]
- Lee JY, Qing X, Xiumin W, et al. Longitudinal monitoring of EGFR mutations in plasma predicts outcomes of NSCLC patients treated with EGFR TKIs: Korean Lung Cancer Consortium (KLCC-12-02). Oncotarget 2016;7:6984-93. [Crossref] [PubMed]
- Zhu YJ, Zhang HB, Liu YH, et al. Estimation of cell-free circulating EGFR mutation concentration predicts outcomes in NSCLC patients treated with EGFR-TKIs. Oncotarget 2017;8:13195-205. [Crossref] [PubMed]
- Ikushima H, Sakatani T, Usui K. Clinical Features of Patients with an Epidermal Growth Factor Receptor T790M Mutation Detected in Circulating Tumor DNA. Oncology 2020;98:23-8. [Crossref] [PubMed]
- Tsao M, Zhang T, Cheema P, et al. P3.01-019 Canadian Multicenter Validation Study of Plasma Circulating Tumor DNA for Epidermal Growth Factor (EGFR) T790M Testing. J Thorac Oncol 2017;12:S2207. [Crossref]
- Stockley T, Souza CA, Cheema PK, et al. Evidence-based best practices for EGFR T790M testing in lung cancer in Canada. Curr Oncol 2018;25:163-9. [Crossref] [PubMed]


