Methylation analyses in liquid biopsy
Introduction
Lung cancer is the second most common cancer in the United States, and is the leading cause of cancer-related deaths worldwide. Patients diagnosed at an early stage have the best chance for survival. Unfortunately, only one third of patients with non-small cell lung cancer (NSCLC), which represents more than 80% of lung cancer cases, are diagnosed with localized, potentially curable disease (1). Screening using low-dose computed tomography (LDCT) has recently been shown to improve the early detection of lung cancer –60% of the cases diagnosed at stage I—and reduce cancer mortality by 20% compared to chest radiography (2). Although very promising, LDCT scanning identifies a high number of nodules that prompt further, invasive testing but do not result in a lung cancer diagnosis (2), detects many lung cancers that seem to be indolent (3), and remains a costly screening method to implement. Changing eligibility criteria for LDCT screening, from heavy smokers (at least 30 pack-years, current smokers or with no more than 15 years since quitting) aged 55 to 80 years, to a risk-based selection of ever-smokers aged 50 to 80 years, has been shown to improve screening effectiveness and efficiency (4). In addition, the systematic computational extraction, mining and interpretation of imaging features found in CT scans of lung cancer patients has led to the development of a prognostic signature (5,6). Radiomics-based biomarkers are now also being evaluated for performance in risk stratification of lung cancer patients diagnosed by LDCT to facilitate individualized patient management (7). Surgery is the main treatment for stage I NSCLC, but some patients with stage IB receive adjuvant chemotherapy to reduce the risk of disease recurrence. Still, 30% of patients experience relapse and die of their disease (8). Developing new markers for the early detection, prognosis and monitoring of lung cancer is therefore urgently needed to increase the efficacy of screening and reduce disease mortality.
Like many types of cancers, lung cancer is characterized by diverse genetic alterations. This diversity poses a challenge for the development of reliable and broadly applicable DNA-based biomarkers. The variability in gene mutations contrasts with the consistency of epigenetic changes that occur during carcinogenesis. Epigenetic abnormalities, comprising alterations in DNA methylation, histone modifications, nucleosome positioning and non-coding RNAs are considered hallmarks of cancer initiation and progression (9). Recent advances in the field of lung cancer epigenetics have revealed promising biomarkers, particularly involving changes in DNA methylation, which is the best studied epigenetic mark in human cancer.
The role of de novo methylation in cancer initiation and progression
DNA methylation occurs at the 5th carbon of the cytosine ring within CpG dinucleotides. This reaction is catalyzed by three main DNA methyltransferases (DNMTs) responsible for de novo methylation during development (DNMT3A and DNMT3B), or the maintenance of methylation following DNA replication (DNMT1 assisted by DNMT3A and DNMT3B). CpG sites are located throughout the genome, but their distribution is uneven and their methylation follows a bimodal profile (10). The vast majority of the genome contains few CpGs (less than 1 CpG per 100 bp), and most of these are methylated in normal cells (11). In contrast, around 2% of the genome contains high CpG density (~1 CpG per 10 bp) in regions referred to as CpG islands (CGIs). CGIs are located at transcription start sites (TSS) within 50–60% of gene promoters and are often unmethylated during normal development or in adult cells (12).
The function of DNA methylation varies according to the localization of the target CpG sites. When it occurs at TSS, DNA methylation is generally a repressive mark that is correlated with the inhibition of transcription initiation (13). Paradoxically, methylation of CpG sites in the gene body appears to stimulate transcription elongation (14). Recent evidence supports a novel role for DNA methylation in regulating gene splicing when it occurs at exon-intron boundaries (15). Intragenic DNA methylation may also silence alternative promoters and suppress the expression of retrotransposon elements, thus maintaining transcription efficiency and ensuring genome stability (16).
Aberrant methylation patterns are hallmarks of many cancers, including lung cancer. Various factors associated with lung cancer have been shown to alter the epigenome, including aging, chronic inflammation and carcinogen exposure, such as cigarette smoking (17). Methylation changes occur early during carcinogenesis and favor tumor progression. The cancer genome is globally hypomethylated, except for the dense methylation at CGIs that is associated with the permanent repression of tumor suppressor genes and other cancer-related genes, which promotes cancer progression (13,18). Typically, 5–10% of the CGIs are hypermethylated in cancer cells (18). Interestingly, studies have shown that these genes were often already silenced in pre-malignant cells and in cancer stem cells (CSCs), through a Polycomb-mediated repressive histone modification (tri-methylated H3K27me3) (19-21). Thus, a silent but reversible state precedes methylation, which serves as a highly stable silencing mark. This epigenetic switch may be responsible for a reduced plasticity of gene expression in adult cancer cells, which may facilitate the proliferation of abnormal cell clones. Finally, spontaneous deamination of 5-methylcytosine residues to thymine is a major cause of cytosine to thymine transition mutations and contributes to the altered genetic profile of cancer cells (22). In NSCLC, CGI hypermethylation is associated with cigarette smoking (23), histological subtype (24,25), progression (26), clinically-relevant molecular subtypes (27-29), and patient prognosis (30-33).
The biological basis of circulating cell-free DNA (cfDNA)
cfDNA is a mixture of nucleic acids originating from different tissues, hematopoietic cells being its main source in healthy subjects (34). The concentration of cfDNA greatly varies between individuals, but is generally very low—often less than 10 ng per mL of plasma (35,36). Elevated level of cfDNA is detected consequent to tissue trauma, inflammation or diseases such as cancer. In the context of cancer, circulating tumor DNA (ctDNA) represents a substantial fraction of cfDNA, ranging from <0.05% (37) to 90% (38). Many factors influence the concentration of ctDNA, most importantly tumor volume, localization and vascularization, but also a patient’s response to chemotherapies, as well as hepatic and renal clearance (39). ctDNA mostly derives from apoptosis and necrosis of the primary tumor and metastatic lesions (38). As a consequence, ctDNA consists of short fragments (180–200 bp), characteristic of the nuclease digestion that occurs during these processes. Recent studies have also reported the contribution of alternative sources of ctDNA, such as circulating tumor cells (CTCs) (40,41), and blood exosomes released by tumor cells (42).
Liquid biopsies have gained increasing attention in recent years, as they are a convenient and minimally invasive means of interrogating tumor DNA. Many studies have shown concordant genetic and epigenetic alterations between ctDNA and corresponding tumor tissue DNA (43-45). ctDNA potentially reflects the intra- and inter-tumor molecular heterogeneity in primary and metastatic cancer. Multiple tumor-specific changes are detectable; including mutations, copy number variations, chromosomal rearrangements and aberrant methylation patterns, which are the scope of this review.
Herein, we provide an overview of candidate DNA methylation-based blood biomarkers for lung cancer reported to date, describe the current technologies to analyze methylated ctDNA, and finally discuss the challenges to implementing these biomarkers for clinical management of lung cancer patients.
Methylated ctDNA as a biomarker for lung cancer
The development of biomarkers based on tumor-specific methylation is a promising approach to improve early detection of lung cancer and disease monitoring. Hereafter we review the state of research in ctDNA methylation biomarkers within the context of their potential application to address the unmet needs of clinical management of lung cancer patients (Figure 1).
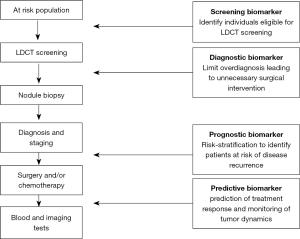
Methylated ctDNA as a screening and diagnostic biomarker
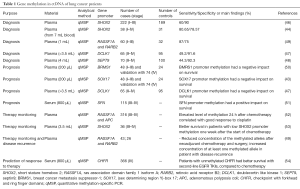
As global hypomethylation of DNA sequences and focal hypermethylation at CGIs are already observed at the early stages of tumorigenesis, methylation constitutes an attractive approach for the early detection of cancer. Several studies have reported the potential of investigating tumor-specific methylations in blood for the screening and diagnosis of lung cancer. Various gene promoters were found to be differentially methylated in ctDNA between patients with lung cancer and controls, including short stature homebox 2 (SHOX2) (44,46), doublecortin like kinase 1 (DCLK1) (47), septin9 (SEPT9) (48), ras association domain family 1 isoform A (RASSF1A) and retinoic acid receptor B2 (RARB2) (49). The sensitivity and the specificity of these candidate biomarkers are reported in Table 1. It is important to note that a large proportion of cases in these studies are late-stage cancers. Therefore, to validate a biomarker useful for the screening and/or diagnosis of lung cancer, inclusion of patients amenable to curative therapy would be necessary.
Full table
Methylated ctDNA as a prognostic biomarker
DNA methylation can be indicative of tumor aggressiveness and risk of cancer recurrence due to residual disease after surgical resection and/or chemotherapy. ctDNA has a short half-life (~2 h), and its persistence in the blood following surgery has been linked to poor prognosis (37). In the context of early stage malignancies, prognostic biomarkers are urgently needed to distinguish patients who are cured with surgery alone, from those at high risk of disease recurrence who may benefit from adjuvant chemotherapy. The prognostic significance of gene promoter ctDNA methylation has been described in several studies, although most of them evaluate late-stage cancers (Table 1). Detection of methylated breast cancer metastasis suppressor-1 (BRMS1) and (sex determining region Y)-box 17 (SOX17) in operable and advanced NSCLC, was shown to have a negative impact on survival (43,50). In contrast, SFN (14-3-3 Sigma) promoter methylation was correlated with a reduced risk of death (51). Interestingly, in addition to its diagnostic value, DCLK1 methylation was also associated with shorter survival (47).
Methylated ctDNA in the prediction and monitoring of response to therapy
One of the most important applications of liquid biopsies is the prediction and monitoring of treatment response. This minimally invasive procedure overcomes the problems of repeated tumor sampling, and allows ‘real-time’ evaluation of tumor dynamics, as ctDNA level is expected to reflect tumor burden. In addition, the abundance of ctDNA may provide an earlier indication of response to drug treatment, compared to imaging or conventional protein-based marker assays. Several studies have reported the use of tumor-specific methylation for tracking a patient’s response to therapy (Table 1). Wang and colleagues reported an elevated level of adenomatous polyposis coli (APC) and RASSF1A promoter methylation in ctDNA within 24 h after cisplatin-based therapy, consistent with chemotherapy-induced cell death (52). Methylation of SHOX2, RASSF1A and RARB2 has shown potential to monitor disease recurrence after surgery and chemotherapy (49,53). The value of methylated ctDNA to predict response to therapy has also been investigated. Patients with unmethylated checkpoint with forkhead and ring finger domains (CHFR) promoter survived longer when receiving EGFR tyrosine kinase inhibitors as second-line treatment, compared to conventional chemotherapy (54). Finally, with the recent demonstration that combined epigenetic therapy has efficacy in lung cancer patients (55), future applications of methylated ctDNA for monitoring the activity of demethylating agents will soon come to the forefront.
Technical aspects associated with analysis of methylated ctDNA
The choice of a specific method for analysis of ctDNA methylation depends essentially on the research question. From studies of a few candidate CGI-associated gene promoters, to analysis of thousands of CpG sites by microarrays, and more recently to whole genome bisulfite sequencing (WGBS), the scope of platform choices for measurement of biomarkers in ctDNA continues to expand. Still, quality and amount of ctDNA—the methylated fraction being even smaller—remain the major limitations that preclude a comprehensive analysis of methylation in ctDNA. Therefore, before giving an overview of the technologies applicable to ctDNA methylation analysis, we will discuss some critical pre-analytical considerations to optimize the cfDNA yield and introduce the general principles associated with distinguishing unmethylated from methylated cytosines.
Pre-analytical parameters
Both plasma and serum have been used to isolate cfDNA, even though it is still debated whether plasma is a better source for cfDNA. When using plasma, EDTA anticoagulant is usually the anticoagulant of choice, even though the type of blood collection tubes is not always detailed. Higher quantity of cfDNA has been reported in serum (36,56). However, several studies have demonstrated that genomic DNA released by the lysis of blood cells during serum collection adversely affects the quality of cfDNA (35). Specifically, DNA released from leukocytes increases in serum within 4 hs, compared to no change in plasma up to 8hs after collection (36). Therefore, same day processing of blood sample is highly recommended, with an immediate serum separation. The starting volume of blood necessary to obtain a sufficient amount of cfDNA will vary depending on the downstream analysis method.
A technical concern arising from the fragmentation of cfDNA is that classical DNA purification methods developed for genomic DNA are not efficient at isolating cfDNA, leading to further DNA loss (57). To overcome this limitation, a broad range of kits specifically developed for cfDNA isolation has become commercially available. A series of publications have compared the performance of several of these extraction kits (57-59). These kits recover low molecular weight nucleic acids with different extraction efficiency and reproducibility. Variations in output DNA may also result from different DNA quantification methods, which makes it difficult to define a ‘gold standard method’ for cfDNA extraction.
Methods to distinguish unmethylated from methylated cytosines
Over the past several years, numerous approaches have been developed that enable the discrimination of methylated from unmethylated CpGs. Treatment of DNA with bisulfite has become the most widely used method, as it can be coupled with a variety of detection technologies including microarrays, next-generation sequencing (NGS) and PCR-based assays. Bisulfite-based methods use a chemical reaction that rapidly deaminates unmethylated cytosines to uracils, whereas methylated cytosines are only slowly converted. Besides transforming an epigenetic mark into point mutation, bisulfite treatment induces random DNA breaks, resulting in short single-stranded DNA fragments. Bisulfite conversion is a harsh process that will dramatically affect DNA recovery, especially for cfDNA which is already highly fragmented and present at very low concentrations. In the past few years, several bisulfite conversion kits with improved recovery of sparse amounts of cfDNA have become commercially available. Increased recovery is mostly attained by reducing the incubation time of DNA with bisulfite conversion reagent. Comparisons of the main features of some of these kits with regards to DNA recovery, DNA fragmentation and conversion efficiency were recently published (60,61). Again, due to the lack of standardization of the quantification methods, DNA yield is inconsistent across studies. Several approaches can be used to assess the amount of converted DNA, such as a UV spectrophotometer (RNA setting on Nanodrop), a fluorimeter (Qubit ssDNA assay kit, Invitrogen), or a qPCR-based assay. For the latter, amplification of a cytosine free sequence will allow the quantification of total amount DNA, including converted and unconverted DNA. Only one of the strands will be amplified and therefore the amount needs to be corrected by a factor 2. An important consideration is that the basis for quantification is different between spectrophotometric and qPCR methods. While spectrophotometric methods assess total DNA, qPCR assays only target amplifiable DNA. In addition, yield depends on amplicon efficiency and the degree of random fragmentation of the amplification template during bisulfite conversion, which makes the DNA recovery measured by qPCR generally lower than that assessed by spectrophotometric methods.
Despite the many advantages of bisulfite conversion, the technique has several drawbacks. Besides substantial DNA degradation, the reduction in sequence complexity resulting from conversion constrains primer design for subsequent PCR amplification and complicates alignment of sequencing reads to a reference genome. In addition, bisulfite treatment does not allow for the distinction between 5-methylcytosines and 5-hydroxymethylcitosines, another base modification, as both are resistant to deamination (62). Therefore, 5-hydroxymethylcytosines may lead to false-positives in downstream analyses. Finally, even though conversion efficiencies of the different commercially available kits are very high (98.7–99%), failure to convert unmethylated cytosines to uracils or inappropriate conversion of methylated cytosines to uracils, will lead to false-positive and false-negative results, respectively, and assay inconsistencies (63).
Bisulfite-independent methods also exist that discriminate methylated CpGs from unmethylated ones. Methylation-Sensitive Restriction Enzymes (MSREs) that solely cut unmethylated DNA have been adapted for the study of cfDNA methylation in serum and plasma from patients with lung (64) or other cancers (65,66). With this method, the rate of false-positives due to incomplete DNA digestion can be prevented using a combination of MSREs. Finally, affinity-based methods such as Methyl-CpG Binding Domain MBD2 proteins (MBD, also termed Methyl Cap) have been developed that take advantage of the interaction of these proteins with methylated sequences and rely on separation of DNA by salt gradients according to CpG methylation density. MBD is applicable to cfDNA studies, even though the very low recovery of methylated DNA (7% to 14.9% with a modified protocol) remains a major limitation for downstream analysis (36,67,68). The method also demonstrates a bias toward CGIs.
Technologies for genome-wide DNA methylation analysis
In recent years, the development of high-throughput techniques to measure DNA methylation has revolutionized our understanding of the role of this epigenetic mark and allowed the discovery of novel gene regulatory mechanisms. Genome-wide evaluation of DNA methylation is generally based on either microarray-hybridization or NGS.
Array-based hybridization for measurement of methylated cytosine
Genome-wide screening of DNA methylation was initially conducted by microarrays featuring an increasing number of targeted CpGs, mostly focused on CGIs. Microarrays hybridization is subsequent to digestion of DNA with MSREs, affinity purification or bisulfite conversion. The Infinium Human Methylation 450 Bead Chip Array (Illumina) is one of the most advanced array-based approaches for methylation profiling, and has been described extensively (69). This array platform can detect the methylation status of 485,000 individual CpGs encompassing 99% of reference genes and 96% of CGIs. It also covers CGI shores (regions about 2 kb upstream of CGIs), miRNA promoters and coding regions. Other microarray platforms are also available to achieve global methylation profiling (70).
The ease of array-based methodologies is ideal for a first pass methylation profiling. However, the large amount of input DNA required (500 ng–1 µg) precludes the widespread assessment of DNA methylation in liquid biopsies. Still, encouraging results from a recent pilot study showed that genome-wide profiles of DNA methylation in ctDNA are consistent with corresponding tumor tissues (71).
NGS
NGS technologies are now more frequently used, as they provide a higher genomic coverage compared to microarrays and a single nucleotide resolution. Protocols have been adapted to allow the methylation analysis of bisulfite converted DNA. Using WGBS, it is theoretically possible to determine the methylation status of the 28 million CpGs contained in the human genome—even though in practice some sites are poorly covered (69). WGBS can be performed with ~30 ng of DNA, and in some cases as little as 125 pg, which makes it an applicable method for analysis of cfDNA. Hence, several recent studies have used WGBS to analyze the methylomes of plasma cfDNA (72-74). Since only a small fraction of the genome is differentially methylated in cancer, enrichment methods can be coupled to bisulfite sequencing to limit the cost associated with WGBS. This also results in increased coverage of specific CpGs. Unfortunately, common enrichment techniques such as reduced representation bisulfite sequencing (RRBS) require high amount of high-quality DNA and are therefore not easily applicable to ctDNA. Finally, online software tools have been designed to specifically analyze bisulfite sequencing data. This step is not straightforward, as converted DNA is difficult to assemble and compare to a reference genome.
Gene-specific DNA methylation assays
The aim of the screening methods described above is to identify methylation differences that will ultimately be confirmed in a locus-specific assay to validate the candidate biomarker and evaluate its clinical utility. So far, studies have essentially focused on the methylation status of CGIs. The most commonly used quantitative approaches include pyrosequencing and PCR assays, both these methods rely on bisulfite-treated DNA.
Pyrosequencing
Pyrosequencing provides real-time quantitative data on the methylation status of multiple CpGs within an amplicon. Distinct from Sanger sequencing, pyrosequencing uses a sequencing-by-synthesis system that relies on the luminometric detection of pyrophosphates released as nucleotides are incorporated into the extended strands. Briefly, CpG-bearing sequences of interest are amplified by PCR, using primers designed specifically to anneal bisulfite-modified DNA. One of the primers is biotinylated, which allows the isolation of the biotin-labeled single strand using streptavidin beads. The DNA template is then added to a pyrosequencing plate containing the pyrosequencing primer, and the reaction is analyzed in a pyrosequencer (75,76). Besides the specific equipment required, one of the main limitations of this method for use with clinical specimens is the length of the sequence read, which decreases the quantitative accuracy of the methylation status of CpG sites distant from the 3’ end of the primer.
Bisulfite-based PCR assays
A number of methods have been developed to allow for ultrasensitive detection of ctDNA. From conventional to quantitative methylation-specific PCR (qMSP, respectively), and more recently digital PCR-based approaches, the current techniques have very high analytical sensitivity to detect rare methylated alleles in a background of unmethylated DNA. Due to the rapid evolution of the technology, PCR assays are not only applicable to patients with high tumor burden, but also to those with early-stage disease.
A critical step common to all these methods is the design of methylation-specific primers and probe. Because bisulfite-treated sequences are highly redundant, primers are more susceptible to bind multiple DNA templates. To reduce false-priming events, a variety of design guidelines and software programs are available (60,77). The different platforms allow users to customize various parameters including primers and probe length, amplicon size, melting temperature, secondary structures, and number and position of CpG sites contained in the sequence.
Methylation-specific PCR (MSP)
MSP is a qualitative assay allowing for the discrimination of the methylation status of a region of interest (78). Two primer sets are used—one binding to the methylated allele and the other to the unmethylated allele—and both are designed to anneal to the same sequence. MSP is generally associated with high false-priming events when high numbers of PCR cycles are used. In addition, a gel electrophoresis is required to detect PCR amplification products, which limits the implementation of MSP in the clinic.
qMSP
The first qMSP assay was named MethyLight, which combined MSP with a Taqman probe (79). In order to determine the methylation status of a region of interest, either the primers, or the probe, or both are designed to overlap CpGs. To maximize specificity towards the methylated allele, primers should contain as many CpGs as possible preferentially close to the 3’ end, and the probe includes three to five CpGs. Only one permutation can be detected at a time, and is generally the fully methylated variant. MethyLight requires a control reaction for input DNA. To amplify only the converted sequence, neither the primers nor the probe contain CpGs. If neither of them overlaps cytosines, then the total amount of DNA is measured, regardless of bisulfite conversion. A standard curve is generated using a serial dilution of a converted fully methylated DNA as a reference. The relative abundance of methylation is then obtained by calculating the percentage of methylated ratio (PMR).
MethyLight overcomes some of the major limitations of MSP, including the real-time detection of PCR amplification and the non-specific priming events, which are reduced by the probe hybridization. The assay is 10-fold more sensitive than MSP. MethyLight enables the detection of a methylated allele in a 10,000-fold excess of unmethylated alleles, which makes it suitable for the analysis of ctDNA. MethyLight can also be multiplexed to detect several regions of interest in one reaction mixture, using different fluorescent-labeled probes.
Digital MethyLight (dMethyLight)
dMethyLight is a compartmentalized MethyLight that allows for the detection and counting of a single methylated allele (80). The DNA sample is diluted and distributed over a large number of reaction wells. In theory, each well contains either one (positive well) or no template molecules, allowing the absolute quantification of the methylated locus so that no calibration is required. Unmethylated DNA is sequestered into negative wells, while the ratio methylated-to-unmethylated is kept high in positive wells. The competition for methylation specific-primers and probe is also reduced, which is an advantage for the rare methylation events in liquid biopsies. The method provides a better detection than MethyLight, and multiplexed dMethyLight assays can also be implemented. However, the sensitivity and the reproducibility of dMethyLight are governed by the number of wells, making it time-consuming and reagent intensive.
Droplet digital MethyLight (ddMethyLight)
ddMethyLight is based on the same compartmentalization principle as dMethyLight, but at a larger scale. Instead of a hundred wells, up to 20,000 droplets are generated in an emulsion. Each single droplet is detected as either positive or negative, depending on the presence of the methylated allele. ddMethyLight has a very high sensitivity (0.001%) and has been shown to be 25-fold more sensitive than MethyLight (81). The small reaction volume (20 µL) makes it more cost-effective than dMethyLight. The fragmented pattern of cfDNA is also an advantage as it improves the performance of ddPCR. However, this technology is low throughput, with only duplex reactions possible.
Other highly sensitive technologies have been developed in recent years, such as Methyl-BEAMing (0.01% sensitivity for rare events) (82) or RainDrop digital PCR (0.0005% sensitivity for rare events) (83), which may in the future offer unique analytical advantages for early-stage biomarkers in liquid biopsies.
Current challenges and future perspectives
As a noninvasive biopsy and potential surrogate for the entire tumor genome, ctDNA is a promising biomarker for early cancer detection, risk stratification, monitoring of tumor dynamics, and prediction of response to therapy. However, a number of challenges still remain before liquid biopsies are fully implemented in the clinical management of cancer patients. The developments of reproducible, sensitive and specific assays, and the demonstration of biomarker performance in prospectively-collected target populations, are the two key hurdles that need to be surpassed before liquid biopsies can become a routine part of patient care.
Despite technological advances in recent years, sensitivity and specificity of currently available assays to detect molecular alterations in ctDNA, are still a major issue of concern for the discovery and the development of biomarkers. Current technologies were essentially adapted to examine methylation in solid biopsies, and most DNA methylation biomarkers are still initially discovered in tumor tissues before being evaluated in blood samples. However, as the sensitivity of analytical tools increases, detection of very low levels of methylated ctDNA from early stage or minimal residual disease may become possible. The development of reliable and reproducible biomarkers will also require standardization of pre-analytical methods. Robust sample processing, from blood collection to ctDNA extraction and preparation for methylation analyses, is essential to improve data quality and accuracy so that results can be compared across studies. Furthermore, validations should be conducted under clinical laboratory improvement amendments (CLIA)-certified laboratory standards (84). CLIA certification is a necessary step for translating a companion diagnostic into a clinical test. Compliance with CLIA federal regulatory standards ensures acceptable test performance characteristics, including accuracy, precision, reportable range, reference interval, analytical sensitivity, and analytical specificity.
As DNA methylation is often considered an early event in carcinogenesis, tumor-specific methylation has a great potential to be used as a screening and/or diagnostic tool in conjunction with LCDT. It could help prioritize individuals to LDCT screening, and limit overdiagnosis that leads to unnecessary surgical procedures. Funding opportunities focused on the integration of imaging and biomarkers are being put forward to stimulate much needed synergy in this area of research. Among them, the National Cancer Institute, NIH is supporting projects that combine established imaging techniques and investigational biomarkers to improve early disease detection (85).
Most candidate biomarkers are discovered in small retrospective cohort or case-control studies, and few are validated in independent studies. To verify that a candidate biomarker performs well for the clinical context and target population for which it was originally developed, it is necessary to validate its clinical utility in statistically well-powered prospective trials. Given that environmental and lifestyle factors (i.e., smoking) influence methylation, the control population should be selected accordingly. Due to financial constraints and/or limited availability of the targeted population, all biomarkers cannot be tested in large trials. Use of blood samples collected as part of a completed prospective trial, or initiatives such as the ‘Biomarker, Imaging and Quality of Life Studies Funding Program’ encouraging the integration of candidate biomarkers to large prospective trials could be an alternative to overcome these obstacles (86).
The first blood test that interrogates ctDNA methylation for cancer screening was approved by the US Food and Drug Administration (FDA) in April 2016 (87). This assay, marketed as Epi proColon (Epigenomics, Inc.), is based on SEPT9 promoter methylation and has been approved for screening and diagnosis of colorectal cancer (88). Proof-of-concept clinical trials of ctDNA methylation as an early indicator of lung cancer (89), prognostic marker (90), or surrogate for DNA demethylating agent activity (91) are currently ongoing. Owing to the wide variety of potential clinical applications of methylated ctDNA and the rapidly advancing technologies, we will soon see a surge in the number of candidate biomarkers. These discoveries should in turn translate into companion diagnostics that address the unmet needs of lung cancer patients.
Acknowledgements
The authors thank Dr. Curtis C. Harris for critical reading of the manuscript and thoughtful suggestions.
Funding: The authors are supported by the Intramural Research Program of the National Cancer Institute, NIH (Z99 CA999999).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin 2015;65:5-29. [Crossref] [PubMed]
- National Lung Screening Trial Research Team, Aberle DR, Adams AM, et al. Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med 2011;365:395-409. [Crossref] [PubMed]
- Patz EF Jr, Pinsky P, Gatsonis C, et al. Overdiagnosis in low-dose computed tomography screening for lung cancer. JAMA Intern Med 2014;174:269-74. [Crossref] [PubMed]
- Katki HA, Kovalchik SA, Berg CD, et al. Development and Validation of Risk Models to Select Ever-Smokers for CT Lung Cancer Screening. JAMA 2016;315:2300-11. [Crossref] [PubMed]
- Aerts HJ, Velazquez ER, Leijenaar RT, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun 2014;5:4006. [PubMed]
- Wang H, Schabath MB, Liu Y, et al. Semiquantitative Computed Tomography Characteristics for Lung Adenocarcinoma and Their Association With Lung Cancer Survival. Clin Lung Cancer 2015;16:e141-63. [Crossref] [PubMed]
- Maldonado F, Duan F, Raghunath SM, et al. Noninvasive Computed Tomography-based Risk Stratification of Lung Adenocarcinomas in the National Lung Screening Trial. Am J Respir Crit Care Med 2015;192:737-44. [Crossref] [PubMed]
- Chansky K, Sculier JP, Crowley JJ, et al. The International Association for the Study of Lung Cancer Staging Project: prognostic factors and pathologic TNM stage in surgically managed non-small cell lung cancer. J Thorac Oncol 2009;4:792-801. [Crossref] [PubMed]
- Sharma S, Kelly TK, Jones PA. Epigenetics in cancer. Carcinogenesis 2010;31:27-36. [Crossref] [PubMed]
- Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat Rev Genet 2009;10:295-304. [Crossref] [PubMed]
- Baylin SB. The cancer epigenome: its origins, contributions to tumorigenesis, and translational implications. Proc Am Thorac Soc 2012;9:64-5. [Crossref] [PubMed]
- Baylin SB, Jones PA. Epigenetic Determinants of Cancer. Cold Spring Harb Perspect Biol 2016;8:a019505. [Crossref] [PubMed]
- Bird A. DNA methylation patterns and epigenetic memory. Genes Dev 2002;16:6-21. [Crossref] [PubMed]
- Jones PA. The DNA methylation paradox. Trends Genet 1999;15:34-7. [Crossref] [PubMed]
- Laurent L, Wong E, Li G, et al. Dynamic changes in the human methylome during differentiation. Genome Res 2010;20:320-31. [Crossref] [PubMed]
- Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet 2012;13:484-92. [Crossref] [PubMed]
- Liu F, Killian JK, Yang M, et al. Epigenomic alterations and gene expression profiles in respiratory epithelia exposed to cigarette smoke condensate. Oncogene 2010;29:3650-64. [Crossref] [PubMed]
- Baylin SB, Jones PA. A decade of exploring the cancer epigenome - biological and translational implications. Nat Rev Cancer 2011;11:726-34. [Crossref] [PubMed]
- Easwaran H, Johnstone SE, Van Neste L, et al. A DNA hypermethylation module for the stem/progenitor cell signature of cancer. Genome Res 2012;22:837-49. [Crossref] [PubMed]
- Ohm JE, McGarvey KM, Yu X, et al. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat Genet 2007;39:237-42. [Crossref] [PubMed]
- Schlesinger Y, Straussman R, Keshet I, et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat Genet 2007;39:232-6. [Crossref] [PubMed]
- Rideout WM 3rd, Coetzee GA, Olumi AF, et al. 5-Methylcytosine as an endogenous mutagen in the human LDL receptor and p53 genes. Science 1990;249:1288-90. [Crossref] [PubMed]
- Sato T, Arai E, Kohno T, et al. Epigenetic clustering of lung adenocarcinomas based on DNA methylation profiles in adjacent lung tissue: Its correlation with smoking history and chronic obstructive pulmonary disease. Int J Cancer 2014;135:319-34. [Crossref] [PubMed]
- Hawes SE, Stern JE, Feng Q, et al. DNA hypermethylation of tumors from non-small cell lung cancer (NSCLC) patients is associated with gender and histologic type. Lung Cancer 2010;69:172-9. [Crossref] [PubMed]
- Carvalho RH, Hou J, Haberle V, et al. Genomewide DNA methylation analysis identifies novel methylated genes in non-small-cell lung carcinomas. J Thorac Oncol 2013;8:562-73. [Crossref] [PubMed]
- Selamat SA, Galler JS, Joshi AD, et al. DNA methylation changes in atypical adenomatous hyperplasia, adenocarcinoma in situ, and lung adenocarcinoma. PLoS One 2011;6:e21443. [Crossref] [PubMed]
- Walter K, Holcomb T, Januario T, et al. DNA methylation profiling defines clinically relevant biological subsets of non-small cell lung cancer. Clin Cancer Res 2012;18:2360-73. [Crossref] [PubMed]
- Shinjo K, Okamoto Y, An B, et al. Integrated analysis of genetic and epigenetic alterations reveals CpG island methylator phenotype associated with distinct clinical characters of lung adenocarcinoma. Carcinogenesis 2012;33:1277-85. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014;511:543-50. [Crossref] [PubMed]
- Brock MV, Hooker CM, Ota-Machida E, et al. DNA methylation markers and early recurrence in stage I lung cancer. N Engl J Med 2008;358:1118-28. [Crossref] [PubMed]
- Sato T, Arai E, Kohno T, et al. DNA methylation profiles at precancerous stages associated with recurrence of lung adenocarcinoma. PLoS One 2013;8:e59444. [Crossref] [PubMed]
- Sandoval J, Mendez-Gonzalez J, Nadal E, et al. A prognostic DNA methylation signature for stage I non-small-cell lung cancer. J Clin Oncol 2013;31:4140-7. [Crossref] [PubMed]
- Robles AI, Arai E, Mathé EA, et al. An Integrated Prognostic Classifier for Stage I Lung Adenocarcinoma Based on mRNA, microRNA, and DNA Methylation Biomarkers. J Thorac Oncol 2015;10:1037-48. [Crossref] [PubMed]
- Lui YY, Chik KW, Chiu RW, et al. Predominant hematopoietic origin of cell-free DNA in plasma and serum after sex-mismatched bone marrow transplantation. Clin Chem 2002;48:421-7. [PubMed]
- El Messaoudi S, Rolet F, Mouliere F, et al. Circulating cell free DNA: Preanalytical considerations. Clin Chim Acta 2013;424:222-30. [Crossref] [PubMed]
- Warton K, Lin V, Navin T, et al. Methylation-capture and Next-Generation Sequencing of free circulating DNA from human plasma. BMC Genomics 2014;15:476. [Crossref] [PubMed]
- Diehl F, Schmidt K, Choti MA, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med 2008;14:985-90. [Crossref] [PubMed]
- Jahr S, Hentze H, Englisch S, et al. DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res 2001;61:1659-65. [PubMed]
- Fleischhacker M, Schmidt B. Circulating nucleic acids (CNAs) and cancer--a survey. Biochim Biophys Acta 2007;1775:181-232.
- Schwarzenbach H, Alix-Panabières C, Müller I, et al. Cell-free tumor DNA in blood plasma as a marker for circulating tumor cells in prostate cancer. Clin Cancer Res 2009;15:1032-8. [Crossref] [PubMed]
- Shaw JA, Brown J, Coombes RC, et al. Circulating tumor cells and plasma DNA analysis in patients with indeterminate early or metastatic breast cancer. Biomark Med 2011;5:87-91. [Crossref] [PubMed]
- Thakur BK, Zhang H, Becker A, et al. Double-stranded DNA in exosomes: a novel biomarker in cancer detection. Cell Res 2014;24:766-9. [Crossref] [PubMed]
- Balgkouranidou I, Chimonidou M, Milaki G, et al. SOX17 promoter methylation in plasma circulating tumor DNA of patients with non-small cell lung cancer. Clin Chem Lab Med 2016;54:1385-93. [Crossref] [PubMed]
- Konecny M, Markus J, Waczulikova I, et al. The value of SHOX2 methylation test in peripheral blood samples used for the differential diagnosis of lung cancer and other lung disorders. Neoplasma 2016;63:246-53. [PubMed]
- Zhang Y, Wang R, Song H, et al. Methylation of multiple genes as a candidate biomarker in non-small cell lung cancer. Cancer Lett 2011;303:21-8. [Crossref] [PubMed]
- Kneip C, Schmidt B, Seegebarth A, et al. SHOX2 DNA methylation is a biomarker for the diagnosis of lung cancer in plasma. J Thorac Oncol 2011;6:1632-8. [Crossref] [PubMed]
- Powrózek T, Krawczyk P, Nicoś M, et al. Methylation of the DCLK1 promoter region in circulating free DNA and its prognostic value in lung cancer patients. Clin Transl Oncol 2016;18:398-404. [Crossref] [PubMed]
- Powrózek T, Krawczyk P, Kucharczyk T, et al. Septin 9 promoter region methylation in free circulating DNA-potential role in noninvasive diagnosis of lung cancer: preliminary report. Med Oncol 2014;31:917. [Crossref] [PubMed]
- Ponomaryova AA, Rykova EY, Cherdyntseva NV, et al. Potentialities of aberrantly methylated circulating DNA for diagnostics and post-treatment follow-up of lung cancer patients. Lung Cancer 2013;81:397-403. [Crossref] [PubMed]
- Balgkouranidou I, Chimonidou M, Milaki G, et al. Breast cancer metastasis suppressor-1 promoter methylation in cell-free DNA provides prognostic information in non-small cell lung cancer. Br J Cancer 2014;110:2054-62. [Crossref] [PubMed]
- Ramirez JL, Rosell R, Taron M, et al. 14-3-3sigma methylation in pretreatment serum circulating DNA of cisplatin-plus-gemcitabine-treated advanced non-small-cell lung cancer patients predicts survival: The Spanish Lung Cancer Group. J Clin Oncol 2005;23:9105-12. [Crossref] [PubMed]
- Wang H, Zhang B, Chen D, et al. Real-time monitoring efficiency and toxicity of chemotherapy in patients with advanced lung cancer. Clin Epigenetics 2015;7:119. [Crossref] [PubMed]
- Schmidt B, Beyer J, Dietrich D, et al. Quantification of cell-free mSHOX2 Plasma DNA for therapy monitoring in advanced stage non-small cell (NSCLC) and small-cell lung cancer (SCLC) patients. PLoS One 2015;10:e0118195. [Crossref] [PubMed]
- Salazar F, Molina MA, Sanchez-Ronco M, et al. First-line therapy and methylation status of CHFR in serum influence outcome to chemotherapy versus EGFR tyrosine kinase inhibitors as second-line therapy in stage IV non-small-cell lung cancer patients. Lung Cancer 2011;72:84-91. [Crossref] [PubMed]
- Juergens RA, Wrangle J, Vendetti FP, et al. Combination epigenetic therapy has efficacy in patients with refractory advanced non-small cell lung cancer. Cancer Discov 2011;1:598-607. [Crossref] [PubMed]
- Lee TH, Montalvo L, Chrebtow V, et al. Quantitation of genomic DNA in plasma and serum samples: higher concentrations of genomic DNA found in serum than in plasma. Transfusion 2001;41:276-82. [Crossref] [PubMed]
- Devonshire AS, Whale AS, Gutteridge A, et al. Towards standardisation of cell-free DNA measurement in plasma: controls for extraction efficiency, fragment size bias and quantification. Anal Bioanal Chem 2014;406:6499-512. [Crossref] [PubMed]
- Mauger F, Dulary C, Daviaud C, et al. Comprehensive evaluation of methods to isolate, quantify, and characterize circulating cell-free DNA from small volumes of plasma. Anal Bioanal Chem 2015;407:6873-8. [Crossref] [PubMed]
- Page K, Guttery DS, Zahra N, et al. Influence of plasma processing on recovery and analysis of circulating nucleic acids. PLoS One 2013;8:e77963. [Crossref] [PubMed]
- Hernández HG, Tse MY, Pang SC, et al. Optimizing methodologies for PCR-based DNA methylation analysis. Biotechniques 2013;55:181-97. [Crossref] [PubMed]
- Holmes EE, Jung M, Meller S, et al. Performance evaluation of kits for bisulfite-conversion of DNA from tissues, cell lines, FFPE tissues, aspirates, lavages, effusions, plasma, serum, and urine. PLoS One 2014;9:e93933. [Crossref] [PubMed]
- Hayatsu H, Shiragami M. Reaction of bisulfite with the 5-hydroxymethyl group in pyrimidines and in phage DNAs. Biochemistry 1979;18:632-7. [Crossref] [PubMed]
- Genereux DP, Johnson WC, Burden AF, et al. Errors in the bisulfite conversion of DNA: modulating inappropriate- and failed-conversion frequencies. Nucleic Acids Res 2008;36:e150. [Crossref] [PubMed]
- Wielscher M, Vierlinger K, Kegler U, et al. Diagnostic Performance of Plasma DNA Methylation Profiles in Lung Cancer, Pulmonary Fibrosis and COPD. EBioMedicine 2015;2:929-36. [Crossref] [PubMed]
- Ellinger J, Bastian PJ, Haan KI, et al. Noncancerous PTGS2 DNA fragments of apoptotic origin in sera of prostate cancer patients qualify as diagnostic and prognostic indicators. Int J Cancer 2008;122:138-43. [Crossref] [PubMed]
- Hauser S, Zahalka T, Fechner G, et al. Serum DNA hypermethylation in patients with kidney cancer: results of a prospective study. Anticancer Res 2013;33:4651-6. [PubMed]
- Lan X, Adams C, Landers M, et al. High resolution detection and analysis of CpG dinucleotides methylation using MBD-Seq technology. PLoS One 2011;6:e22226. [Crossref] [PubMed]
- Nair SS, Coolen MW, Stirzaker C, et al. Comparison of methyl-DNA immunoprecipitation (MeDIP) and methyl-CpG binding domain (MBD) protein capture for genome-wide DNA methylation analysis reveal CpG sequence coverage bias. Epigenetics 2011;6:34-44. [Crossref] [PubMed]
- Kurdyukov S, Bullock M. DNA Methylation Analysis: Choosing the Right Method. Biology (Basel) 2016;5:E3. [Crossref] [PubMed]
- Huang YW, Huang TH, Wang LS. Profiling DNA methylomes from microarray to genome-scale sequencing. Technol Cancer Res Treat 2010;9:139-47. [Crossref] [PubMed]
- Zhai R, Zhao Y, Su L, et al. Genome-wide DNA methylation profiling of cell-free serum DNA in esophageal adenocarcinoma and Barrett esophagus. Neoplasia 2012;14:29-33. [Crossref] [PubMed]
- Chan KC, Jiang P, Chan CW, et al. Noninvasive detection of cancer-associated genome-wide hypomethylation and copy number aberrations by plasma DNA bisulfite sequencing. Proc Natl Acad Sci U S A 2013;110:18761-8. [Crossref] [PubMed]
- Lun FM, Chiu RW, Sun K, et al. Noninvasive prenatal methylomic analysis by genomewide bisulfite sequencing of maternal plasma DNA. Clin Chem 2013;59:1583-94. [Crossref] [PubMed]
- Legendre C, Gooden GC, Johnson K, et al. Whole-genome bisulfite sequencing of cell-free DNA identifies signature associated with metastatic breast cancer. Clin Epigenetics 2015;7:100. [Crossref] [PubMed]
- Delaney C, Garg SK, Yung R. Analysis of DNA Methylation by Pyrosequencing. Methods Mol Biol 2015;1343:249-64. [Crossref] [PubMed]
- Dupont JM, Tost J, Jammes H, et al. De novo quantitative bisulfite sequencing using the pyrosequencing technology. Anal Biochem 2004;333:119-27. [Crossref] [PubMed]
- Ostrow KL, Hoque MO, Loyo M, et al. Molecular analysis of plasma DNA for the early detection of lung cancer by quantitative methylation-specific PCR. Clin Cancer Res 2010;16:3463-72. [Crossref] [PubMed]
- Herman JG, Graff JR, Myöhänen S, et al. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci U S A 1996;93:9821-6. [Crossref] [PubMed]
- Eads CA, Danenberg KD, Kawakami K, et al. MethyLight: a high-throughput assay to measure DNA methylation. Nucleic Acids Res 2000;28:E32. [Crossref] [PubMed]
- Weisenberger DJ, Trinh BN, Campan M, et al. DNA methylation analysis by digital bisulfite genomic sequencing and digital MethyLight. Nucleic Acids Res 2008;36:4689-98. [Crossref] [PubMed]
- Yu M, Carter KT, Makar KW, et al. MethyLight droplet digital PCR for detection and absolute quantification of infrequently methylated alleles. Epigenetics 2015;10:803-9. [Crossref] [PubMed]
- Li M, Chen WD, Papadopoulos N, et al. Sensitive digital quantification of DNA methylation in clinical samples. Nat Biotechnol 2009;27:858-63. [Crossref] [PubMed]
- Day E, Dear PH, McCaughan F. Digital PCR strategies in the development and analysis of molecular biomarkers for personalized medicine. Methods 2013;59:101-7. [Crossref] [PubMed]
- Clinical Laboratory Improvement Amendments (CLIA). Available online: http://wwwn.cdc.gov/clia/
- Imaging and Biomarkers for Early Detection of Aggressive Cancer. Available online: http://grants.nih.gov/grants/guide/pa-files/PAR-16-089.html
- Biomarker, Imaging and Quality of Life Studies Funding Program (BIQSFP). Available online: http://www.cancer.gov/about-nci/organization/ccct/funding/biqsfp
- Epi proColon Approval Order. Available online: http://www.accessdata.fda.gov/cdrh_docs/pdf13/P130001A.pdf
- Payne SR. From discovery to the clinic: the novel DNA methylation biomarker (m)SEPT9 for the detection of colorectal cancer in blood. Epigenomics 2010;2:575-85. [Crossref] [PubMed]
- Characterization of Methylation Patterns in Lung Cancer. Available online: https://clinicaltrials.gov/ct2/show/NCT02373917
- Biomarkers in Tissue and Blood Samples From Patients With Early-Stage Non-Small Cell Lung Cancer. Available online: https://clinicaltrials.gov/ct2/show/NCT01139944
- Residual Hypermethylation in Early Stage Non-Small Cell Lung Cancer (NSCLC) As Part of Adjuvant Therapy and Preventive Strategy. Available online: https://clinicaltrials.gov/ct2/show/NCT01209520


