Digital detection of T790M—yes or no to an ultrasensitive assay
In the management of advanced non-small cell lung cancer it is now routine clinical practice to screen diagnostic specimens for activating mutations in epidermal growth factor receptor (EGFR) and KRAS and translocations involving ALK and ROS. When detected, these mutations directly inform clinical decision-making. In particular, patients with one of the common EGFR mutations, exon 19 deletion or L858R, often respond extremely well to EGFR tyrosine kinases inhibitors (TKIs). However, almost inevitably patients will relapse and the most common mechanism of resistance is a secondary mutation in EGFR that alters the affinity of the binding pocket to ATP—the T790M mutation (1).
This raises a number of interesting and clinically important questions. First, is the T790M mutation present in TKI-naïve biopsies or does it emerge during therapy? Second, what is the best way to detect the T790M mutation? Third, how should this information be used to guide clinical decision-making?
Watanabe and colleagues in a recent publication in Clinical Cancer Research have made a significant contribution to the field (2). They demonstrate the potential to use droplet digital polymerase chain reaction (ddPCR) as an ultrasensitive tool for the detection of rare variants in a large cohort of patients undergoing surgical resection, and they show that approximately 80% of TKI-naïve patients have detectable T790M in mainly early stage disease.
Watanabe and colleagues chose to use a digital PCR assay. ddPCR is a relatively recent refinement of digital PCR; a protocol which has been around for over 20 years but has been relatively under-utilized (3). However, recent advances in platform development, notably the development of microdroplet protocols have led to a marked increase in its popularity (4,5). Digital PCR is itself a very simple concept (Figure 1). It involves diluting DNA into aliquots, which can be chambers in microfluidic devices or microdroplets (6). The aim is usually to dilute DNA so that each droplet has on average less than a single copy of the target DNA locus. Then the droplets are tested for the presence or absence of the target DNA. This binary outcome—positive or negative is what gives the technique the name “digital”. For point mutations differently labeled probes are used in a single reaction so that each microdroplet is tested for both the mutant and wild type species.
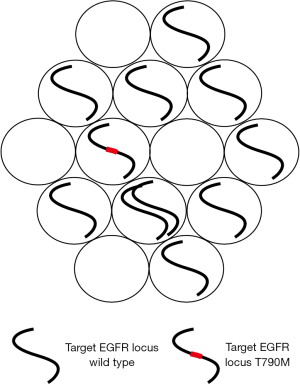
Digital PCR, although having a less broad dynamic range, has certain advantages over standard quantitative PCR protocols in a number of clinically relevant scenarios (4). It can be used to measure the absolute number of copies of an individual DNA species in any given sample and it performs extremely well in the detection and quantification of rare variants (6,7). It is accurate and precise and the data produced is straightforward to interpret.
The authors used the RainDance platform, in which DNA from each sample is diluted into over 1 million picoliter-sized drops also containing PCR master mix buffer, primers and probes labeled with different fluorophores—one that will recognize wild type EGFR; the other that recognizes the mutated sequence (EGFR, 2369C>T). The microdroplets are channeled past a laser excitation/detection reader apparatus equivalent to a flow cytometer. Each microdroplet will fluoresce differently dependent on whether or not they contain the wild type or mutated PCR amplicon.
A clear objective in this study was to devise an assay capable of detecting rare variant mutations that may be present at a mutant allele frequency (MAF) of much less than 1%. The authors were therefore painstaking in their approach to establishing the sensitivity of the assay and the lower limit of valid detection of the T790M mutation. The lower limit of detection [what the authors termed the limit of blank (LOB)] is a measure of how wild type DNA performs in an assay and it defines the level above which an assay can be deemed positive. These attributes (sensitivity/LOB) will vary with the platform used and the primers/probe sets and need to be established and validated for all rare variant assays.
Through a series of control experiments using a variety of assumed wild type template DNAs, including cell lines that had been fixed and paraffin-embedded, the authors determined that ten or more positive droplets equated to a positive result. They went on to show, using spike-in experiments, that the measured concentration of the mutation matched the expected concentration down to a MAF of 0.01%; and further showed that they were able to detect the mutant allele down to a MAF of 0.001%. Importantly, a subgroup of assays performed on patient biopsy DNA, was reproduced by a different operator on a different day and delivered almost identical results.
As regards the critical potential issue of FFPE-mediated bias (8), this was addressed by sampling normal tissue adjacent to tumor in 16 patients—8 with EGFR mutated disease and eight with EGFR—wild type disease and demonstrating no excess of T790M mutations in either group.
In summary, this represents a very sensitive, reproducible and robust assay for the detection of EGFR T790M in patient-derived specimens.
To put the detection sensitivity in context, the published results of a competing technology—targeted next generation sequencing—range from 0.4–2% in the analysis of cfDNA (9,10), although reports on small numbers of patients indicate that may be improved (11). The commonly used MiSeq platform was used as a comparator in this study with an estimated threshold of detection of 1.4%.
Watanabe et al. went on to apply their validated protocol to the largest series of patients reported to date. Of the total of 373 patients, 89.2% had stage I or II disease, consistent with this being a surgical series. All patients had a mutation in EGFR with 94.9% being the commonly reported mutations—L858R or exon 19 deletion. Using the ddPCR assay a surprising 79.9% of patients were positive for the T790M mutation at baseline. In the vast majority of these individuals, and 75.3% of all patients, the T790M bearing cells represented a rare subclone with a MAF in the interval 0.01–0.1%.
It has been known for some time that T790M is present pre-treatment in a proportion of TKI-naïve EGFR mutant lung cancer. As assay sensitivity has increased, then so too has the reported prevalence of T790M—with the overall detection rate ranging from less than 10% using Sanger sequencing to 78.9% using colony PCR in a small cohort (12,13). The concern that very sensitive assays may yield false positives has been addressed in this study by the careful assay validation discussed above. Indeed one reasonable hypothesis may be that if a more sensitive protocol were developed or limitless tumor-derived DNA available, that an even higher proportion of tumors may be positive for T790M. This hypothesis is consistent with the observation that T790M was detectable in all EGFR-mutated specimens (but not controls) in which >150 ng of genomic DNA was analyzed.
On the face of it this data suggests that in most cases, even in relatively early stage EGFR-mutant disease, that the resistance conferring T790M mutation is already present at the point of diagnosis. This is somewhat controversial for two reasons: first, the concerns raised that the detection of T790M rare variants in formalin-fixed paraffin-embedded specimens is biased by artifactual C>T transitions (8); and second, recent in vitro experiments suggest only a proportion of T790M-mediated resistance is present prior to treatment (14,15). These carefully performed studies identify a second group of “late” TKI-resistance clones. The authors suggest a model whereby T790M negative cells tolerate the TKI for a period (persister cells) before a clone with T790M or another resistance-conferring mutation emerges.
There are many publications detailing the common and more infrequent mechanisms of resistance to TKIs in EGFR-mutant lung cancer. These have usually been identified via rebiopsy of progressive disease and generally in the advanced disease setting rather than in early disease (1). The proportion of TKI-resistance generally ascribed to T790M mutation is near to 50%, which at first glance is at odds with the 80% prevalence reported in this study. However, multiple mechanisms of resistance can coexist at both the cellular and subclonal levels, and the presence of multiple subclones may be missed by the standard clinical practice of performing a single rebiopsy procedure on disease progression (1). It is standard practice to rebiopsy EGFR-mutant disease when there is progression on a TKI as establishing the mechanism of resistance often necessitates histological assessment (small cell transformation) or a FISH assay (MET amplification). It will be important in future studies to correlate results from rebiopsy specimens with cfDNA analyses (16). For example, it is a formal possibility that in the clinic T790M is actually much more common at relapse post-TKI than reported, but that in many individuals a separate dominant subclone outcompetes the minor T790M subclone.
So what will be the clinical utility of detection of “ultra” rare variants? It is difficult with current knowledge to integrate the detection of a subclone at 0.01% into clinical decision-making, particularly in resected disease. It is worth speculating that patients with markedly different T790M mutational loads (such as >10% vs. 0.01%) may have different outcomes, particularly in the advanced setting in which a selective pressure will be exerted by TKIs.
The 3rd generation TKIs has potent in vitro activity against the activating mutations—L858R and exon 19 deletion, and against double mutants with either of these mutations plus T790M. These drugs now have proven clinical efficacy in T790M positive disease, albeit in early phase trials in the advanced setting. Should they be used as first-line therapies in advanced EGFR-mutant disease in which rare T790M events can be defined?
The jury is firmly out on this suggestion. There are competing rationales. We know that initial responses to 1st and 2nd line TKIs are often impressive and that the most common resistance mechanism (T790M) is therapeutically tractable. We now also know that tertiary mutations and resistance mechanisms emerge in response to the 3rd generation TKIs, and these may not yet be therapeutically tractable (16,17). If such mutations were to emerge earlier as a result of upfront 3rd generation TKIs then overall outcomes may be poorer. However, the calculation may be different for those patients with a relatively high T790M MAF pretreatment. Further, the distinction between pre-existing T790M mutations and the “persister cell” model could have profound implications for the choreography of treatment of EGFR-mutated lung cancer (14,15).
A separate question is whether decisions on adjuvant therapy could be informed by the detection of T790M in a surgical resection specimen. The role for adjuvant TKIs in EGFR mutated disease is not yet established (18).
There is therefore much scope for further research in this field that would be facilitated by a very sensitive, validated and robust assay for T790M status.
Finally, the authors reported associations between T790M mutation and various subgroups. Perhaps unsurprisingly the larger tumors and those with TP53 mutations were more likely to have a T790M mutation. More intriguing was the association between PIK3CA mutation and T790M mutation, albeit in small numbers.
This study is an important one because it sets the current benchmark in terms of the combination of EGFR T790M assay sensitivity and cohort size. It cements microdroplet digital PCR as a rational choice for robust, reproducible, easily interpreted assays. As with many good translational studies it raises as many questions as it addresses. First is it reproducible by other groups and in other cohorts? Is the community reassured that these ultrasensitive techniques are not delivering false positive results? Will this assay be translatable to cfDNA and urine? Will stratification not only by the presence or absence of an activating mutation but also by T790M mutational load be clinically informative? Critically, when should 3rd generation TKIs be given to patients with detectable T790M? There are not always binary “digital”—answers to these questions.
Acknowledgements
Funding: This work is supported by the Wellcome Trust and the Roy Castle Lung Cancer Foundation. FM is a Wellcome Trust Intermediate Clinical Fellow (WT097143MA). This work is also supported by King’s Health Partners Challenge Fund and the National Institute for Health Research (NIHR) Biomedical Research Centre based at Guy’s and St Thomas’ NHS Foundation Trust and King’s College London. The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR or the Department of Health.
Footnote
Provenance: This is a Guest Editorial commissioned by Section Editor Hongbing Liu, MD, PhD (Department of Respiratory Medicine, Jinling Hospital, Nanjing University School of Medicine, Nanjing, China).
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Chong CR, Jänne PA. The quest to overcome resistance to EGFR-targeted therapies in cancer. Nat Med 2013;19:1389-400. [Crossref] [PubMed]
- Watanabe M, Kawaguchi T, Isa S, et al. Ultra-Sensitive Detection of the Pretreatment EGFR T790M Mutation in Non-Small Cell Lung Cancer Patients with an EGFR-Activating Mutation Using Droplet Digital PCR. Clin Cancer Res 2015;21:3552-60. [Crossref] [PubMed]
- Vogelstein B, Kinzler KW. Digital PCR. Proc Natl Acad Sci U S A 1999;96:9236-41. [Crossref] [PubMed]
- Day E, Dear PH, McCaughan F. Digital PCR strategies in the development and analysis of molecular biomarkers for personalized medicine. Methods 2013;59:101-7. [Crossref] [PubMed]
- Ivey A, Hills RK, Simpson MA, et al. Assessment of Minimal Residual Disease in Standard-Risk AML. N Engl J Med 2016;374:422-33. [Crossref] [PubMed]
- Hindson BJ, Ness KD, Masquelier DA, et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal Chem 2011;83:8604-10. [Crossref] [PubMed]
- Sanders R, Huggett JF, Bushell CA, et al. Evaluation of digital PCR for absolute DNA quantification. Anal Chem 2011;83:6474-84. [Crossref] [PubMed]
- Ye X, Zhu ZZ, Zhong L, et al. High T790M detection rate in TKI-naive NSCLC with EGFR sensitive mutation: truth or artifact? J Thorac Oncol 2013;8:1118-20. [Crossref] [PubMed]
- Paweletz CP, Sacher AG, Raymond CK, et al. Bias-Corrected Targeted Next-Generation Sequencing for Rapid, Multiplexed Detection of Actionable Alterations in Cell-Free DNA from Advanced Lung Cancer Patients. Clin Cancer Res 2016;22:915-22. [Crossref] [PubMed]
- Forshew T, Murtaza M, Parkinson C, et al. Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Sci Transl Med 2012;4:136ra68. [Crossref] [PubMed]
- Newman AM, Bratman SV, To J, et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med 2014;20:548-54. [Crossref] [PubMed]
- Su KY, Chen HY, Li KC, et al. Pretreatment epidermal growth factor receptor (EGFR) T790M mutation predicts shorter EGFR tyrosine kinase inhibitor response duration in patients with non-small-cell lung cancer. J Clin Oncol 2012;30:433-40. [Crossref] [PubMed]
- Fujita Y, Suda K, Kimura H, et al. Highly sensitive detection of EGFR T790M mutation using colony hybridization predicts favorable prognosis of patients with lung cancer harboring activating EGFR mutation. J Thorac Oncol 2012;7:1640-4. [Crossref] [PubMed]
- Ramirez M, Rajaram S, Steininger RJ, et al. Diverse drug-resistance mechanisms can emerge from drug-tolerant cancer persister cells. Nat Commun 2016;7:10690. [Crossref] [PubMed]
- Hata AN, Niederst MJ, Archibald HL, et al. Tumor cells can follow distinct evolutionary paths to become resistant to epidermal growth factor receptor inhibition. Nat Med 2016;22:262-9. [Crossref] [PubMed]
- Thress KS, Paweletz CP, Felip E, et al. Acquired EGFR C797S mutation mediates resistance to AZD9291 in non-small cell lung cancer harboring EGFR T790M. Nat Med 2015;21:560-2. [Crossref] [PubMed]
- Piotrowska Z, Niederst MJ, Karlovich CA, et al. Heterogeneity Underlies the Emergence of EGFRT790 Wild-Type Clones Following Treatment of T790M-Positive Cancers with a Third-Generation EGFR Inhibitor. Cancer Discov 2015;5:713-22. [Crossref] [PubMed]
- Kelly K, Altorki NK, Eberhardt WE, et al. Adjuvant Erlotinib Versus Placebo in Patients With Stage IB-IIIA Non-Small-Cell Lung Cancer (RADIANT): A Randomized, Double-Blind, Phase III Trial. J Clin Oncol 2015;33:4007-14. [Crossref] [PubMed]


