
Microbiota dysbiosis in lung cancer: evidence of association and potential mechanisms
Introduction
Human body is inhabited by trillions of microbes (i.e., bacteria, archaea, fungi, protists, and viruses) that are increasingly considered critical to human health (1,2). Symbiotic communities in our bodies are involved in degradation of nutrients, fight against invasion by xenobiotics, elimination of pathogens, and maturation of our immune system (3). Restrained by the development of experimental methods and technology, our knowledge of this vast microbiome seems to be very limited previously. With the development and spread of sequencing technology, we have gained a deep understanding of the human microbiome; thus, our understanding of the role of microbiome in human health and disease has greatly increased (4,5). On the basis of the key features of the microbiome including microbial diversity, relative abundance, and microbial gene richness, human microbiome project consortium studies have demonstrated that healthy individuals have high bacterial diversity and distinct individual variability at the species level (6,7). Recent studies have illustrated complex interaction between the human microbiome and different disease statuses, including cancers (8-11). For example, Helicobacter pylori, a common Proteobacteria residing in the upper gastrointestinal tract, significantly increases the risk of gastric cancer and pancreatic carcinogenesis (12,13). Furthermore, Enterotoxigenic Bacteroides fragilis, as one of the most prevalent pathobionts detected in colorectal cancer patients, has been shown to induce murine colon tumorigenesis by generating DNA mutagens, such as the genotoxin, superoxide, and hydrogen peroxide (14). In addition to the studies that focus on single pathogenic species, increasing studies have identified changes in the composition of microbiota in various anatomic sites associated with carcinogenesis (15). Furthermore, hard evidence suggests that physiological and environmental factors including diet, smoking, alcohol consumption, and air pollution significantly alter the composition of microbiota in various anatomic sites (16). These factors are often associated with carcinogenesis. Thus, extensive studies are required to further identify the link between microbiota and cancer development. Lung cancer, as the leading cause of cancer-related deaths worldwide in men and women, shows an increase in incidence in developing countries (17). It has a grim prognosis, that is, over half of people diagnosed with lung cancer die within one year of diagnosis, and the five-year survival rate is less than 18% (18). The two main histological groups of lung cancer are small cell lung carcinoma (SCLC, 15% of all lung cancers) and non-small cell lung carcinoma (NSCLC, 85% of all lung cancers). NSCLC is further classified into squamous cell carcinoma, adenocarcinoma, and large cell carcinoma (19). Although well characterized in etiology, morphology, and intrinsic molecular character, little is known about the relationship of lung cancer with the lung microbiota. In this review, we mainly summarize the current research progress of the lung microbiota and the role of lung microbiome in various lung diseases and other related diseases, especially in lung cancer. In addition, we focus on the change in lung microbiome and the potential pathogenic mechanisms to provide a theoretical basis for treating this deadly disease.
Lung microbiota in healthy humans and lung diseases
Although lungs are connected with outside air, the lungs of healthy people were even previously regarded as sterile for a long time. Recently, a great number of studies confirm the presence of lung microbiota in healthy people. Culture-independent molecular techniques were used in previous studies to analyze the most common bacterial phyla including Bacteroidetes, Firmicutes, and Proteobacteria, and the prominent genera include Prevotella, Veillonella, and Streptococcus (20-23). These genera are also detected in oral samples. Furthermore, lungs contain specific bacteria such as Enterobacteriaceae, Haemophilus, Methylobacterium, Ralstonia, and Tropheryma species (21,24). In healthy lungs, microbial density is low, containing 103 cells/g to 105 cells/g of tissue (for comparison, intestinal microbiota reaches a density of 1011 to 1012 cells/g) (25,26). The composition of lung microbiota is determined by the balance between microbial immigration from the upper respiratory tract and microbe elimination (e.g., by coughing or immune defenses), with relatively few contributions from regional growth of the microbes themselves (27-29). Microbial immigration from the upper respiratory tract occurs mainly through microaspiration. This view can be supported by the high similarity between the lower airway and oropharynx microbiota rather than nasopharynx. Microaspiration is a passive process involving the oral and pharyngeal muscles; it mainly occurs during sleep (8). Nevertheless, a study using 16S rRNA gene sequencing shows similarity of the lung microbiota to the oropharynx and nasopharynx microbiota in young children (23). This finding might be due to the different anatomical structure of upper respiratory tract in children and adults. The balance between microbial immigration (e.g., via microaspiration) and microbial elimination (e.g., mucociliary clearance, cough, and host immune defenses) determines the composition of the lung microbiota.
Furthermore, another layer of complexity is added by the impact of environmental conditions (e.g., pH, temperature, nutrient, oxygen tension, and activation of host inflammatory cells) on this process. Thus, the geographical, physiological, and immunological diversities also shape the composition of the lung microbiota (30,31). Recent studies have indicated that the atmospheric concentration (µg/m3) of particulate matter with diameters of 10 and 2.5 micrometers (PM10 and PM2.5) might affect the lung microbiota and respiratory functions (32-34). In addition, household air pollution plays a role in the composition of lung microbiome (35,36). Smoking, as the risk factor for lung diseases, has been reported to alter microbial diversities and communities in the lower respiratory tract of mice and human trials (37,38). Antibiotic is another important factor influencing lung microbiota; it has attracted a great deal of attention in recent years. However, the influence of antibiotics on microbiota is mainly focused on intestinal trials; less is known about the influence on lung microbiome composition (39-41). A recent study has shown that after one year of azithromycin treatment, bacterial diversity decreased in patients with asthma (42). Similarly, treatment with azithromycin reduced alpha diversity in patients with chronic obstructive pulmonary disease (COPD), but did not change the total bacterial burden (43).
In patients with lung disease, the balance between immigration and elimination is disturbed. Thus, the lung microbiota is altered, the abundance of symbiotic bacteria is decreased, and pathogenic bacteria predominate. This change leads to a decrease in diversity of lung microbiota and is associated with the progression of chronic lung diseases, such as COPD (44-54), cystic fibrosis (55-58), asthma (59-62), and idiopathic pulmonary fibrosis (63-65) (Table 1).
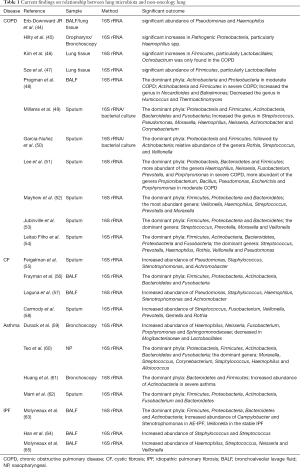
Full table
In summary, homeostasis of the lung microbiota is associated with the balance between immune defense of pathogens and immune tolerance of the commensals. On one hand, host lungs have established three major pathways to defend against the invasion of pathogens. First, Mucus, mainly secreted by goblet and club cells, provides an effective defense against epithelial injury and limits the migration of pathogens to epithelial cells. Thus, the systematic spread of microbes in the body is prevented; inflammation is also prevented, and homeostasis of the microbiota and host is protected (66,67). Second, alveolar surfactant containing sIgA covers the surface of lung alveolar epithelial cells and participates in lung innate immunity (68). Finally, the epithelial cell layer is not only a structural barrier, but also a component of the innate host defense. Epithelial cells express several pattern recognition receptors (PRRs) and secrete antimicrobial molecules and mucins to defend against invading pathogens (69). The PRRs, including toll-like receptor (TLR) and nucleotide-binding oligomerization domain-like receptors can prevent the overload of pathogens or metabolites by identifying pathogen-associated molecular patterns or cell damage-associated molecular patterns (70,71). Then, the pathogens are further eliminated by activating the downstream inflammatory signaling pathway (72). On the other hand, immune tolerance of commensals is regulated by anti-inflammatory macrophages in alveoli by inhibiting inflammatory pathways and adaptive immune responses (73,74).
Relationship between gut microbiome and lung microbiome
Recently, many studies have begun to focus on the two-way manner between gut and lungs, which is known as the “gut–lung axis” (75). This theory is based on “gut–lymph” theory of Samuelson et al. (76). According to the theory, many macrophages and other immune cells are present in the intestinal submucosa or the mesenteric lymph nodes that contain many translocating bacteria. If they are not cleared by the first line of defense, then the surviving bacteria, cell wall fragments, or protein fragments of dead bacteria would escape along with the cytokines and chemokines produced in the intestine and travel along the mesenteric lymphatic system to the cisterna chyli and then into the circulatory system. One way is access to pulmonary circulation, leading to local activation of dendritic cells and macrophages and the initiation and differentiation of T cells. Another way is activation of immune cells, which affect the lung area through their own migration, in the first contact with the antigen in the intestinal mucosa. Therefore, gut microbiomes influence the lung microbiome partly through inhalations of the gastroesophageal content, swallowing of the sputum, and most importantly through modulation of host immune.
Studies have shown that cigarette smoke, as a crucial risk of COPD, can also change the composition of intestinal microbiota and reduce the diversity of intestinal bacteria (77), and the change in composition of microbiota is closely related to many inflammatory diseases, including intestinal inflammation and inflammatory bowel disease (IBD) (75,78,79). Many respiratory infections are often accompanied by gastrointestinal symptoms given that the “gut–lung axis” is bidirectional (80). This finding has also been demonstrated in animal models with respiratory infection (81,82). However, direct evidence for the influence of lung microbiota and its products and their circulation is lacking. An animal experiment showed that nonabsorbable tracer deposited into the nasal cavity can be found in the gastrointestinal tract subsequently (83). Further research is needed to validate and extend these findings.
Microbiome and lung cancer
Oral microbiome and lung cancer
The oral microbiome is highly correlated with the lung microbiome because lungs are directly connected with the oral cavity. More recently, the association of oral microbiome with lung cancer has received considerable attention. Several prospective cohorts have consistently shown that periodontal diseases, known to alter the oral microbiota, are associated with increased lung cancer risk after controlling potential confounding factors including smoking (84-88). 16S rRNA gene sequencing results of salivary microbiome suggest an elevated abundance of Capnocytophaga and Veillonella with a reduced number of Neisseria (89). Another study in nonsmoking female with lung cancer indicates that genera Blastomonas and Sphingomonas were significantly increased in the oral microbiota of patients with lung cancer, whereas Acinetobacter and Streptococcus were higher in controls (90). Although the association between periodontal disease and lung cancer is generally considered strong, the causality remains a large problem, and further research is needed to evaluate potential mechanisms.
Lung microbiome and lung cancer
Studies have shown that patients with lung cancer may have similar lung microecology. A summary overview of the relationship between lung microbiota and lung cancer is shown in Table 2 (30,91-102). Hasegawa et al. collected intraoperative bronchial fluids using a microsampling probe from nine subjects with pulmonary carcinoma and cultured anaerobically on blood agar plates. Predominant isolates from intraoperative bronchial fluids are commonly indigenous to the oral cavity, namely, Streptococcus, Veillonella, Gemella, Porphyromonas, Olsenella, and Eikenella. These findings indicate that intraoperative bronchial fluids contain bacteria probably derived from the oral microbiota (93). The result of another study on airway brushing samples showed that the prominent phyla or genera were also dominated in oral samples of patients with lung cancer (94). Simon et al. collected sputum samples from 10 patients with possible LC, four of which were eventually diagnosed with LC (LC+), and six had no LC after one year (LC−). Among the seven bacterial species found in all samples, Streptococcus viridans was significantly higher in LC+. Among the five species having significantly higher abundances in LC+, Granulicatella adiacens showed the highest level of abundance change. Moreover, G. adiacens abundance was correlated with six other bacterial species only in LC+ samples, namely, Enterococcus sp. 130, Streptococcus intermedius, Escherichia coli, S. viridans, Acinetobacter junii, and Streptococcus sp. 6 that could be related to LC stage. The results in this study showed that the spontaneous sputum would be a viable source of bacterial biomarkers for LC status and stage (91). Another study on sputum samples from a nonsmoking female patient with LC showed the enrichment of Granulicatella, Abiotrophia, and Streptococcus (92). Lee et al. studied bronchoalveolar lavage fluid from 28 patients and found that Acteroidetes, Firmicutes, and Proteobacteria were the most common phyla, and Prevotella, Streptococcus, and Neisseria were the most common genera in both groups. The relative abundance of two phyla (Firmicutes and TM7) and four genera (Veillonella, Megasphaera, Atopobium, and Selenomonas) were significantly increased in patients with lung cancer. Furthermore, the combination of the two genera (Veillonella and Megasphaera) showed a higher receiver operating characteristic value than the individual genus in predicting lung cancer. Thus, this combination could be used as biomarker for lung cancer. Another noteworthy finding was that smoking patients with lung cancer have a significantly higher ratio of Firmicutes to Bacteroidetes than nonsmoking patients (92). A similar result is also shown in a study of lung tissue samples with COPD (47). In addition, an increase in the phylum TM7 was reported in COPD (48). These results further support the view about the strong relationship between COPD and lung cancer, as indicated in other studies (103,104).
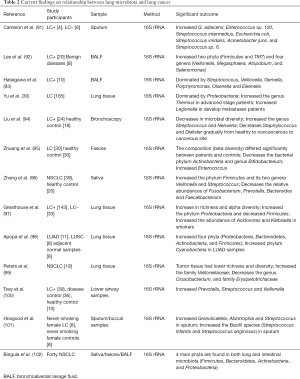
Full table
Most previous studies on lung microbiota used oral, sputum, or bronchoscopic brushing samples. A common concern with these samples is that they may be contaminated by the upper respiratory or oral microbiota. Some studies have suggested that the bacteria in lung carcinogenesis may be associated with aspiration of oral or pharyngeal bacteria. However, a research on 165 nonmalignant lung tissue samples from cancer patients showed that the lung microbiota has distinct features that differ from those of the oral cavity and other body sites. In fact, it is dominated by Proteobacteria. Similar results from other studies show the domination of Proteobacteria in lung tissue samples with lung cancer (97,98,102). Furthermore, the genus Thermus is more abundant in tissue from advanced stage patients, and Legionella is high in patients with metastases (30). Moreover, the lung microbiota is affected by exposure to air pollution and tobacco smoking.
In summary, patients with lung cancer show changes in the relative abundance of multiple genera. Consistent conclusions from all recent studies are limited due to the small sample size of most studies and the heterogeneity of lung cancer. However, most studies indicate that Streptococcus and Proteobacteria may be the key bacteria of lung cancer. Nevertheless, further large-scale studies are needed to verify certain microbial biomarkers for patients with lung cancer.
Possible mechanisms mediating lung microbiota and lung cancer
Dysbiosis of the microbiome is mainly manifested by the decrease in symbiotic bacteria and the increase in pathogenic bacteria, and then inducement of carcinogenesis at multiple levels, including metabolism alteration, inflammation, and immune response (105,106). Present studies on the mechanisms of microbiota and cancer are mainly focused on intestinal flora and colon cancer (107-109). When dysregulated, the intestinal microbiota can contribute to colorectal cancer development through the modulation of immune function and the production of microbial-derived metabolites (110). The increase in pathogenic bacteria can lead to chronic inflammation through the persistent generation of inflammatory mediators, thereby affecting cell apoptosis and increasing mutations. Moreover, the metabolites of bacteria, such as reactive oxygen and nitrogen, through direct DNA damage or modification of cellular signaling generate a pro-carcinogenic environment. Bacteria influence cellular signaling and/or induce mucosal inflammation to initiate or promote colon tumorigenesis through producing a variety of oncogenic toxins to directly damage DNA(111).
However, studies on the mechanism of lung microbiota and lung cancer are few. Great effort is exerted to identify and characterize a potential causal relationship from the lung cancer and lung microbe interaction.
A large cohort study by Boursi et al. demonstrated that recurrent exposure to certain antibiotics (penicillin, cephalosporins, or macrolides) may increase lung cancer risk (112). Antibiotic-induced dysbiosis not only alter bacterial abundance, composition, and diversity in animal models, but also accelerate Lewis LC that progressed on the host side (113). The dysbiosis of lung microbiota may promote the development of lung cancer by releasing cancer-promoting bacterial metabolites and inducting host inflammatory pathways (114). The possible mechanisms mediating lung microbiota and lung cancer are illustrated in Figure 1.
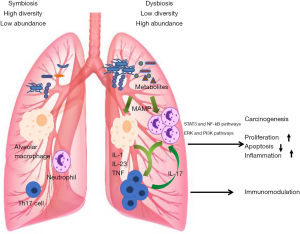
Metabolism
Recent studies have implicated that the metabolites of bacteria may be involved in the development of lung cancer. Cytolethal distending toxin (CDT) as a bacterial genotoxin produced by variety of gram-negative bacteria, such as Actinobacillus, can induce apoptosis in human lung adenocarcinoma A549 cell line (115). G. adiacens was found to be associated with lung cancer. In addition, the research on functional capacity demonstrated that G. adiacens was involved in the metabolism of polyamine (91). Interestingly, elevated levels of polyamines, such as putrescine and gamma-aminobutyric acid, have been associated with a range of cancers including lung malignancies (116). Apopa et al. found that the abundance of Cyanobacteria was significantly increased in lung adenocarcinoma, and the functional analysis suggested that the Cyanobacteria toxin (i.e., microcystin) might be related to the increase in procyclic acidic repetitive protein 1 (PARP1), thereby enhancing inflammation and leading to cancer. The result was further confirmed in microcystin-challenged lung adenocarcinoma (A427) cell lines (98).
In addition, some bacteria in the intestinal microbiome can increase the bioavailability of anticancer drugs. Niu suggested that Bacteroidaceae and Prevotellaceae contain species capable of hydrolyzing ginsenosides present in red ginseng extract (RGE), enhancing the effect of RGE in the prevention and treatment of lung cancer (117). This finding indicates that the decrease in symbiotic bacteria could accelerate LC progression.
Inflammatory pathways and immune response
In recent years, considerable studies have shown that chronic inflammation plays an important role in the development of several forms of cancer, including lung cancer. Dysbiosis of the microbiota can activate the inflammatory pathway to trigger the proliferation and survival of epithelial cells under certain conditions, promoting the development of tumors. TLR4, as a member of pattern recognition receptor, initiates natural immunity in the early stage of pathogen invasion. Increasing evidence shows that it plays a crucial role in the development of tumor microenvironment and has been increasingly investigated. TLRs promote carcinogenic effects by activating nuclear factor κB (NF-κB) pathway, releasing inflammatory factor, and activating transcription 3 (STAT3) (118). TLR4 is expressed more strongly in lung cancer tissue than in paracancer tissue (119).
Ochoa et al. found that exposure of the airway to smoke particulates and nontypeable Haemophilus influenzae (NTHi) promoted lung cancer cell proliferation by release of IL-6 and TNF, which further activated the STAT3 and NF-kB pathways in airway epithelium (120). Another study demonstrated that IL-6 blockade significantly inhibited lung cancer promotion, tumor cell intrinsic STAT3 activation, tumor cell proliferation, and angiogenesis markers (121).
In addition, Th17 cell-mediated inflammation has been identified to play a critical role in lung tumorigenesis (122). Jungnickel et al. indicated that the epithelial cytokine IL-17C mediates the tumor-promoting effect of bacteria, such as NTHi, through neutrophilic inflammation (123). Recently, growing awareness of the importance of NTHi in the pathophysiology of COPD has been observed, and COPD-like airway inflammation induced by NTHi provides a tumor microenvironment that favors lung tumor promotion and progression (124-126). Thus, NTHi may act as a bridge between COPD and lung cancer.
Furthermore, accumulating evidence indicated that upregulation of the PI3K pathway played a central role in the cell proliferation, survival, and tissue invasion of early lung cancer (127). Tsay et al. reported that the oral taxa (Streptococcus and Veillonella) enriched in the lower airways of patients with lung cancer were associated with upregulation of the ERK and PI3K signaling pathways, and the same signaling pathways were upregulated in vitro exposure of airway epithelial cells to Veillonella, Prevotella, and Streptococcus (128). These studies preliminarily indicated that pulmonary bacteria up-regulate the expression of inflammatory mediators and cytokines by acting on microbe-associated molecular pattern recognition receptors (MAMP), thereby affecting the development of lung cancer.
Application of lung microbiome in clinical trial
Most experimental studies analyzing the application of lung microbiota in the clinical therapy mainly focused on animal models. The relevant content theme has been covered in several reviews (11,129,130). Most commonly studied microorganisms in the context of lung disease are known probiotics, such as Lactobacillus and Bifidobacterium. Their beneficial role in animal models of lung diseases has been well demonstrated (130). For instance, orally or intranasal administered Lactobacillus and Bifidobacterium were shown to protect mice against lung infection by augmenting antibody production, enhancing natural killer cell activity, and IFN-γ production, as well as increasing secretion of IL-10. In addition, Lactobacillus and Bifidobacterium were shown to confer beneficial effects on allergic airway inflammation by inducing Treg cells and TH1 cells. Currently, experimental studies on the application of lung microbiome in lung cancer are relatively limited. Orally administered Lactobacillus acidophilus on mice lung cancer model was shown to reduce tumor size and increase survival rate after receiving cisplatin treatment (131). Moreover, administered Enterococcus hirae and Barnesiella intestinihominis in combination with chemoimmunotherapy can significantly improve efficacy in patients with advanced lung cancer (132).
Conclusions
In summary, accumulating evidence for specific bacteria as biomarkers of lung cancer presence is found. However, the precise mechanism of lung microbiota on the regulation of lung cancer is still partly unclear. Future research of the causal role of these bacteria in lung carcinogenesis will be beneficial for our understanding of the interactions between the lung microbiota and lung function, which is also valuable in ultimately providing therapeutic targets for lung cancer prevention and therapy.
Acknowledgments
Funding: This research was supported by the National Natural Science Foundation of China (81871734, 81471994, 81700463, 81700389), Jiangsu Provincial Medical Talent (ZDRCA2016053), Six talent peaks project of Jiangsu Province (WSN-135), Advanced health talent of six-one project of Jiangsu Province (LGY2016042).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tlcr-20-156). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Barton W, O'Sullivan O, Cotter PD. Metabolic phenotyping of the human microbiome. F1000Res 2019;8:F1000 Faculty Rev-1956.
- Sender R, Fuchs S, Milo R. Revised Estimates for the Number of Human and Bacteria Cells in the Body. PLoS Biol 2016;14:e1002533. [Crossref] [PubMed]
- Thomas RM, Zajac-Kaye M. Microbial marauders: pancreatic microbiota and its impact on carcinogenesis. Ann Transl Med 2018;6:S63. [Crossref] [PubMed]
- Chakravorty S, Helb D, Burday M, et al. A detailed analysis of 16S ribosomal RNA gene segments for the diagnosis of pathogenic bacteria. J Microbiol Methods 2007;69:330-9. [Crossref] [PubMed]
- Van Treuren W, Dodd D. Microbial Contribution to the Human Metabolome: Implications for Health and Disease. Annu Rev Pathol 2020;15:345-69. [Crossref] [PubMed]
- Arumugam M, Raes J, Pelletier E, et al. Enterotypes of the human gut microbiome. Nature 2011;473:174-80. [Crossref] [PubMed]
- Human Microbiome Project C. Structure, function and diversity of the healthy human microbiome. Nature 2012;486:207-14. [Crossref] [PubMed]
- Chen J, Domingue JC, Sears CL. Microbiota dysbiosis in select human cancers: Evidence of association and causality. Semin Immunol 2017;32:25-34. [Crossref] [PubMed]
- Cong J, Zhang XC. How human microbiome talks to health and disease. Eur J Clin Microbiol Infect Dis 2018;37:1595-601. [Crossref] [PubMed]
- Orr MR, Kocurek KM, Bakos YJ, et al. A restoration ecology perspective on the treatment of inflammatory bowel disease. Evol Med Public Health 2019.217-20. [Crossref] [PubMed]
- Fabbrizzi A, Amedei A, Lavorini F, et al. The lung microbiome: clinical and therapeutic implications. Intern Emerg Med 2019;14:1241-50. [Crossref] [PubMed]
- Guo Y, Liu W, Wu J. Helicobacter pylori infection and pancreatic cancer risk: A meta-analysis. J Cancer Res Ther 2016;12:C229-32. [Crossref] [PubMed]
- Trikudanathan G, Philip A, Dasanu C, et al. Association Between Helicobacter pylori Infection and Pancreatic Cancer A Meta-Analysis. Am J Gastroenterol 2010;105:S48-S.
- Wu SG, Rhee KJ, Albesiano E, et al. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat Med 2009;15:1016-22. [Crossref] [PubMed]
- Schwabe RF, Jobin C. The microbiome and cancer. Nat Rev Cancer 2013;13:800-12. [Crossref] [PubMed]
- Casey SC, Vaccari M, Al-Mulla F, et al. The effect of environmental chemicals on the tumor microenvironment. Carcinogenesis 2015;36:S160-83. [Crossref] [PubMed]
- Shankar A, Saini D, Dubey A, et al. Feasibility of lung cancer screening in developing countries: challenges, opportunities and way forward. Transl Lung Cancer Res 2019;8:S106-21. [Crossref] [PubMed]
- Zappa C, Mousa SA. Non-small cell lung cancer: current treatment and future advances. Transl Lung Cancer Res 2016;5:288-300. [Crossref] [PubMed]
- Yoneyama R, Saji H, Kato Y, et al. Clinicopathological characteristics and treatment strategies for young lung cancer patients. Ann Transl Med 2019;7:100. [Crossref] [PubMed]
- Segal LN, Alekseyenko AV, Clemente JC, et al. Enrichment of lung microbiome with supraglottic taxa is associated with increased pulmonary inflammation. Microbiome 2013;1:19. [Crossref] [PubMed]
- Morris A, Beck JM, Schloss PD, et al. Comparison of the Respiratory Microbiome in Healthy Nonsmokers and Smokers. Am J Respir Crit Care Med 2013;187:1067-75. [Crossref] [PubMed]
- Bassis CM, Erb-Downward JR, Dickson RP, et al. Analysis of the Upper Respiratory Tract Microbiotas as the Source of the Lung and Gastric Microbiotas in Healthy Individuals. mBio 2015;6:e00037. [Crossref] [PubMed]
- Marsh RL, Kaestli M, Chang AB, et al. The microbiota in bronchoalveolar lavage from young children with chronic lung disease includes taxa present in both the oropharynx and nasopharynx. Microbiome 2016;4:37. [Crossref] [PubMed]
- Dickson RP, Erb-Downward JR, Freeman CM, et al. Spatial Variation in the Healthy Human Lung Microbiome and the Adapted Island Model of Lung Biogeography. Ann Am Thorac Soc 2015;12:821-30. [Crossref] [PubMed]
- Mathieu E, Escribano-Vazquez U, Descamps D, et al. Paradigms of Lung Microbiota Functions in Health and Disease, Particularly, in Asthma. Front Physiol 2018;9:1168. [Crossref] [PubMed]
- Maddi A, Sabharwal A, Violante T, et al. The microbiome and lung cancer. J Thorac Dis 2019;11:280-91. [Crossref] [PubMed]
- Dickson RP, Erb-Downward JR, Huffnagle GB. Towards an ecology of the lung: new conceptual models of pulmonary microbiology and pneumonia pathogenesis. Lancet Respir Med 2014;2:238-46. [Crossref] [PubMed]
- Dickson RP, Martinez FJ, Huffnagle GB. The role of the microbiome in exacerbations of chronic lung diseases. Lancet 2014;384:691-702. [Crossref] [PubMed]
- Venkataraman A, Bassis CM, Beck JM, et al. Application of a Neutral Community Model To Assess Structuring of the Human Lung Microbiome. mBio 2015;6:e02284-14. [Crossref] [PubMed]
- Yu G, Gail MH, Consonni D, et al. Characterizing human lung tissue microbiota and its relationship to epidemiological and clinical features. Genome Biol 2016;17:163. [Crossref] [PubMed]
- Ni L, Chuang CC, Zuo L. Fine particulate matter in acute exacerbation of COPD. Front Physiol 2015;6:294. [Crossref] [PubMed]
- Wang L, Cheng HX, Wang DB, et al. Airway microbiome is associated with respiratory functions and responses to ambient particulate matter exposure. Ecotoxicol Environ Saf 2019;167:269-77. [Crossref] [PubMed]
- Qin T, Zhang FR, Zhou HJ, et al. High-Level PM2.5/PM10 Exposure Is Associated With Alterations in the Human Pharyngeal Microbiota Composition. Front Microbiol 2019;10:54. [Crossref] [PubMed]
- Li X, Sun Y, An YH, et al. Air pollution during the winter period and respiratory tract microbial imbalance in a healthy young population in Northeastern China. Environmental Pollution 2019;246:972-9. [Crossref] [PubMed]
- Rylance J, Kankwatira A, Nelson DE, et al. Household air pollution and the lung microbiome of healthy adults in Malawi: a cross-sectional study. BMC Microbiol 2016;16:182. [Crossref] [PubMed]
- Hosgood HD 3rd, Sapkota AR, Rothman N, et al. The potential role of lung microbiota in lung cancer attributed to household coal burning exposures. Environ Mol Mutagen 2014;55:643-51. [Crossref] [PubMed]
- Zhang R, Chen L, Cao L, et al. Effects of smoking on the lower respiratory tract microbiome in mice. Respir Res 2018;19:253. [Crossref] [PubMed]
- Huang C, Shi GC. Smoking and microbiome in oral, airway, gut and some systemic diseases. J Transl Med 2019;17:225. [Crossref] [PubMed]
- Zimmermann P, Curtis N. The effect of antibiotics on the composition of the intestinal microbiota - a systematic review. J Infect 2019;79:471-89. [Crossref] [PubMed]
- Kim S, Covington A, Pamer EG. The intestinal microbiota: Antibiotics, colonization resistance, and enteric pathogens. Immunol Rev 2017;279:90-105. [Crossref] [PubMed]
- Hufnagl K, Pali-Scholl I, Roth-Walter F, et al. Dysbiosis of the gut and lung microbiome has a role in asthma. Semin Immunopathol 2020;42:75-93. [Crossref] [PubMed]
- Taylor SL, Leong LEX, Mobegi FM, et al. Long-Term Azithromycin Reduces Haemophilus influenzae and Increases Antibiotic Resistance in Severe Asthma. Am J Respir Crit Care Med 2019;200:309-17. [Crossref] [PubMed]
- Segal LN, Clemente JC, Wu BG, et al. Randomised, double-blind, placebo-controlled trial with azithromycin selects for anti-inflammatory microbial metabolites in the emphysematous lung. Thorax 2017;72:13-22. [Crossref] [PubMed]
- Erb-Downward JR, Thompson DL, Han MK, et al. Analysis of the lung microbiome in the "healthy" smoker and in COPD. PLoS One 2011;6:e16384. [Crossref] [PubMed]
- Hilty M, Burke C, Pedro H, et al. Disordered microbial communities in asthmatic airways. PLoS One 2010;5:e8578. [Crossref] [PubMed]
- Kim HJ, Kim YS, Kim KH, et al. The microbiome of the lung and its extracellular vesicles in nonsmokers, healthy smokers and COPD patients. Exp Mol Med 2017;49:e316. [Crossref] [PubMed]
- Sze MA, Dimitriu PA, Hayashi S, et al. The lung tissue microbiome in chronic obstructive pulmonary disease. Am J Respir Crit Care Med 2012;185:1073-80. [Crossref] [PubMed]
- Pragman AA, Kim HB, Reilly CS, et al. The lung microbiome in moderate and severe chronic obstructive pulmonary disease. PLoS One 2012;7:e47305. [Crossref] [PubMed]
- Millares L, Ferrari R, Gallego M, et al. Bronchial microbiome of severe COPD patients colonised by Pseudomonas aeruginosa. Eur J Clin Microbiol Infect Dis 2014;33:1101-11. [Crossref] [PubMed]
- Garcia-Nuñez M, Millares L, Pomares X, et al. Severity-related changes of bronchial microbiome in chronic obstructive pulmonary disease. J Clin Microbiol 2014;52:4217-23. [Crossref] [PubMed]
- Lee SW, Kuan CS, Wu LS, et al. Metagenome and Metatranscriptome Profiling of Moderate and Severe COPD Sputum in Taiwanese Han Males. PLoS One 2016;11:e0159066. [Crossref] [PubMed]
- Mayhew D, Devos N, Lambert C, et al. Longitudinal profiling of the lung microbiome in the AERIS study demonstrates repeatability of bacterial and eosinophilic COPD exacerbations. Thorax 2018;73:422-30. [Crossref] [PubMed]
- Jubinville E, Veillette M, Milot J, et al. Exacerbation induces a microbiota shift in sputa of COPD patients. PLoS One 2018;13:e0194355. [Crossref] [PubMed]
- Leitao Filho FS, Alotaibi NM, Ngan D, et al. Sputum Microbiome Is Associated with 1-Year Mortality after Chronic Obstructive Pulmonary Disease Hospitalizations. Am J Respir Crit Care Med 2019;199:1205-13. [Crossref] [PubMed]
- Feigelman R, Kahlert CR, Baty F, et al. Sputum DNA sequencing in cystic fibrosis: non-invasive access to the lung microbiome and to pathogen details. Microbiome 2017;5:20. [Crossref] [PubMed]
- Frayman KB, Armstrong DS, Carzino R, et al. The lower airway microbiota in early cystic fibrosis lung disease: a longitudinal analysis. Thorax 2017;72:1104-12. [Crossref] [PubMed]
- Laguna TA, Wagner BD, Williams CB, et al. Airway Microbiota in Bronchoalveolar Lavage Fluid from Clinically Well Infants with Cystic Fibrosis. PLoS One 2016;11:e0167649. [Crossref] [PubMed]
- Carmody LA, Zhao J, Schloss PD, et al. Changes in cystic fibrosis airway microbiota at pulmonary exacerbation. Ann Am Thorac Soc 2013;10:179-87. [Crossref] [PubMed]
- Durack J, Lynch SV, Nariya S, et al. Features of the bronchial bacterial microbiome associated with atopy, asthma, and responsiveness to inhaled corticosteroid treatment. J Allergy Clin Immunol 2017;140:63-75. [Crossref] [PubMed]
- Teo SM, Mok D, Pham K, et al. The infant nasopharyngeal microbiome impacts severity of lower respiratory infection and risk of asthma development. Cell Host Microbe 2015;17:704-15. [Crossref] [PubMed]
- Huang YJ, Nariya S, Harris JM, et al. The airway microbiome in patients with severe asthma: Associations with disease features and severity. J Allergy Clin Immunol 2015;136:874-84. [Crossref] [PubMed]
- Marri PR, Stern DA, Wright AL, et al. Asthma-associated differences in microbial composition of induced sputum. J Allergy Clin Immunol 2013;131:346-52.e1-3.
- Molyneaux PL, Cox MJ, Wells AU, et al. Changes in the respiratory microbiome during acute exacerbations of idiopathic pulmonary fibrosis. Respir Res 2017;18:29. [Crossref] [PubMed]
- Han MK, Zhou Y, Murray S, et al. Lung microbiome and disease progression in idiopathic pulmonary fibrosis: an analysis of the COMET study. Lancet Respir Med 2014;2:548-56. [Crossref] [PubMed]
- Molyneaux PL, Cox MJ, Willis-Owen SA, et al. The role of bacteria in the pathogenesis and progression of idiopathic pulmonary fibrosis. Am J Respir Crit Care Med 2014;190:906-13. [Crossref] [PubMed]
- Fahy JV, Dickey BF. Airway mucus function and dysfunction. N Engl J Med 2010;363:2233-47. [Crossref] [PubMed]
- Evans CM, Raclawska DS, Ttofali F, et al. The polymeric mucin Muc5ac is required for allergic airway hyperreactivity. Nat Commun 2015;6:6281. [Crossref] [PubMed]
- LeVine AM, Whitsett JA, Gwozdz JA, et al. Distinct effects of surfactant protein A or D deficiency during bacterial infection on the lung. J Immunol 2000;165:3934-40. [Crossref] [PubMed]
- Leiva-Juárez MM, Kolls JK, Evans SE. Lung epithelial cells: therapeutically inducible effectors of antimicrobial defense. Mucosal Immunol 2018;11:21-34. [Crossref] [PubMed]
- Medzhitov R. Recognition of microorganisms and activation of the immune response. Nature 2007;449:819-26. [Crossref] [PubMed]
- Mariathasan S, Monack DM. Inflammasome adaptors and sensors: intracellular regulators of infection and inflammation. Nat Rev Immunol 2007;7:31-40. [Crossref] [PubMed]
- Jahnsen FL, Strickland DH, Thomas JA, et al. Accelerated antigen sampling and transport by airway mucosal dendritic cells following inhalation of a bacterial stimulus. J Immunol 2006;177:5861-7. [Crossref] [PubMed]
- Westphalen K, Gusarova GA, Islam MN, et al. Sessile alveolar macrophages communicate with alveolar epithelium to modulate immunity. Nature 2014;506:503-6. [Crossref] [PubMed]
- Kopf M, Schneider C, Nobs SP. The development and function of lung-resident macrophages and dendritic cells. Nat Immunol 2015;16:36-44. [Crossref] [PubMed]
- Budden KF, Gellatly SL, Wood DL, et al. Emerging pathogenic links between microbiota and the gut-lung axis. Nat Rev Microbiol 2017;15:55-63. [Crossref] [PubMed]
- Samuelson DR, Welsh DA, Shellito JE. Regulation of lung immunity and host defense by the intestinal microbiota. Front Microbiol 2015;6:1085. [Crossref] [PubMed]
- Budden KF, Shukla SD, Rehman SF, et al. Functional effects of the microbiota in chronic respiratory disease. Lancet Respir Med 2019;7:907-20. [Crossref] [PubMed]
- Kabeerdoss J, Jayakanthan P, Pugazhendhi S, et al. Alterations of mucosal microbiota in the colon of patients with inflammatory bowel disease revealed by real time polymerase chain reaction amplification of 16S ribosomal ribonucleic acid. Indian J Med Res 2015;142:23-32. [Crossref] [PubMed]
- Schwab C, Berry D, Rauch I, et al. Longitudinal study of murine microbiota activity and interactions with the host during acute inflammation and recovery. ISME J 2014;8:1101-14. [Crossref] [PubMed]
- Dilantika C, Sedyaningsih ER, Kasper MR, et al. Influenza virus infection among pediatric patients reporting diarrhea and influenza-like illness. BMC Infect Dis 2010;10:3. [Crossref] [PubMed]
- Wang J, Li F, Wei H, et al. Respiratory influenza virus infection induces intestinal immune injury via microbiota-mediated Th17 cell-dependent inflammation. J Exp Med 2014;211:2397-410. [Crossref] [PubMed]
- Samuelson DR, Charles TP, de la Rua NM, et al. Analysis of the intestinal microbial community and inferred functional capacities during the host response to Pneumocystis pneumonia. Exp Lung Res 2016;42:425-39. [Crossref] [PubMed]
- Southam DS, Dolovich M, O'Byrne PM, et al. Distribution of intranasal instillations in mice: effects of volume, time, body position, and anesthesia. Am J Physiol Lung Cell Mol Physiol 2002;282:L833-9. [Crossref] [PubMed]
- Yoon HS, Wen W, Long J, et al. Association of oral health with lung cancer risk in a low-income population of African Americans and European Americans in the Southeastern United States. Lung Cancer 2019;127:90-5. [Crossref] [PubMed]
- Zeng XT, Xia LY, Zhang YG, et al. Periodontal Disease and Incident Lung Cancer Risk: A Meta-Analysis of Cohort Studies. J Periodontol 2016;87:1158-64. [Crossref] [PubMed]
- Michaud DS, Lu J, Peacock-Villada AY, et al. Periodontal Disease Assessed Using Clinical Dental Measurements and Cancer Risk in the ARIC Study. J Natl Cancer Inst 2018;110:843-54. [Crossref] [PubMed]
- Mai X, LaMonte MJ, Hovey KM, et al. Periodontal disease severity and cancer risk in postmenopausal women: the Buffalo OsteoPerio Study. Cancer Causes Control 2016;27:217-28. [Crossref] [PubMed]
- Fernández-Plata R, Olmedo-Torres D, Martinez-Briseno D, et al. Prevalence of severe periodontal disease and its association with respiratory disease in hospitalized adult patients in a tertiary care center. Gac Med Mex 2015;151:608-13. [PubMed]
- Yan X, Yang M, Liu J, et al. Discovery and validation of potential bacterial biomarkers for lung cancer. Am J Cancer Res 2015;5:3111-22. [PubMed]
- Yang J, Mu X, Wang Y, et al. Dysbiosis of the Salivary Microbiome Is Associated With Non-smoking Female Lung Cancer and Correlated With Immunocytochemistry Markers. Front Oncol 2018;8:520. [Crossref] [PubMed]
- Cameron SJS, Lewis KE, Huws SA, et al. A pilot study using metagenomic sequencing of the sputum microbiome suggests potential bacterial biomarkers for lung cancer. PLoS One 2017;12:e0177062. [Crossref] [PubMed]
- Lee SH, Sung JY, Yong D, et al. Characterization of microbiome in bronchoalveolar lavage fluid of patients with lung cancer comparing with benign mass like lesions. Lung Cancer 2016;102:89-95. [Crossref] [PubMed]
- Hasegawa A, Sato T, Hoshikawa Y, et al. Detection and identification of oral anaerobes in intraoperative bronchial fluids of patients with pulmonary carcinoma. Microbiol Immunol 2014;58:375-81. [Crossref] [PubMed]
- Liu HX, Tao LL, Zhang J, et al. Difference of lower airway microbiome in bilateral protected specimen brush between lung cancer patients with unilateral lobar masses and control subjects. Int J Cancer 2018;142:769-78. [Crossref] [PubMed]
- Zhuang H, Cheng L, Wang Y, et al. Dysbiosis of the Gut Microbiome in Lung Cancer. Front Cell Infect Microbiol 2019;9:112. [Crossref] [PubMed]
- Zhang W, Luo JW, Dong XP, et al. Salivary Microbial Dysbiosis is Associated with Systemic Inflammatory Markers and Predicted Oral Metabolites in Non-Small Cell Lung Cancer Patients. J Cancer 2019;10:1651-62. [Crossref] [PubMed]
- Greathouse KL, White JR, Vargas AJ, et al. Interaction between the microbiome and TP53 in human lung cancer. Genome Biol 2018;19:123. [Crossref] [PubMed]
- Apopa PL, Alley L, Penney RB, et al. PARP1 Is Up-Regulated in Non-small Cell Lung Cancer Tissues in the Presence of the Cyanobacterial Toxin Microcystin. Front Microbiol 2018;9:1757. [Crossref] [PubMed]
- Peters BA, Hayes RB, Goparaju C, et al. The Microbiome in Lung Cancer Tissue and Recurrence-Free Survival. Cancer Epidemiol Biomarkers Prev 2019;28:731-40. [Crossref] [PubMed]
- Tsay JJ, Wu BG, Badri MH, et al. Airway Microbiota Is Associated with Upregulation of the PI3K Pathway in Lung Cancer. Am J Respir Crit Care Med 2018;198:1188-98. [Crossref] [PubMed]
- Hosgood HD, Sapkota AR, Rothman N, et al. The Potential Role of Lung Microbiota in Lung Cancer Attributed to Household Coal Burning Exposures. Environ Mol Mutagen 2014;55:643-51. [Crossref] [PubMed]
- Bingula R, Filaire M, Radosevic-Robin N, et al. Characterisation of gut, lung, and upper airways microbiota in patients with non-small cell lung carcinoma Study protocol for case-control observational trial. Medicine (Baltimore) 2018;97:e13676. [Crossref] [PubMed]
- Seijo LM, Zulueta JJ. Understanding the Links Between Lung Cancer, COPD, and Emphysema: A Key to More Effective Treatment and Screening. Oncology (Williston Park) 2017;31:93-102. [PubMed]
- Dai J, Yang P, Cox A, et al. Lung cancer and chronic obstructive pulmonary disease: From a clinical perspective. Oncotarget 2017;8:18513-24. [Crossref] [PubMed]
- Russo E, Taddei A, Ringressi MN, et al. The interplay between the microbiome and the adaptive immune response in cancer development. Therap Adv Gastroenterol 2016;9:594-605. [Crossref] [PubMed]
- Rajagopala SV, Vashee S, Oldfield LM, et al. The Human Microbiome and Cancer. Cancer Prev Res (Phila) 2017;10:226-34. [Crossref] [PubMed]
- Hold GL. Gastrointestinal Microbiota and Colon Cancer. Dig Dis 2016;34:244-50. [Crossref] [PubMed]
- Birt DF, Phillips GJ. Diet, genes, and microbes: complexities of colon cancer prevention. Toxicol Pathol 2014;42:182-8. [Crossref] [PubMed]
- Rescigno M. The pathogenic role of intestinal flora in IBD and colon cancer. Curr Drug Targets 2008;9:395-403. [Crossref] [PubMed]
- De Almeida CV, de Camargo MR, Russo E, et al. Role of diet and gut microbiota on colorectal cancer immunomodulation. World J Gastroenterol 2019;25:151-62. [Crossref] [PubMed]
- Dejea C, Wick E, Sears CL. Bacterial oncogenesis in the colon. Future Microbiol 2013;8:445-60. [Crossref] [PubMed]
- Boursi B, Mamtani R, Haynes K, et al. Recurrent antibiotic exposure may promote cancer formation--Another step in understanding the role of the human microbiota? Eur J Cancer 2015;51:2655-64. [Crossref] [PubMed]
- Jenkins SV, Robeson MS 2nd, Griffin RJ, et al. Gastrointestinal Tract Dysbiosis Enhances Distal Tumor Progression through Suppression of Leukocyte Trafficking. Cancer Res 2019;79:5999-6009. [Crossref] [PubMed]
- Holmes I, Harris K, Quince C. Dirichlet multinomial mixtures: generative models for microbial metagenomics. PLoS One 2012;7:e30126. [Crossref] [PubMed]
- Yaghoobi H, Bandehpour M, Kazemi B. Apoptotic Effects of the B Subunit of Bacterial Cytolethal Distending Toxin on the A549 Lung Cancer Cell Line. Asian Pac J Cancer Prev 2016;17:299-304. [Crossref] [PubMed]
- Nowotarski SL, Woster PM, Casero RA Jr. Polyamines and cancer: implications for chemotherapy and chemoprevention. Expert Rev Mol Med 2013;15:e3. [Crossref] [PubMed]
- Niu T, Smith DL, Yang Z, et al. Bioactivity and bioavailability of ginsenosides are dependent on the glycosidase activities of the A/J mouse intestinal microbiome defined by pyrosequencing. Pharm Res 2013;30:836-46. [Crossref] [PubMed]
- Herfs M, Hubert P, Delvenne P. Epithelial metaplasia: adult stem cell reprogramming and (pre)neoplastic transformation mediated by inflammation? Trends Mol Med 2009;15:245-53. [Crossref] [PubMed]
- Wang K, Wang J, Wei F, et al. Expression of TLR4 in Non-Small Cell Lung Cancer Is Associated with PD-L1 and Poor Prognosis in Patients Receiving Pulmonectomy. Front Immunol 2017;8:456. [Crossref] [PubMed]
- Ochoa CE, Mirabolfathinejad SG, Ruiz VA, et al. Interleukin 6, but not T helper 2 cytokines, promotes lung carcinogenesis. Cancer Prev Res (Phila) 2011;4:51-64. [Crossref] [PubMed]
- Caetano MS, Zhang H, Cumpian AM, et al. IL6 Blockade Reprograms the Lung Tumor Microenvironment to Limit the Development and Progression of K-ras-Mutant Lung Cancer. Cancer Res 2016;76:3189-99. [Crossref] [PubMed]
- Chang SH, Mirabolfathinejad SG, Katta H, et al. T helper 17 cells play a critical pathogenic role in lung cancer. Proc Natl Acad Sci U S A 2014;111:5664-9. [Crossref] [PubMed]
- Jungnickel C, Schmidt LH, Bittigkoffer L, et al. IL-17C mediates the recruitment of tumor-associated neutrophils and lung tumor growth. Oncogene 2017;36:4182-90. [Crossref] [PubMed]
- King PT, Sharma R. The Lung Immune Response to Nontypeable Haemophilus influenzae (Lung Immunity to NTHi). J Immunol Res 2015;2015:706376.
- Moghaddam SJ, Ochoa CE, Sethi S, et al. Nontypeable Haemophilus influenzae in chronic obstructive pulmonary disease and lung cancer. Int J Chron Obstruct Pulmon Dis 2011;6:113-23. [Crossref] [PubMed]
- Sriram KB, Cox AJ, Sivakumaran P, et al. Non-typeable Haemophilus Influenzae detection in the lower airways of patients with lung cancer and chronic obstructive pulmonary disease. Multidiscip Respir Med 2018;13:11. [Crossref] [PubMed]
- Gustafson AM, Soldi R, Anderlind C, et al. Airway PI3K pathway activation is an early and reversible event in lung cancer development. Sci Transl Med 2010;2:26ra25. [Crossref] [PubMed]
- Tsay JJ, Wu BG, Badri MH, et al. Airway Microbiota Is Associated with Upregulation of the PI3K Pathway in Lung Cancer. Am J Respir Crit Care Med 2018;198:1188-98. [Crossref] [PubMed]
- Sommariva M, Le Noci V, Bianchi F, et al. The lung microbiota: role in maintaining pulmonary immune homeostasis and its implications in cancer development and therapy. Cell Mol Life Sci 2020. [Epub ahead of print]. [Crossref] [PubMed]
- Wypych TP, Wickramasinghe LC, Marsland BJ. The influence of the microbiome on respiratory health. Nat Immunol 2019;20:1279-90. [Crossref] [PubMed]
- Sivan A, Corrales L, Hubert N, et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science 2015;350:1084-9. [Crossref] [PubMed]
- Daillère R, Vetizou M, Waldschmitt N, et al. Enterococcus hirae and Barnesiella intestinihominis Facilitate Cyclophosphamide-Induced Therapeutic Immunomodulatory Effects. Immunity 2016;45:931-43. [Crossref] [PubMed]


