
High integrin α3 expression is associated with poor prognosis in patients with non-small cell lung cancer
Introduction
Non-small cell lung cancer (NSCLC) accounts for 80–85% of lung cancer, which is the leading cause of cancer-related deaths worldwide (1,2). Despite advances in early detection and treatment, the prognosis for patients with NSCLC remains poor (3). Novel therapeutic targets and treatment strategies are needed to improve the clinical outcomes for NSCLC patients, especially those diagnosed with unresectable, locally advanced or metastatic NSCLC. α3β1 integrin is a promising cancer biomarker and drug target in NSCLC (4). Like other integrins, α3β1 integrin is a heterodimeric transmembrane glycoprotein receptor without a cytoplasmic kinase domain, and binds to various ligands such as fibronectin, laminin, collagen, epiligrin, thrombospondin and chondroitin sulfate proteoglycan 4 (CSPG4) (5). α3β1 integrin mediates the adhesion, cytoskeleton function, and metastasis of NSCLC via cross-talk with EGFR and other receptor tyrosine kinase (RTK) (4,6-8). The integrin α3 (ITGA3) subunit binds to the integrin β1 (ITGB1) subunit as its exclusive binding partner. α3β1 integrin is weakly detected in the basement membranes of normal alveolar walls but is highly expressed in primary lung cancer cells (9,10) and tumor-derived exosomes (4,11). Overexpression of α3β1 integrin has been detected in multiple tumor types and is associated with tumorigenesis, invasion, metastasis, as well as resistance to cancer treatment in several cancer types, including NSCLC (4,12), breast cancer (13), cervical cancer (14), glioma (15), and other cancer type metastasis to the lung (16,17). Almost all these studies were done on preclinical models. Due to the poor sensitivity and specificity of early commercially available antibodies, limited study has been reported on the clinical significance of ITGA3 expression using archival formalin fixed, paraffin embedded (FFPE) patient samples (14). Furthermore, there are conflicting data on the prognosis of ITGA3 expression in NSCLC (12,18). We have recently generated and characterized LXY30, a high affinity peptide ligand for targeting α3β1 integrin-expressing, live tumor cells and tumor-derived exosomes in NSCLC, regardless of histology and tumor genotypes (4,19). Of note, LXY30 has been optimized to bind to live tumor cells but not to live normal cells (4). However, LXY30 does not bind to tumor cells on the archival, FFPE specimens. In the current study, we determined the expression pattern and prognosis of ITGA3 expression by immunohistochemistry (IHC) using commercially available antibodies on archived NSCLC tumors.
Methods
Tissue microarrays (TMAs)
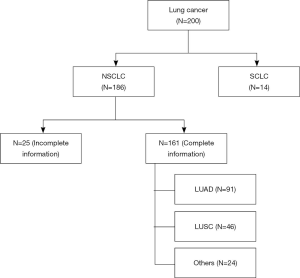
TMAs were prepared from 200 FFPE tissue blocks using an IRB approved protocol (IRB 293828, UC Davis Cancer Center Biorepository) (Figure 1). Three 0.6 mm (diameter) by 0.4 mm (length) cylindrical cores of tumor and adjacent non-malignant (control) lung tissues were collected from each case and placed in the same block (quick-ray manual tissue microarrayer). Three cores of normal liver and kidney tissues were also included on the same block as normal controls. One TMA section was stained with hematoxylin and eosin (H&E) to visualize the presence of tumor and normal lung tissue. The deidentified database for the TMAs has been annotated with patient demographics (age at diagnosis, gender, race/ethnicity), stage, histopathology, and survival outcomes.
IHC stain
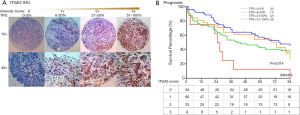
TMA slides were stained by IHC for ITGA3 integrin expression. 4-µm FFPE tissue sections were cut for each TMA. Slides were deparaffinized and pretreated with Heat-Induced-Epitope-Retrieval solution (pH 9) (DAKO). Slides are preincubated in Dual Endogenous Enzyme Block (DEEB) blocking solution in a DAKO Autostainer Link48. Primary antibody for ITGA3 (ab131055, dilution 1:200; Abcam) were used for IHC. The slides were incubated for 30 minutes and then washed, which was followed by detection by EDL solution (DAKO EnVision and Dual Link System-HRP). Hematoxylin was used as the counterstain. IHC slides were scored by two readers (QL and WM) and verified by two pathologists (TW and CZ) blinded to clinicopathologic information. Staining for ITGA3 was semi-quantified and assessed using both intensity and percentage of positive cells. Staining intensity was graded as 0= negative (no cells stained); 1= weak; 2= moderate; 3= strong. The percentage of positive cells was defined as 0 (0–5%), 1+ (6–20%), 2+ (21–50%), and 3+ (51–100%) (Figure 2A). After the initial analysis, study pathologists recommended to measure the ITGA3 expression by tumor proportion score (TPS), i.e., the percentage of viable tumor cells showing partial or complete membrane staining at any intensity. The specimen was considered to have ITGA3 expression if TPS >5% and no ITGA3 expression if TPS ≤5%.
Statistical analysis
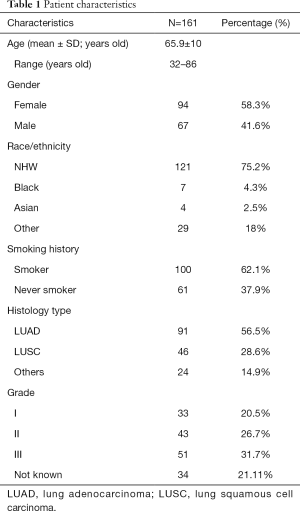
All baseline demographics and patient characteristics were compared between different ITGA3 expression level groups by Pearson chi-square tests except for age, which was considered a continuous variable and therefore analyzed with Kruskal-Wallis test. Multivariate logistic regression with backward variable section was applied to patient and tumor characteristics to examine factors associated with expression (TPS >5%) versus no (TPS ≤5%) ITGA3 expression levels.
The Kaplan-Meier method and log-rank test were first used to compare survival outcomes between different ITGA3 expression levels. Univariate survival analysis and multivariate analysis were performed with Cox proportional hazards models using overall survival (OS) as outcomes. Factors found to be significant in univariate analysis were included and selected by backward method in multivariate analysis (except ITGA3 IHC expression which was always included). To ensure the proportional hazards assumption holds, the assumption was examined using Schoenfeld residuals. To account for potential confounding and covariate imbalances between different ITGA3 expression groups, we used propensity score-weighted Kaplan-Meier estimator, incorporating the inverse of the propensity score (20). The propensity score was used to balance the covariate distribution between groups for studies with either causal or non-causal purposes (21). For survival outcomes, this method has been shown to produce less biased treatment effect estimates than stratification or covariate adjustment based on the propensity score (22). Significant covariates in univariate and multivariate survival analyses were used in the propensity score model using logistic regression. We also performed subgroup analyses for different patient characteristics groups. As there was high proportion of unknown metastasis status (63% of all patients) in the database, and the above analyses created an additional level for patients with unknown metastasis status, we performed additional sensitivity analyses using the multiple imputation method for unknown metastasis based on missing at random (MAR) assumption (23). All analyses were performed using SAS 9.4 (SAS Institute Inc., Cary, NC, USA) and R 3.6.0 (R Foundation for Statistical Computing, Vienna, Austria). All P values were two-sided, and a P≤0.05 was considered statistically significant.
TCGA data access and analysis
The transcriptome expression data from NSCLC patients were accessed on September 9th, 2019 in The Cancer Genome Atlas (TCGA) database (http://www.cancergenome.nih.gov) including 500 lung adenocarcinoma (LUAD) and 494 lung squamous cell carcinoma (LUSC) samples. Fragments per kilobase of exon per million reads mapped (FPKM) values were chosen as the representative measure of gene expression (24). Survival data was obtained for the individual patient barcodes matched to these FPKM values and was used to separate NSCLC subtypes into high and low expression groups, on which log-rank tests for significant difference (P<0.05) were conducted.
Results
Patient characteristics
Figure 1 illustrates the distribution of the 200 cases of lung cancer on the TMA. Excluding those cases with small cell lung cancer (SCLC) and NSCLC without complete clinical information, the expression of ITGA3 was analyzed in 161 cases. All the statistics for baseline demographics and patient characteristics are summarized in Table 1.
Full table
High ITGA3 expression was associated with poor prognosis
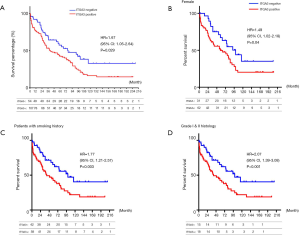
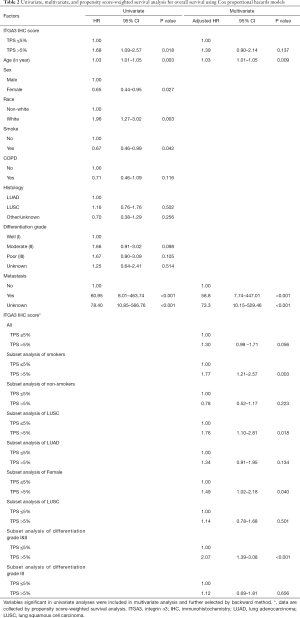
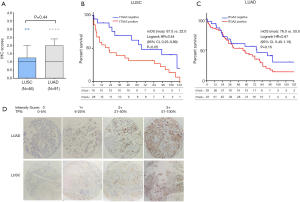
ITGA3 IHC expression was detected in 107/161 (66.5%) of the NSCLC samples. Weak (1+), moderate (2+) and strong (3+) ITGA3 expression was detected in 66 (41.0%), 33 (20.5%), and 8 (5.0%) cases, respectively (Figure 2A). Kaplan-Meier curves indicated that weak (1+) and strong (3+) ITGA3 expression was significantly associated with poor OS in NSCLC patients (P<0.05) (Figure 2B). Overall, NSCLC patients whose tumors expressed ITGA3 by IHC (1+ to 3+) was associated with poorer prognosis compared to those patients whose tumors did not express ITGA3 (Figure 3A). We also performed propensity-score-weighted survival analyses between different ITGA3 expression levels for different patient characteristics groups. We found that association between ITGA3 IHC expression (weak to strong) and poor prognosis is significant in subgroups of female patients (HR =1.49, 95% CI: 1.02–2.18, P=0.04) (Figure 3B), smokers (HR =1.77, 95% CI: 1.21–2.57, P=0.003) (Figure 3C) and differentiation grade I & II (HR =2.07, 95% CI: 1.39–3.08, P<0.001) (Figure 3D), but not in male patients (P=0.50), non-smokers (P=0.22), differentiation grade III (P=0.66) (Table 2). Sensitivity analyses using multiple imputation demonstrated similar conclusions for these subgroups.
Multivariate survival analysis demonstrated that poor prognosis is associated with older age and metastasis (Table 2). Propensity-score-weighted survival analysis demonstrated that the significance of association between ITGA3 IHC expression (1+ to 3+) and prognosis is near the edge (HR =1.30, 95% CI: 0.99–1.71, P=0.056) for all NSCLC patients after adjusting for confounders (Table 3).
Full table

Full table
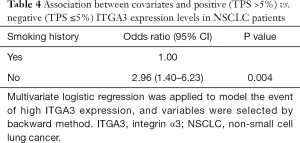
Due to a high proportion of unknown metastasis, sensitivity analyses using multiple imputation method for unknown metastasis were performed and demonstrated that ITGA3 IHC expression (1+ to 3+) was significantly associated with poor prognosis (HR =1.67, 95% CI: 1.05–2.64, P=0.029) for all NSCLC patients after adjusting for confounders and imputing for unknown metastasis (Table 4).
Full table
ITGA3 expression in histology subgroups of NSCLC
We found the median score of ITGA3 IHC was not significantly higher in LUAD than LUSC (P=0.44) (Figure 4A), and high ITGA3 IHC score was associated with a statistically significant difference in LUSC (P<0.05) (Figure 4B) but not LUAD (P=0.16) (Figure 4C). Representative ITGA3 IHC stains on LUAC and LUSC are showed (Figure 4D). After adjusting for confounders, propensity-score-weighted survival analysis demonstrated that the association of ITGA3 IHC expression (weak to strong) and poor prognosis was statistically significant in LUSC patients (HR =1.76, 95% CI: 1.10–2.81, P=0.018) but not in LUAD patients (P=0.13).
Poor prognosis was associated with tumor metastasis
We found that ITGA3 expression was significantly higher in the NSCLC tumors from patients with metastatic tumors (N=19) compared to those patients without metastatic tumors (N=41). Due to a high proportion of unknown metastasis status (63% of all patients) in the database, the wide confidence interval of HR for metastasis was observed, although the association is still significant. We performed additional sensitivity analysis using multiple imputation method for unknown metastasis, and the significance of metastasis on poor prognosis was confirmed by sensitive analysis (HR =2.89, 95% CI: 1.48–5.65, P=0.003) (Table 3).
ITGA3 expression was associated with no smoking history
Multivariate logistic regression with backward variable section was applied to patient and tumor characteristics to examine factors associated with expression (TPS >5%) versus no (TPS ≤5%) ITGA3 expression levels (Table 4). We found ITGA3 expression was associated with no smoking history (OR =2.96, 95% CI: 1.40–6.23, P=0.004), but not significantly associated with other factors.
High α3 integrin expression by transcriptome sequencing was associated with poor prognosis in patients with NSCLC
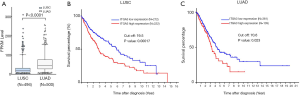
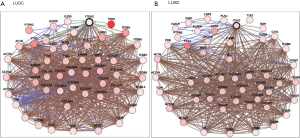
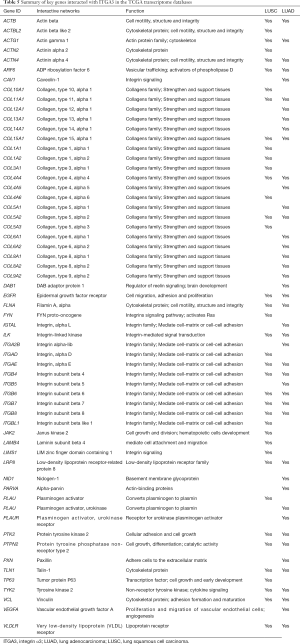
We also validated our findings in an independent cohort of NSCLC tumors. We analyzed the RNA expression of ITGA3 gene in 494 LUSC and 500 LUAD samples using the transcriptome expression data in TCGA database. We found that the median RNA expression level of ITGA3 was significantly higher in LUAD than LUSC (43.80 vs. 17.40, P<0.0001) (Figure 5A). Similar to our finding in IHC, high ITGA3 RNA expression was associated with a statistically significant poor prognosis in LUSC (P<0.05) (Figure 5B). Using a higher cutoff than LUSC (70.6 vs. 19.5 FPKM), high ITGA3 RNA expression was also associated with poor prognosis in LUAD (P=0.023) (Figure 5C). ITGA3 shared many interactive genes mediating cell adhesion and motility in both LUSC and LUAD (Figure 6A,B, Table 5), respectively. Furthermore, ITGA3 interacted with many key genes regulating epithelial to mesenchymal transition, angiogenesis, invasion and metastasis in both LUAD and LUSC as illustrated in Figure 7.
Full table
Discussion
α3β1 integrin is a promising biomarker for lung cancer detection and a potential drug target, being one of the most commonly expressed integrin subtypes found on tumor cells mediating metastasis and treatment resistance (6). Using a novel peptide ligand LXY30 that has higher affinity and longer half-life than natural integrin (4,19), we recently showed that ITGA3 was expressed in about 90% of live tumor cells and exosomes isolated from patients with metastatic NSCLC (4). However, LXY30 does not work in FFPE specimens. We have screened several commercially available anti-ITGA3 antibodies and selected the Abcam ab131055 antibody in this study. We found that ITGA3 was expressed in the majority (66.5%) of NSCLC specimens by IHC (Figure 2). We further categorized the ITGA3 IHC expression in our study as weak (1+), moderate (2+) and strong (3+), which were detected in 66 (41.0%), 33 (20.5%), and 8 (5.0%) cases, respectively (Figure 2). The frequencies and patterns of ITGA3 expression in our study are within the range of two different antibodies in the Human Protein Atlas (https://www.proteinatlas.org) (Table 6). These data further support our effort in targeting ITGA3 using LXY30 in patients with live NSCLC. Propensity-score-weighted survival analysis demonstrated that the significance of association between ITGA3 IHC expression (1+ to 3+) and prognosis is near the edge (HR =1.30, 95% CI: 0.99–1.71, P=0.056) after adjusting for confounders. However, propensity-score-weighted survival analysis demonstrated that the association of ITGA3 IHC expression and poor prognosis was not statistically significant in either LUSC patients (P=0.11) or LUAD patients (P=0.07). Thus, histology alone should not be used to select NSCLC for ITGA3 expression.

Full table
The expression of specific integrin subtypes has been linked to organotropic metastasis of circulating tumor cells and exosomes in epithelial tumors (11,12,17). ITGA3 and β1 subunits were detected in 13 (54%) and 24 (100%) of exosomes isolated from 24 human metastatic cell lines, while the normal tissues or PBMCs did not express any integrin (4). Although there are a significant number of patients with an unknown metastasis status, we found that ITGA3 IHC expression was only significantly associated with non-smoking history in both univariate and multivariate analyses.
The TCGA datasets are publicly accessible resources containing sequencing data of human tumors and normal tissues using NGS and other clinically relevant modern genomic technologies. Since its conception, TCGA datasets have been proven invaluable tools to cancer research community to study the role of specific genes and genomic changes in the biology and prognosis of specific cancer types (https://cancergenome.nih.gov). Using the TCGA RNASeq dataset, we found a similar pattern of high ITGA3 expression in LUAD compared to LUSC. The interactive networking analysis showed that LUAD and LUSC shared many cell adhesion and motility genes while distinct genes interaction was also noted for LUAD and LUSC, respectively (Figure 6). Similar to our recent report, ITGA3 interacts with EGFR in LUAD (4). Furthermore, TP63 and PTK2 (FAK) modulates the ITGA3 expression in LUSC, which warrant further validation.
Our study has several translational potentials. First, we have recently shown that a novel, potent peptide LXY30 has high affinity for binding to α3β1 integrin, which can increase the sensitivity of cancer detection, molecular diagnosis and in vivo targeted delivery of imaging dye and cancer drugs in α3β1 integrin-expression NSCLC (4). The ITGA3 IHC can be used to identify candidate NSCLC patients using archived FFPE tissue specimens for targeted therapy although the caution should be excised to select the primary antibody for ITGA3. Second, we have confirmed that ITGA3 expression is associated with metastasis of NSCLC tumors. Analysis of TCGA databases showing that ITGA3 significantly interacted with many key genes regulating cell adhesion, motility, epithelial to mesenchymal transition, angiogenesis, invasion and metastasis in both LUAD and LUSC (Table 5 and Figure 7). Further study is warranted to delineate the mechanism by which ITGA3 may mediate these malignant processes, and to develop therapeutic strategies for NSCLC patients.
Our study has several limitations. First, we could not explain the impact of sex, histology and smoking status on ITGA3 expression. Second, there were no molecular and immune biomarker data (such as PD-L1 IHC, tumor mutation burden) of tumors available for this dataset, which included NSCLC specimens from before the era of precision oncology. We plan to study the association of ITGA3 expression with molecular and immune biomarkers in future studies.
In conclusion, we have shown that the expression of ITGA3 subunit in patients with NSCLC is associated with poor prognosis and tumor metastasis. Further research is warranted for targeting α3β1 integrin in NSCLC.
Acknowledgments
The authors would like to thank Dr. Regina Gandour Edwards and Irmgard Feldman at the UC Davis Cancer Center Biorepository for the acquisition of the lung cancer tissue microarray.
Funding: This research was supported by the University of California Cancer Research Coordinating Committee grant (3-444964-34912) and Personalized Cancer Therapy Gift Fund (TL), the Biostatistics Shared Resource funded by the UC Davis Comprehensive Cancer Center Support Grant (CCSG) awarded by the National Cancer Institute (NCI P30CA093373) (SC). QL was also supported by a research and training scholarship from Shanghai Jiaotong University Affiliated Sixth People’s Hospital.
Footnote
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tlcr-19-633
Peer Review File: Available at http://dx.doi.org/10.21037/tlcr-19-633
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tlcr-19-633). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was approved by the institutional review board (IRB) at the University of California, Davis (Protocol No. 226210) and conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- DeSantis CE, Miller KD, Goding Sauer A, et al. Cancer statistics for African Americans, 2019. CA Cancer J Clin 2019;69:211-33. [Crossref] [PubMed]
- Li T, Kung HJ, Mack PC, et al. Genotyping and genomic profiling of non-small-cell lung cancer: implications for current and future therapies. J Clin Oncol 2013;31:1039-49. [Crossref] [PubMed]
- Postmus PE, Kerr KM, Oudkerk M, et al. Early and locally advanced non-small-cell lung cancer (NSCLC): ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 2017;28:iv1-21. [Crossref]
- Xiao W, Ma W, Wei S, et al. High-affinity peptide ligand LXY30 for targeting alpha3beta1 integrin in non-small cell lung cancer. J Hematol Oncol 2019;12:56. [Crossref] [PubMed]
- Millard M, Odde S, Neamati N. Integrin targeted therapeutics. Theranostics 2011;1:154-88. [Crossref] [PubMed]
- Desgrosellier JS, Cheresh DA. Integrins in cancer: biological implications and therapeutic opportunities. Nat Rev Cancer 2010;10:9-22. [Crossref] [PubMed]
- Legate KR, Wickstrom SA, Fassler R. Genetic and cell biological analysis of integrin outside-in signaling. Genes Dev 2009;23:397-418. [Crossref] [PubMed]
- Morse EM, Brahme NN, Calderwood DA. Integrin cytoplasmic tail interactions. Biochemistry 2014;53:810-20. [Crossref] [PubMed]
- Bartolazzi A, Cerboni C, Flamini G, et al. Expression of alpha 3 beta 1 integrin receptor and its ligands in human lung tumors. Int J Cancer 1995;64:248-52. [Crossref] [PubMed]
- Guo L, Zhang F, Cai Y, et al. Expression profiling of integrins in lung cancer cells. Pathol Res Pract 2009;205:847-53. [Crossref] [PubMed]
- Hoshino A, Costa-Silva B, Shen TL, et al. Tumour exosome integrins determine organotropic metastasis. Nature 2015;527:329-35. [Crossref] [PubMed]
- Yoshimasu T, Sakurai T, Oura S, et al. Increased expression of integrin alpha3beta1 in highly brain metastatic subclone of a human non-small cell lung cancer cell line. Cancer Sci 2004;95:142-8. [Crossref] [PubMed]
- Shirakihara T, Kawasaki T, Fukagawa A, et al. Identification of integrin alpha3 as a molecular marker of cells undergoing epithelial-mesenchymal transition and of cancer cells with aggressive phenotypes. Cancer Sci 2013;104:1189-97. [Crossref] [PubMed]
- Du Q, Wang W, Liu T, et al. High Expression of Integrin alpha3 Predicts Poor Prognosis and Promotes Tumor Metastasis and Angiogenesis by Activating the c-Src/Extracellular Signal-Regulated Protein Kinase/Focal Adhesion Kinase Signaling Pathway in Cervical Cancer. Front Oncol 2020;10:36. [Crossref] [PubMed]
- Nakada M, Nambu E, Furuyama N, et al. Integrin alpha3 is overexpressed in glioma stem-like cells and promotes invasion. Br J Cancer 2013;108:2516-24. [Crossref] [PubMed]
- Morini M, Mottolese M, Ferrari N, et al. The alpha 3 beta 1 integrin is associated with mammary carcinoma cell metastasis, invasion, and gelatinase B (MMP-9) activity. Int J Cancer 2000;87:336-42. [Crossref] [PubMed]
- Zhou B, Gibson-Corley KN, Herndon ME, et al. Integrin alpha3beta1 can function to promote spontaneous metastasis and lung colonization of invasive breast carcinoma. Mol Cancer Res 2014;12:143-54. [Crossref] [PubMed]
- Adachi M, Taki T, Huang C, et al. Reduced integrin alpha3 expression as a factor of poor prognosis of patients with adenocarcinoma of the lung. J Clin Oncol 1998;16:1060-7. [Crossref] [PubMed]
- Xiao W, Li T, Bononi FC, et al. Discovery and characterization of a high-affinity and high-specificity peptide ligand LXY30 for in vivo targeting of alpha3 integrin-expressing human tumors. EJNMMI Res 2016;6:18. [Crossref] [PubMed]
- Imbens GW. The role of the propensity score in estimating dose-response functions. Biometrika 2000;87:706-10. [Crossref]
- Li F, Zaslavsky AM, Landrum MB. Propensity score weighting with multilevel data. Stat Med 2013;32:3373-87. [Crossref] [PubMed]
- Austin PC. The performance of different propensity score methods for estimating marginal hazard ratios. Stat Med 2013;32:2837-49. [Crossref] [PubMed]
- Rubin DB. Multiple Imputation for Nonresponse in Surveys. 1987.
- Wagner GP, Kin K, Lynch VJ. Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theory Biosci 2012;131:281-5. [Crossref] [PubMed]


