Circulating tumor cells versus circulating tumor DNA in lung cancer—which one will win?
Introduction
Lung cancer is the leading cause of cancer-related mortality among men and women worldwide, with more than 1.8 million estimated new cases each year (1). Despite advances in biomedical research and improvements in both the diagnostic tools and therapeutic options that have become available over the past few decades, lung cancer still has a 5-year overall survival (OS) rate of 18% for all stages (2). The main reason for this poor outcome for this cancer type is late diagnosis: a high percentage of the patients are diagnosed at advanced stages when curative surgery is no longer possible. The clinical management of lung cancer in advanced stages is also changing; better understanding and descriptions of the molecular abnormalities present in lung cancer have opened up new therapeutic options in specific disease subsets.
Lung cancer: driver alterations, predictive biomarkers, and intratumor heterogeneity
The development of a new generation of molecular techniques has led to substantial advances in the knowledge of cancer genomes, and specifically in lung cancer, facilitating the discovery of oncogenic-driver mutations/alterations that cause aberrant signaling and proliferation in certain tumor subsets. These findings have allowed the development of new treatment strategies based on molecular targets and thus have reshaped tumor classification from classical histological methods towards molecular pathology approaches. Indeed, non-small cell lung cancer (NSCLC) is one of the most genomically diverse tumor types and there is a wide variety of molecularly-defined patient subsets characterized by specific driver-mutations, involving different genes such as epidermal growth factor receptor (EGFR), anaplastic lymphoma kinase (ALK), or V-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog (KRAS) (3-7) among others.
The identification of driver mutations located in the tyrosine kinase domain of EGFR as the primary oncogenic event in a subset of lung adenocarcinomas (8-13) led to the development of specific tyrosine-kinase inhibitors (TKIs) for this receptor, resulting in a dramatic change in the treatment of advanced NSCLC patients harboring EGFR mutations. The use of EGFR-TKIs has produced a particularly large increase in progression-free survival (PFS) with a negligible toxicity profile as well as an increase in OS to more than 24 months (11,14-18). Unfortunately, the effect of TKIs is limited over time because of the emergence of drug resistance. A number of molecular mechanisms underlying acquired resistance to EGFR-TKIs have been reported, including the secondary EGFR Thr790Met (T790M) mutation found in around 50% of patients, loss of the EGFR mutant allele, MET amplification, hepatocyte growth factor (HGF) overexpression, phosphatase and tensin homolog (PTEN) downregulation, epithelial-mesenchymal transition (EMT), BRAF mutations, and other mechanisms (19-22). Resistance is frequently related to the emergence of a clone thought to be initially present at a low percentage in the tumor, thus emphasizing the role of intratumour heterogeneity as one way of explaining resistance mechanisms to targeted agents (23,24).
Other driver mutations in lung cancer have also been targeted by these new drugs but, again, the emergence of resistance is a common event in patients treated with targeted-therapies (25,26). In addition, metastases are responsible for roughly 90% of cancer-associated patient mortality, through largely unknown mechanisms. Therefore, future studies must aim to directly analyze metastatic cells in order to better understand the mechanisms of cancer spread (27). However, metastasis biopsy is an invasive procedure limited to certain locations, and additionally, recent work has shown that different metastatic sites harbor different genomic aberrations and so biopsy of only one or two accessible metastases may not be enough to represent the whole tumor genome (24,28). Finally, solid tumors also exhibit temporal heterogeneity, evolving over time under selection-pressure with treatment (29-32). Even though, and considering the heterogeneity of the tumors, it becomes difficult to have a complete scenario of the whole tumor based on the information obtained from small biopsies, and in several cases from a restringed number of tumor cells which, in turn, could lead to erroneous therapeutic decisions.
Since the beginning of the era of targeted therapies, there has been a clear need to understand the mechanisms of resistance and therefore rebiopsies at the time of clinical progression or the emergence of treatment resistance were gradually incorporated into clinical practice. Considering all the above-mentioned facts about lung cancer: presence of driver mutations, tumor heterogeneity, tumor dynamics, drug sensitivity, drug resistance, it is of remarkable importance the development of a non-invasive way to obtain this valuable biologic information, which includes the ability to diagnose, prognose and monitor lung cancer evolution. At this point, liquid biopsies seem to be the approach that covers all these requirements. In lung cancer, blood samples are the most explored type of liquid biopsy, and have been used to improve the diagnosis as well as for the searching prognostic and predictive biomarkers. Liquid biopsies have several advantages: (I) they allow rapid biomarker assessment in lung cancer patients for whom solid biopsies are impossible because of restricted or extremely risky access possibilities; (II) they can easily be repeated during cancer patient follow-up in order to control treatment efficiency; and/or (III) they can be used to detect genomic alterations occurring as result of resistance to therapy.
Liquid biopsies in lung cancer
In lung cancer liquid biopsies, blood samples are mainly used as a sample source for analyzing circulating tumor cells (CTCs) or circulating tumor DNA (ctDNA), in addition to other biomarkers of interest, such as circulating microRNAs, circulating RNA, platelets, plasma/serum metabolites, or exosomes (Figure 1). To explore the data available regarding the above in this review we used MEDLINE to perform a comprehensive literature search for original and review articles related to the terms “liquid biopsy” and “CTCs”, “ctDNA” and “NSCLC”, or “lung cancer”. Furthermore, to complete our search we also reviewed the bibliographies of these articles; only those published in English and in peer-reviewed journals were included. Our purpose was to describe the current contribution of CTC and ctDNA detection and analysis in lung cancer patients and to compare the advantages and disadvantages of these two approaches.
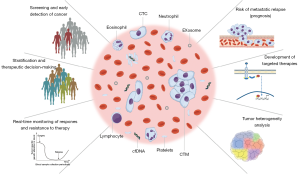
CTCs versus ctDNA: which one will win?
There are an increasing number of scientific reports pointing out the advantages of, and difficulties in, both detecting mutations and isolating CTCs and ctDNA in both metastatic and non-metastatic lung cancer (Table 1). The discrepancies in sensitivity, reproducibility, and concordance with tissue biopsies are likely due to the different approaches and methodologies used as well as their clinical settings.

Full table
CTCs
CTCs are tumor cells from solid tumors that spread via blood and/or lymphatic vessels. CTCs are shed into the vasculature from primary tumors and are postulated to contain subpopulations of cells with the potential to spread and initiate distant metastases (33). They were observed for the first time by Thomas Ashworth in 1869 in the blood of a man with metastatic cancer (34), but they only became relevant in modern cancer research a couple of decades ago with the demonstration of their presence early in the course of malignant disease (35). Several models have been suggested to describe the dissemination process whereby tumor cells leave the primary tumor to colonize distant organs, either when they become competent to metastasize or because of physical tumor extension (27,36).
CTCs can circulate in the bloodstream of lung cancer patients as single cells or as aggregates known as circulating tumor microemboli (CTM) (37-39). In this regard, the phenotype of single or aggregated CTCs can be different and so may present different levels of potential aggressiveness (37,38,40,41). Similar to single migratory mesenchymal-like CTCs, CTMs appear to be enriched in mesenchymal markers, an indicator of increased potential plasticity, which in turn seems to be related to more aggressive behavior, thus supporting their role in both tumor cell dissemination and the initiation of metastatic outgrowth (38,42-44). The presence of CTMs has been reported as a negative prognostic factor in lung cancer patients (38,40,45).
Isolation and detection of CTCs
Although many technologies have been developed over the past few years to detect and isolate CTCs in the peripheral blood of NSCLC patients (44,46-48), this task remains challenging (Figure 2). In advanced lung cancer patients, CTCs are rare cells present in very low concentrations in the bloodstream; approximately 1 mL of whole peripheral blood contains 1–10 CTCs against a background of 106–107 nucleated blood cells and around 109 red blood cells (49). Therefore, to reach the extreme sensitivity required to detect CTCs, an enrichment step is often required to increase their concentration before trying to detect or capture them. In this context, different methods for cellular enrichment, characterization, and identification of lung CTCs have been developed, including CTC microchips, filtration devices, quantitative reverse-transcription PCR assays, automated microscopy systems, etc. (46,50,51).
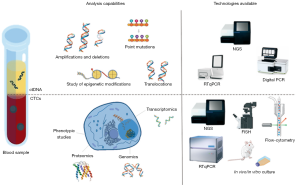
CTC detection or capture methods can be broadly categorized as: (I) label-dependent, based on positive enrichment involving cell surface markers such as epithelial cell adhesion molecule (EpCAM); or (II) label-independent, based on negative selection, such as size, or other differential biophysical properties of CTCs. Besides these two main approaches, other strategies include direct CTC imaging and functional assays (46,47,52-54). The first label-dependent studies tried to detect lung CTCs based on the assumption that circulating tumoral cells maintain the same characteristics as their tissue of origin, therefore most lung CTC categorization was based on the expression of epithelial-specific markers such as cytokeratin (CK) or intermediate filaments (IF) (41,55-60). Therefore, positive enrichment methods define lung CTCs as nucleated cells present in the bloodstream that express epithelial CKs and EpCAM and do not express the white blood cell surface antigen CD45 (59,61,62).
One of these methodologies is the CellSearch® system (Veridex, Raritan, NJ, USA) which has been approved by the Food and Drug Administration (FDA) for monitoring metastatic breast cancer (63), castration-resistant prostate cancer (64), and colon cancer patients (65). It has also been shown to be of prognostic significance for small cell lung cancer (SCLC) but the test has not yet been FDA-approved for this cancer type (40,66,67). The method is based on an initial enrichment of EpCAM positive cells followed by immunofluorescent staining using epithelial markers (CK 8, 18, 19), a leucocyte marker (CD45), and 4’, 6-diamidino-2-phenylindole (DAPI) for nuclear staining. The CellSearch® definition of CTCs is any intact EpCAM+/CK+/CD45− cell at least 4 µm in size and with a nucleus occupying at least 50% of the cytoplasm (41,55-61). Using EpCAM-dependent assays, CTCs can be detected in approximately 20–40% of NSCLC patients (41,55-58,60). Unfortunately, technologies relying on EpCAM positive selection cannot detect CTCs that have undergone EMT or any cancer stem cells that have not yet started epithelial differentiation. In lung cancer, CK-negative CTCs, which potentially represent tumor cells undergoing EMT, have also been identified. Consequently, the use of EpCAM as a positive selection marker should be carefully considered when trying to detect CTCs in NSCLC patients. Unfortunately, so far no reliable surface markers, which distinguish lung CTCs from normal epithelial cells and can be used for such label-dependent approaches, have yet been identified.
Label-independent approaches to CTC detection in lung cancer are an attractive alternative. One such method, Isolation by Size of Epithelial Tumor cell (ISET®, developed by Rarecells Company, France), used for cells isolated from lung cancer patients, involves a CTC enrichment step based on size by using a filtration device followed by cytological characterization. This system has been used to detect lung CTCs in both metastatic and non-metastatic NSCLC patients and has shown increased sensitivity in a wider range of patients compared to label-dependent methods such as CellSearch® (41,55,68-75).
Another direct technology, known as ScreenCell®, is also based on the size of CTCs but, in addition, allows their isolation so that they can be subjected to further morphological and molecular studies (76,77). Interestingly, it has been shown that lung CTCs isolated with different systems, can be cultured in vitro which is of particular interest for generating in vitro and in vivo models. As a first step towards this goal, data has already been generated for successful short-term cultures (up to 28 days) of CTCs isolated from patients with lung cancers (78-80). Such model systems could be used to study drug susceptibility or to generate sufficient numbers of cells for systematic deep analysis of their molecular profiles or biological behavior (52,81). Several recent studies have reported the development of mouse xenografts generated directly from CTCs or from breast, colorectal, prostate, hepatocellular, small cell lung, or gastric cancer CTC cultures (82-87). In particular, CTCs enriched from blood samples from SCLC patients were subcutaneously implanted into immunocompromised mice as CTC-derived explants (CDX); the CTCs were tumorigenic at densities of more than 53 CTCs/1 mL of blood, however, such large numbers of CTCs are not always obtained from advanced patients, thus highlighting one of the biggest challenges associated with these approaches (86).
Current models generated either in vitro or in vivo are also potentially limited if the clones they are grown from do not accurately reflect the true heterogeneity of the tumor (e.g., there may be a selective advantage for highly aggressive clones). Furthermore, in vivo xenograft models do not recapitulate tumor-host interactions that may play a role in drug resistance. Direct comparison between label-dependent and label-independent CTC isolation methods shows that both approaches have pros and cons. Label-dependent methods are more specific but they are rendered ineffective when antigen expression is lost in certain CTC subpopulations, and the cells become less viable after isolation. On the other hand, label-independent approaches are less specific but do not depend on CTC phenotype, and seem to better preserve CTC viability for downstream applications. There are currently many other technological developments focused on exhaustive lung CTC characterization in the pipeline at several diagnostic companies.
CTCs: clinical applications
CTC analysis is considered an interesting approach for early diagnosis, prognosis assessment, prediction of treatment efficacy, and early detection of lung cancer relapse. The most relevant lung cancer CTC studies are summarized in Table 2.
Full table
Larger numbers of CTCs are recovered from SCLC patients than from NSCLC patients (40,56). In addition, some researches have reported a positive association between the number of CTCs and clinical stage or the presence of distant metastases in primary lung cancer (56,57,73,88,93), whereas other studies failed to find any significant differences (50,92). Regarding other clinical applications, the majority of articles on CTCs in lung cancer focus on their prognostic role. In a large population of NSCLC patients, one group showed that isolation of more than 50 CTCs per 10 mL sample (using ISET®) is of prognostic value and is associated with shorter OS and disease-free survival (DFS) (89). Similar results were reported by Krebs et al. showing that PFS and OS was significantly better in advanced NSCLC patients with fewer CTCs (41). However, in another NSCLC patient cohort it was reported that although the median survival time tends to be shorter in CTC-positive than in CTC-negative patients, the difference was not significant (92).
In SCLC a significant association between higher numbers of CTCs and shorter survival has been described, and at least one study has reported that CTCs are a better predictor of survival than disease stage and tumor response (40,66,90). Moreover, a reduction in the number of CTCs after chemotherapy was also significantly associated with better outcomes in SCLC (66,94). Regarding the role of CTCs as biomarkers for therapeutic monitoring in NSCLC, comparisons between studies performed on samples collected before and after chemotherapy have consistently found that survival rates were significantly worse for patients with CTC counts that remained positive during treatment (95,96). In a group of patients treated with erlotinib and pertuzumab, the authors found that the decrease in CTC count upon treatment were significantly associated with disease response (91).
One of the main difficulties of working with CTCs in the field of lung cancer is their use as a theranostic tool for detecting somatic mutations (97). However, in 2008 Maheswaran and Sequist identified the presence of EGFR-activating mutations in 11 out of 12 (92%) of CTCs isolated from EGFR-mutated patients. During follow-up the authors detected the T790M mutation (which confers drug resistance) in CTCs collected from patients who progressed to TKI treatment (98). Moreover, EGFR mutations in CTCs from NSCLC patients were recently successfully specifically assessed using sensitive next generation sequencing (NGS) (59). Similarly, in 2012 Paul Hofman’s group published results from ALK-specific fluorescence in situ hybridization (FISH) analyses performed on archived lung cancer patient CTC samples. Their blind analysis of CTCs and corresponding tumor tissue showed a perfect match: 5 patients positive for ALK rearrangement in both CTC and tumor tissue were found while 82 were negative for this mutation in both CTC and tumor tissue (99).
In summary, there are currently 343 studies registered on “ClinicalTrials.gov” concerning CTCs and lung cancer, but despite the numerous scientific publications on this topic, these cells are still not used in routine clinical practice. This can be explained by the large number of methods available for CTC detection, and by the difficulty of selecting a reliable lung CTC marker. Despite the efforts made by the scientific community in the CTC field to try to improve lung cancer management, the analytical specificity and clinical utility of these methods must still be demonstrated in large prospective multicenter studies in order to obtain the evidence required for their introduction into the daily management of lung cancer patients.
ctDNA
The field of ctDNA analysis originally started almost 70 years ago (100); higher levels of circulating free DNA (cfDNA) were identified in cancer patients compared to healthy controls, suggesting that this correlated with malignancy and tumor stage (101-103). To date, two main mechanisms for releasing ctDNA have been postulated: “passive” and “active”. The passive mechanism involves the release of nucleic acids into the bloodstream, either directly from apoptotic and necrotic tumor cells or indirectly by necrotic tumor cells engulfed by macrophages (104). Data about the size distribution of cfDNA has revealed an enrichment in 150–180 bp fragments which reflects the nucleosomal pattern of DNA fragmentation characteristic of apoptotic processes (105-107). In contrast, fragments of tumoral nucleic acid can also be actively released into the circulation by living cells. One potential explanation hypothesizes that cancer cells release nucleic acids to transform the targeted recipient cells at distant locations, although the mechanisms are not completely understood (108). It is important to consider that ctDNA may represent a small proportion of the total cfDNA, at levels corresponding to one genome equivalent in 5 mL of plasma (0.01% allele fraction), and thus it may be undetectable with routine sampling (103,109). Apart from this, ctDNA levels can vary according to tumor burden and stage, anatomical proximity to vasculature, and biological features like apoptotic rate and metastatic potential (110,111).
Detection and quantification of ctDNA
The most common sample source used for ctDNA isolation is plasma collected in standard EDTA tubes. However, considering the low percentage of ctDNA present within total isolated cfDNA, it is important to control both the analytical and pre-analytical steps that can significantly affect ctDNA detection in blood samples (112). Plasma samples should be processed and stored immediately after blood collection to avoid increases in genomic DNA released from white blood cell lysis that could modify the relative levels of ctDNA. Therefore, the uses of standardized protocols in conjunction with specialized preservative-containing tubes (e.g., Streck Cell-Free DNA BCT) are strongly recommended (113).
The amounts of ctDNA present in in lung cancer patient samples give important diagnostic and prognostic information about the disease (114,115). However, the most important advantage of this technology is that it enables such analyses via easily obtained, minimally-invasive samples which are likely to reflect any genomic abnormalities present in the original neoplasm, giving insights into the types of mutations, indels, chromosomal rearrangements, chromosomal region gains or losses, and epigenetic modifications present (54,116-118). Given the small proportion of ctDNA present in the total cfDNA samples obtained, it is important to select the correct methods for its isolation and analysis; several highly sensitive techniques have been developed for the latter, ranging from PCR-based to more complex methodologies using NGS (summarized in Figure 2).
In lung cancer a variety of methods have been used for ctDNA analysis, many of them based on real-time PCR, although these approaches are more applicable when a limited number of loci are evaluated. Such systems include peptide nucleic acid (PNA) or locked nucleic acid (LNA) mutant-enriched PCR (ME-PCR) (119-121), amplification-refractory mutation system (ARMS) (122), digital PCR (including droplet-based systems) (123), and the beads, emulsification, amplification, and magnetics (BEAMing) system (124,125). Moreover, recently developed NGS technologies have also shown that it is possible to detect many cancer-associated mutations in single lung cancer patient blood samples (126,127). There are also protocols specifically intended to improve the sensitivity of NGS ctDNA sample analysis; these include tagged-amplicon deep sequencing (TAm-Seq) (128), Safe-Sequencing System (Safe-SeqS) (129), and cancer personalized profiling by deep sequencing (CAPP-seq) (109,130,131), among others.
ctDNA: clinical applications in lung cancer
The clinical applications of ctDNA can be divided into two main categories: (I) quantification of circulating DNA for early diagnosis, prognosis, and response prediction; and (II) analysis of ctDNA in order to profile and characterize molecular tumor alterations (Table 3).
Full table
Lung cancer patients have increased plasma cfDNA levels compared with control individuals (142,143) and the amount of cfDNA has been associated with tumor stage and burden in lung cancer (109). However, there are data demonstrating that this absolute cfDNA amount is limited as diagnostic tool in the absence of contextual knowledge of any associated tumor mutations (114). High levels of cfDNA have been reported as an indicator of poor outcome in lung cancer patients (115,138,142), but in other studies pretreatment levels of total cfDNA were not prognostic of survival (144,145). One possible restriction of these approaches is that cfDNA is also present at high levels in the blood of patients with benign diseases such as hepatic disorders, diabetes, cardiovascular diseases, non-neoplastic lung diseases, or infections (75).
Regarding the prognostic information provided by ctDNA, monitoring tumor-specific alterations present in ctDNA isolated from plasma from early stage NSCLC patients following surgical resection identified patients with residual disease and was able to detect disease recurrence (109,145). However, despite the reporting of some controversial results, when KRAS mutations in ctDNA were assessed as a prognostic marker in NSCLC patients (135,146,147) KRAS status in plasma ctDNA was associated with poor tumor response to EGFR-TKIs in NSCLC patients and so it could be used as a predictive marker for selecting appropriate NSCLC patients for such treatments (148-150). Is in this latest aspect, the presence of circulating DNA containing tumor-specific sequences, where we find the most widespread and important applications of ctDNA.
Several reports have analyzed the concordance between genomic alterations (such as EGFR mutations) present in lung cancer tissues and corresponding ctDNA samples (119,125,132-134,137): depending on the technology used, the agreement ranges between 60% to more than 90% (122,123,125,138,139,149,151,152). The EURTAC trial was one of the first to explore the feasibility of using ctDNA as a surrogate for tumor biopsy and to correlate mutations in plasma with PFS and OS (120). Since then, several other clinical trials in lung cancer have incorporated the analysis of plasma as a sample source for studying genomic tumor alterations (138).
In the context of metastatic disease the use of ctDNA is particularly useful for patients with tumors that are difficult to biopsy, those with contraindications for biopsy procedures, or where tumor samples have been exhausted; in these cases the possibility of determining the presence of genomic tumor alterations in ctDNA have brought forward the prospect of implementing precision oncology. In addition, ctDNA can be used for real-time monitoring of therapeutic responses to targeted-agents (132,136,152-154) as a valid surrogate for the current use of invasive rebiopsies. In this respect, a number of research groups have recently shown that ctDNA analysis can sensitively and specifically detect T790M clones early, i.e., before therapy or their emergence during EGFR-TKI treatments, demonstrating that this approach represents also an elegant way to overcome the problems arising from tumor heterogeneity(19,29,140,141,155).
For broader applicability, ctDNA analysis platforms focus on not only maximizing analytical sensitivity, but also on providing sufficient genomic coverage to be able to analyze multiple molecular markers in the same sample, thus providing the possibility of anticipating the molecular alterations expected as tumors evolve. Therefore, ongoing and future prospective studies should aim to test if treatment strategies informed by the unique data provided by ctDNA could yield superior clinical outcomes compared to tissue-based approaches.
The war: strengths, and limitations
There are many studies aiming to detect and/or characterize CTCs or ctDNA in lung cancer patients; the question is which of these two approaches will become the eventual gold standard for managing these patients in the era of precision oncology. In this “war” the usefulness of CTCs for ex vivo models, including in functional studies such as cultures, mouse xenografts, or real-time in vitro assays for drug sensitivity evaluation, is undisputed. CTC enumeration as a prognostic biomarker in lung cancer research has not been as successful as it was in breast, prostate, and colon cancers for which there is a FDA-approved CTC method; even so, the adoption of this approach in these cancers in daily oncological practice remains low.
Reports on comparative mutation analyses of CTCs and cfDNA have shown an interesting relationship between them in cancer patient blood samples (57,91). Maheswaran et al. analyzed EGFR mutations in CTCs and ctDNA obtained from the same NSCLC patients and determined that genotyping seemed to be more sensitive in CTCs than in cfDNA and that the associated CTC quantification provided an important context in which to interpret these genotyping results (98). Thus, with the emergence of extremely sensitive technologies, complete genomic and transcriptomic profiles, drug sensitivity testing in CTC-derived cell cultures or in single cells might soon become a reality. Until now, the use of ctDNA has remained limited to research scenarios. However, an EGFR plasma test (TheraScreen® EGFR plasma PCR kit) has recently been approved in Europe and China for screening advanced NSCLC patients where it is impossible to obtain a tumor biopsy, allowing subsequent treatment with gefitinib in appropriate cases. Hence, new perspectives for implementing ctDNA in clinical settings are starting to open up (122).
There is mounting scientific evidence supporting the use of ctDNA for profiling and characterizing lung tumor molecular alterations as well as for monitoring therapies and identifying mutations associated with acquired drug resistance (91,118,119,122,126,144,149). In this context ctDNA, rather than CTC analysis, is more appealing because plasma samples can be collected and analyzed without requiring prior enrichment and there is no need to isolate a rare cell population. Although pre-analytical conditions for ctDNA analysis must be further standardized, it seems that ctDNA, therefore, is likely to be the preferred option for genotyping and treatment-response monitoring. However, one important limitation of working with these samples is that in situ and morphological analyses using FISH and ICC (of particular interest in lung cancer for assessing ALK or ROS1 status) cannot be performed with these samples (49,156). Another drawback of ctDNA is that, because of the high sensitivity of the different methodologies used, it also detects clinically irrelevant molecular changes.
In order to fully incorporate liquid biopsies into clinical practice some critical points must still be addressed: (I) a consensus is required regarding the best matrix of detection (CTC or ctDNA) for each required application; (II) a consensus regarding the ideal technical approach for each application is mandatory; (III) the pre-analytical phase should be standardized to obtain robust and reproducible results; (IV) investment and uptake of the currently available techniques is required in order to bring down prices which presently limit accessibility to patients.
Conclusions
Liquid biopsy has great potential for the management of lung cancer patients. Despite the numerous techniques and experimental approaches that have been established in this field, the common objective of all of them is to develop a useful, sensitive, specific, and real-time prognostic, predictive, and monitoring system using minimally-invasive samples, which can be easily transferred into the clinical practice. From our point of view, ctDNA analysis should be chosen for analysis of mutations, copy number aberrations, and DNA methylation changes, whereas CTC analysis provides the unique opportunity to study whole cells, thus allowing DNA, RNA, and protein-based molecular profiling, as well as use in vivo studies. It is likely that both CTCs and ctDNA will have complementary roles as cancer biomarkers and might be used in parallel for earlier lung cancer diagnosis, prediction of treatment responses, or detection of disease progression. Taking all of these arguments into account, we consider the real victory in this “war” is the genuine possibility these technologies create for translating the concept of precision oncology into clinical practice. Liquid biopsies represent an important advance in the management of lung cancer in which CTCs and ctDNA are both expected to play complementary roles based on their relative strengths and limitations.
Acknowledgements
Funding: SC is supported by a grant from the Spanish Ministry of Economy and Competitiveness and the National Program for Research Aimed at the Challenges of Society, RETOS-Colaboración [RTC-2014-1532-1]. Funding was also provided from the Red Temática de Investigación Cooperativa en Cáncer (RTICC), Instituto de Salud Carlos III (ISCIII), the Spanish Ministry of Economy and Competitiveness, and the European Regional Development Fund (ERDF) “Una manera de hacer Europa” [RD12/0036/0025].
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Torre LA, Bray F, Siegel RL, et al. Global Cancer Statistics, 2012. CA Cancer J Clin 2015;65:87-108. [Crossref] [PubMed]
- Siegel RL, Miller KDK, Jemal A. Cancer Statistics, 2016. CA Cancer J Clin 2016;66:7-30. [Crossref] [PubMed]
- Ladanyi M. Targeted therapy of cancer: new roles for pathologists. Mod Pathol 2008;21 Suppl 2:S1. [Crossref] [PubMed]
- Hirsch FR, Wynes MW, Gandara DR, et al. The tissue is the issue: personalized medicine for non-small cell lung cancer. Clin Cancer Res 2010;16:4909-11. [Crossref] [PubMed]
- Pao W, Girard N. New driver mutations in non-small-cell lung cancer. Lancet Oncol 2011;12:175-80. [Crossref] [PubMed]
- Kwak EL, Bang Y-J, Camidge DR, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med 2010;363:1693-703. [Crossref] [PubMed]
- Slebos RJ, Kibbelaar RE, Dalesio O, et al. K-ras oncogene activation as a prognostic marker in adenocarcinoma of the lung. N Engl J Med 1990;323:561-5. [Crossref] [PubMed]
- Kosaka T, Yatabe Y, Endoh H, et al. Mutations of the epidermal growth factor receptor gene in lung cancer: biological and clinical implications. Cancer Res 2004;64:8919-23. [Crossref] [PubMed]
- Tokumo M, Toyooka S, Kiura K, et al. The relationship between epidermal growth factor receptor mutations and clinicopathologic features in non-small cell lung cancers. Clin Cancer Res 2005;11:1167-73. [PubMed]
- Sordella R, Bell DW, Haber DA, et al. Gefitinib-sensitizing EGFR mutations in lung cancer activate anti-apoptotic pathways. Science 2004;305:1163-7. [Crossref] [PubMed]
- Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med 2004;350:2129-39. [Crossref] [PubMed]
- Sequist L V, Joshi VA, Janne PA, et al. Epidermal growth factor receptor mutation testing in the care of lung cancer patients. Clin Cancer Res 2006;12:4403s-4408s. [Crossref] [PubMed]
- Paez J, Sellers WR. PI3K/PTEN/AKT pathway. A critical mediator of oncogenic signaling. Cancer Treat Res 2003;115:145-67. [Crossref] [PubMed]
- Sequist L V, Martins RG, Spigel D, et al. First-line gefitinib in patients with advanced non-small-cell lung cancer harboring somatic EGFR mutations. J Clin Oncol 2008;26:2442-9. [Crossref] [PubMed]
- Mok TS, Wu Y-L, Thongprasert S, et al. Gefitinib or Carboplatin–Paclitaxel in Pulmonary Adenocarcinoma. N Engl J Med 2009;361:947-57. [Crossref] [PubMed]
- Rosell R, Carcereny E, Gervais R, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol 2012;13:239-46. [Crossref] [PubMed]
- Keedy VL, Temin S, Somerfield MR, et al. Epidermal growth factor receptor (EGFR) mutation testing for patients with advanced non-small-cell lung cancer considering first-line EGFR tyrosine kinase inhibitor therapy. J Clin Oncol 2011;29:2121-7. [Crossref] [PubMed]
- Fukuoka M, Wu YL, Thongprasert S, et al. Biomarker analyses and final overall survival results from a phase III, randomized, open-label, first-line study of gefitinib versus carboplatin/paclitaxel in clinically selected patients with advanced non-small-cell lung cancer in Asia (IPASS). J Clin Oncol 2011;29:2866-74. [Crossref] [PubMed]
- Kobayashi S, Boggon TJ, Dayaram T, et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med 2005;352:786-92. [Crossref] [PubMed]
- Gazdar AF. Activating and resistance mutations of EGFR in non-small-cell lung cancer: role in clinical response to EGFR tyrosine kinase inhibitors. Oncogene 2009;28 Suppl 1:S24-31. [Crossref] [PubMed]
- Lee JK, Shin J-Y, Kim S, et al. Primary resistance to epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors (TKIs) in patients with non-small-cell lung cancer harboring TKI-sensitive EGFR mutations: an exploratory study. Ann Oncol 2013;24:2080-7. [Crossref] [PubMed]
- Zhou W, Ercan D, Chen L, et al. Novel mutant-selective EGFR kinase inhibitors against EGFR T790M. Nature 2009;462:1070-4. [Crossref] [PubMed]
- Aparicio S, Caldas C. The Implications of Clonal Genome Evolution for Cancer Medicine. N Engl J Med 2013;368:842-51. [Crossref] [PubMed]
- Gerlinger M, Rowan AJ, Horswell S, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med 2012;366:883-92. [Crossref] [PubMed]
- Shaw AT, Kim D-W, Mehra R, et al. Ceritinib in ALK-Rearranged Non–Small-Cell Lung Cancer. N Engl J Med 2014;370:1189-97. [Crossref] [PubMed]
- Shu CA, Rizvi NA. Into the Clinic With Nivolumab and Pembrolizumab. Oncologist 2016;21:527-8. [Crossref] [PubMed]
- Valastyan S, Weinberg RA. Tumor Metastasis : Molecular Insights and Evolving Paradigms. Cell 2011;147:275-92. [Crossref] [PubMed]
- Chong CR, Janne PA. The quest to overcome resistance to EGFR-targeted therapies in cancer. Nat Med 2013;19:1389-400. [Crossref] [PubMed]
- Piotrowska Z, Niederst MJ, Karlovich CA, et al. Heterogeneity Underlies the Emergence of EGFRT790 Wild-Type Clones Following Treatment of T790M-Positive Cancers with a Third-Generation EGFR Inhibitor. Cancer Discov 2015;5:713-22. [Crossref] [PubMed]
- Marusyk A, Almendro V, Polyak K. Intra-tumour heterogeneity: a looking glass for cancer? Nat Rev Cancer 2012;12:323-34. [Crossref] [PubMed]
- Burrell RA, McGranahan N, Bartek J, et al. The causes and consequences of genetic heterogeneity in cancer evolution. Nature 2013;501:338-45. [Crossref] [PubMed]
- Piotrowska Z, Sequist LV. Treatment of EGFR-Mutant Lung Cancers After Progression in Patients Receiving First-Line EGFR Tyrosine Kinase Inhibitors: A Review. JAMA Oncol 2016;2:948-54. [Crossref] [PubMed]
- Pantel K, Brakenhoff RH, Brandt B. Detection, clinical relevance and specific biological properties of disseminating tumour cells. Nat Rev Cancer 2008;8:329-40. [Crossref] [PubMed]
- Ashworth TR. A case of cancer in which cells similar to those in the tumours were seen in the blood after death. Aust Med J 1869;14:146-9.
- Alix-Panabières C, Pantel K. Circulating tumor cells: liquid biopsy of cancer. Clin Chem 2013;59:110-8. [Crossref] [PubMed]
- Kim MY, Oskarsson T, Acharyya S, et al. Tumor Self-Seeding by Circulating Cancer Cells. Cell 2009;139:1315-26. [Crossref] [PubMed]
- Carlsson A, Nair VS, Luttgen MS, et al. Circulating tumor microemboli diagnostics for patients with non-small-cell lung cancer. J Thorac Oncol 2014;9:1111-9. [Crossref] [PubMed]
- Hou J-MM, Krebs M, Ward T, et al. Circulating Tumor Cells as a Window on Metastasis Biology in Lung Cancer. Am J Pathol 2011;178:989-96. [Crossref] [PubMed]
- Hou J-M, Greystoke A, Lancashire L, et al. Evaluation of circulating tumor cells and serological cell death biomarkers in small cell lung cancer patients undergoing chemotherapy. Am J Pathol 2009;175:808-16. [Crossref] [PubMed]
- Hou JM, Krebs MG, Lancashire L, et al. Clinical significance and molecular characteristics of circulating tumor cells and circulating tumor microemboli in patients with small-cell lung cancer. J Clin Oncol 2012;30:525-32. [Crossref] [PubMed]
- Krebs MG, Sloane R, Priest L, et al. Evaluation and prognostic significance of circulating tumor cells in patients with non-small-cell lung cancer. J Clin Oncol 2011;29:1556-63. [Crossref] [PubMed]
- Chen X-X, Bai F. Single-cell analyses of circulating tumor cells. Cancer Biol Med 2015;12:184-92. [PubMed]
- Lim LS, Hu M, Huang MC, et al. Microsieve lab-chip device for rapid enumeration and fluorescence in situ hybridization of circulating tumor cells. Lab Chip 2012;12:4388-96. [Crossref] [PubMed]
- O’Flaherty JD, Gray S, Richard D, et al. Circulating tumour cells, their role in metastasis and their clinical utility in lung cancer. Lung Cancer 2012;76:19-25. [Crossref] [PubMed]
- Funaki S, Sawabata N, Abulaiti A, et al. Significance of tumour vessel invasion in determining the morphology of isolated tumour cells in the pulmonary vein in non-small-cell lung cancer. Eur J Cardiothorac Surg 2013;43:1126-30. [Crossref] [PubMed]
- Parkinson DR, Dracopoli N, Petty BG, et al. Considerations in the development of circulating tumor cell technology for clinical use. J Transl Med 2012;10:138. [Crossref] [PubMed]
- Yap TA, Lorente D, Omlin A, et al. Circulating tumor cells: A multifunctional biomarker. Clin Cancer Res 2014;20:2553-8. [Crossref] [PubMed]
- Pantel K, Alix-Panabières C. Real-time liquid biopsy in cancer patients: Fact or fiction? Cancer Res 2013;73:6384-8. [Crossref] [PubMed]
- Alama A, Truini A, Coco S, et al. Prognostic and predictive relevance of circulating tumor cells in patients with non-small-cell lung cancer. Drug Discov Today 2014;19:1671-6. [Crossref] [PubMed]
- Wendel M, Bazhenova L, Boshuizen R, et al. Fluid biopsy for circulating tumor cell identification in patients with early-and late-stage non-small cell lung cancer: a glimpse into lung cancer biology. Phys Biol 2012;9:016005. [Crossref] [PubMed]
- Andree KC, van Dalum G, Terstappen LWMM. Challenges in circulating tumor cell detection by the CellSearch system. Mol Oncol 2016;10:395-407. [Crossref] [PubMed]
- Ozkumur E, Shah AM, Ciciliano JC, et al. Inertial focusing for tumor antigen-dependent and -independent sorting of rare circulating tumor cells. Sci Transl Med 2013;5:179ra47. [Crossref] [PubMed]
- Joosse SA, Gorges TM, Pantel K. Biology, detection, and clinical implications of circulating tumor cells. EMBO Mol Med 2014;7:1-11. [Crossref] [PubMed]
- Ignatiadis M, Lee M, Jeffrey SS. Circulating tumor cells and circulating tumor DNA: Challenges and opportunities on the path to clinical utility. Clin Cancer Res 2015;21:4786-800. [Crossref] [PubMed]
- Hofman V, Ilie MI, Long E, et al. Detection of circulating tumor cells as a prognostic factor in patients undergoing radical surgery for non-small-cell lung carcinoma: Comparison of the efficacy of the CellSearch AssayTM and the isolation by size of epithelial tumor cell method. Int J Cancer 2011;129:1651-60. [Crossref] [PubMed]
- Tanaka F, Yoneda K, Kondo N, et al. Circulating tumor cell as a diagnostic marker in primary lung cancer. Clin Cancer Res 2009;15:6980-6. [Crossref] [PubMed]
- Isobe K, Hata Y, Kobayashi K, et al. Clinical significance of circulating tumor cells and free DNA in non-small cell lung cancer. Anticancer Res 2012;32:3339-44. [PubMed]
- Hirose T, Murata Y, Oki Y, et al. Relationship of circulating tumor cells to the effectiveness of cytotoxic chemotherapy in patients with metastatic non-small-cell lung cancer. Oncol Res 2012;20:131-7. [Crossref] [PubMed]
- Marchetti A, Del Grammastro M, Felicioni L, et al. Assessment of EGFR mutations in circulating tumor cell preparations from NSCLC patients by next generation sequencing: Toward a real-time liquid biopsy for treatment. PLoS One 2014;9:e103883. [Crossref] [PubMed]
- Devriese LA, Bosma AJ, van de Heuvel MM, et al. Circulating tumor cell detection in advanced non-small cell lung cancer patients by multi-marker QPCR analysis. Lung Cancer 2012;75:242-7. [Crossref] [PubMed]
- Muinelo-Romay L, Vieito M, Abalo A, et al. Evaluation of circulating tumor cells and related events as prognostic factors and surrogate biomarkers in advanced NSCLC patients receiving first-line systemic treatment. Cancers (Basel) 2014;6:153-65. [Crossref] [PubMed]
- de Wit S, van Dalum G, Lenferink AT, et al. The detection of EpCAM(+) and EpCAM(-) circulating tumor cells. Sci Rep 2015;5:12270. [Crossref] [PubMed]
- Cristofanilli M, Hayes DF, Budd GT, et al. Circulating tumor cells: a novel prognostic factor for newly diagnosed metastatic breast cancer. J Clin Oncol 2005;23:1420-30. [Crossref] [PubMed]
- de Bono JS, Scher HI, Montgomery RB, et al. Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin Cancer Res 2008;14:6302-9. [Crossref] [PubMed]
- Cohen SJ, Punt CJ, Iannotti N, et al. Relationship of circulating tumor cells to tumor response, progression-free survival, and overall survival in patients with metastatic colorectal cancer. J Clin Oncol 2008;26:3213-21. [Crossref] [PubMed]
- Hiltermann TJN, Pore MM, Van den Berg A, et al. Circulating tumor cells in small-cell lung cancer: A predictive and prognostic factor. Ann Oncol 2012;23:2937-42. [Crossref] [PubMed]
- Krebs MG, Metcalf RL, Carter L, et al. Molecular analysis of circulating tumour cells-biology and biomarkers. Nat Rev Clin Oncol 2014;11:129-44. [Crossref] [PubMed]
- Farace F, Massard C, Vimond N, et al. A direct comparison of CellSearch and ISET for circulating tumour-cell detection in patients with metastatic carcinomas. Br J Cancer 2011;105:847-53. [Crossref] [PubMed]
- Krebs MG, Hou JM, Sloane R, et al. Analysis of circulating tumor cells in patients with non-small cell lung cancer using epithelial marker-dependent and -independent approaches. J Thorac Oncol 2012;7:306-15. [Crossref] [PubMed]
- Mascalchi M, Falchini M, Maddau C, et al. Prevalence and number of circulating tumour cells and microemboli at diagnosis of advanced NSCLC. J Cancer Res Clin Oncol 2016;142:195-200. [Crossref] [PubMed]
- Hofman V, Long E, Ilie M, et al. Morphological analysis of circulating tumour cells in patients undergoing surgery for non-small cell lung carcinoma using the isolation by size of epithelial tumour cell (ISET) method. Cytopathology 2012;23:30-8. [Crossref] [PubMed]
- Pailler E, Adam J, Barthélémy A, et al. Detection of circulating tumor cells harboring a unique ALK rearrangement in ALK-positive non-small-cell lung cancer. J Clin Oncol 2013;31:2273-81. [Crossref] [PubMed]
- Pailler E, Auger N, Lindsay CR, et al. High level of chromosomal instability in circulating tumor cells of ROS1-rearranged non-small-cell lung cancer. Ann Oncol 2015;26:1408-15. [PubMed]
- Faugeroux V, Pailler E, Auger N, et al. Clinical Utility of Circulating Tumor Cells in ALK-Positive Non-Small-Cell Lung Cancer. Front Oncol 2014;4:281. [Crossref] [PubMed]
- Ilie M, Hofman V, Long E, et al. Current challenges for detection of circulating tumor cells and cell-free circulating nucleic acids, and their characterization in non-small cell lung carcinoma patients. What is the best blood substrate for personalized medicine? Ann Transl Med 2014;2:107. [PubMed]
- Fiorelli A, D’Andrilli A, Anile M, et al. Sequential Bilateral Bronchoscopic Lung Volume Reduction With One-Way Valves for Heterogeneous Emphysema. Ann Thorac Surg 2016;102:287-94. [Crossref] [PubMed]
- Chudasama D, Rice A, Soppa G, et al. Circulating tumour cells in patients with lung cancer undergoing endobronchial cryotherapy. Cryobiology 2015;71:161-3. [Crossref] [PubMed]
- Kolostova K, Spicka J, Matkowski R, et al. Isolation, primary culture, morphological and molecular characterization of circulating tumor cells in gynecological cancers. Am J Transl Res 2015;7:1203-13. [PubMed]
- Zhang Z, Shiratsuchi H, Lin J, et al. Expansion of CTCs from early stage lung cancer patients using a microfluidic co-culture model. Oncotarget 2014;5:12383-97. [Crossref] [PubMed]
- Baccelli I, Schneeweiss A, Riethdorf S, et al. Identification of a population of blood circulating tumor cells from breast cancer patients that initiates metastasis in a xenograft assay. Nat Biotechnol 2013;31:539-44. [Crossref] [PubMed]
- Maheswaran S, Haber DA. Ex Vivo Culture of CTCs: An Emerging Resource to Guide Cancer Therapy. Cancer Res 2015;75:2411-5. [Crossref] [PubMed]
- Yu M, Bardia A, Aceto N, et al. Cancer therapy. Ex vivo culture of circulating breast tumor cells for individualized testing of drug susceptibility. Science 2014;345:216-20. [Crossref] [PubMed]
- Cayrefourcq L, Mazard T, Joosse S, et al. Establishment and characterization of a cell line from human circulating colon cancer cells. Cancer Res 2015;75:892-901. [Crossref] [PubMed]
- Rossi E, Rugge M, Facchinetti A, et al. Retaining the long-survive capacity of Circulating Tumor Cells (CTCs) followed by xeno-transplantation: not only from metastatic cancer of the breast but also of prostate cancer patients. Oncoscience 2013;1:49-56. [Crossref] [PubMed]
- Sun Y-F, Xu Y, Yang X-R, et al. Circulating stem cell-like epithelial cell adhesion molecule-positive tumor cells indicate poor prognosis of hepatocellular carcinoma after curative resection. Hepatology 2013;57:1458-68. [Crossref] [PubMed]
- Hodgkinson CL, Morrow CJ, Li Y, et al. Tumorigenicity and genetic profiling of circulating tumor cells in small-cell lung cancer. Nat Med 2014;20:897-903. [Crossref] [PubMed]
- Yuan D, Chen L, Li M, et al. Isolation and characterization of circulating tumor cells from human gastric cancer patients. J Cancer Res Clin Oncol 2015;141:647-60. [Crossref] [PubMed]
- Sienel W, Seen-Hibler R, Mutschler W, et al. Tumour cells in the tumour draining vein of patients with non-small cell lung cancer: detection rate and clinical significance. Eur J Cardiothorac Surg 2003;23:451-6. [Crossref] [PubMed]
- Hofman V, Bonnetaud C, Ilie MI, et al. Preoperative circulating tumor cell detection using the isolation by size of epithelial tumor cell method for patients with lung cancer is a new prognostic biomarker. Clin Cancer Res 2011;17:827-35. [Crossref] [PubMed]
- Naito T, Tanaka F, Ono A, et al. Prognostic impact of circulating tumor cells in patients with small cell lung cancer. J Thorac Oncol 2012;7:512-9. [Crossref] [PubMed]
- Punnoose EA, Atwal S, Liu W, et al. Evaluation of circulating tumor cells and circulating tumor DNA in non-small cell lung cancer: association with clinical endpoints in a phase II clinical trial of pertuzumab and erlotinib. Clin Cancer Res 2012;18:2391-401. [Crossref] [PubMed]
- Juan O, Vidal J, Gisbert R, et al. Prognostic significance of circulating tumor cells in advanced non-small cell lung cancer patients treated with docetaxel and gemcitabine. Clin Transl Oncol 2014;16:637-43. [Crossref] [PubMed]
- Hashimoto M, Tanaka F, Yoneda K, et al. Significant increase in circulating tumour cells in pulmonary venous blood during surgical manipulation in patients with primary lung cancer. Interact Cardiovasc Thorac Surg 2014;18:775-83. [Crossref] [PubMed]
- Bevilacqua S, Gallo M, Franco R, et al. A “live” biopsy in a small-cell lung cancer patient by detection of circulating tumor cells. Lung Cancer 2009;65:123-5. [Crossref] [PubMed]
- Li J, Shi S-B, Shi W-L, et al. LUNX mRNA-positive cells at different time points predict prognosis in patients with surgically resected nonsmall cell lung cancer. Transl Res 2014;163:27-35. [Crossref] [PubMed]
- Yamashita J, Matsuo A, Kurusu Y, et al. Preoperative evidence of circulating tumor cells by means of reverse transcriptase-polymerase chain reaction for carcinoembryonic antigen messenger RNA is an independent predictor of survival in non-small cell lung cancer: a prospective study. J Thorac Cardiovasc Surg 2002;124:299-305. [Crossref] [PubMed]
- Costa DB. Identification of somatic genomic alterations in circulating tumors cells: Another step forward in non-small-cell lung cancer? J Clin Oncol 2013;31:2236-9. [Crossref] [PubMed]
- Maheswaran S, Sequist L V, Nagrath S, et al. Detection of Mutations in EGFR in Circulating Lung-Cancer Cells. N Engl J Med 2008;359:366-77. [Crossref] [PubMed]
- Ilie M, Long E, Butori C, et al. ALK-gene rearrangement: A comparative analysis on circulating tumour cells and tumour tissue from patients with lung adenocarcinoma. Ann Oncol 2012;23:2907-13. [Crossref] [PubMed]
- Mandel P, Metais P. Les acides nucléiques du plasma sanguin chez l'homme. C R Seances Soc Biol Fil 1948;142:241-3. [PubMed]
- Gormally E, Caboux E, Vineis P, et al. Circulating free DNA in plasma or serum as biomarker of carcinogenesis: practical aspects and biological significance. Mutat Res 2007;635:105-17. [Crossref] [PubMed]
- Anker P, Mulcahy H, Chen XQ, et al. Detection of circulating tumour DNA in the blood (plasma/serum) of cancer patients. Cancer Metastasis Rev 1999;18:65-73. [Crossref] [PubMed]
- Bettegowda C, Sausen M, Leary RJ, et al. Detection of Circulating Tumor DNA in Early- and Late-Stage Human Malignancies. Sci Transl Med 2014;6:224ra24. [Crossref] [PubMed]
- Crowley E, Di Nicolantonio F, Loupakis F, et al. Liquid biopsy: monitoring cancer-genetics in the blood. Nat Rev Clin Oncol 2013;10:472-84. [Crossref] [PubMed]
- Jahr S, Hentze H, Englisch S, et al. DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res 2001;61:1659-65. [PubMed]
- Diehl F, Li M, Dressman D, et al. Detection and quantification of mutations in the plasma of patients with colorectal tumors. Proc Natl Acad Sci U S A 2005;102:16368-73. [Crossref] [PubMed]
- Heitzer E, Ulz P, Geigl JB. Circulating tumor DNA as a liquid biopsy for cancer. Clin Chem 2015;61:112-23. [Crossref] [PubMed]
- Stroun M, Lyautey J, Lederrey C, et al. Alu repeat sequences are present in increased proportions compared to a unique gene in plasma/serum DNA: evidence for a preferential release from viable cells? Ann N Y Acad Sci 2001;945:258-64. [Crossref] [PubMed]
- Newman AM, Bratman S, To J, et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med 2014;20:548-54. [Crossref] [PubMed]
- Kidess E, Jeffrey SS. Circulating tumor cells versus tumor-derived cell-free DNA: rivals or partners in cancer care in the era of single-cell analysis? Genome Med 2013;5:70. [Crossref] [PubMed]
- Diehl F, Schmidt K, Choti MA, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med 2008;14:985-90. [Crossref] [PubMed]
- El Messaoudi S, Rolet F, Mouliere F, et al. Circulating cell free DNA: Preanalytical considerations. Clin Chim Acta 2013;424:222-30. [Crossref] [PubMed]
- Qin J, Williams TL, Fernando MR. A novel blood collection device stabilizes cell-free RNA in blood during sample shipping and storage. BMC Res Notes 2013;6:380. [Crossref] [PubMed]
- Sozzi G, Conte D, Mariani L, et al. Analysis of circulating tumor DNA in plasma at diagnosis and during follow-up of lung cancer patients. Cancer Res 2001;61:4675-8. [PubMed]
- Sirera R, Bremnes RM, Cabrera A, et al. Circulating DNA is a useful prognostic factor in patients with advanced non-small cell lung cancer. J Thorac Oncol 2011;6:286-90. [Crossref] [PubMed]
- Ignatiadis M, Dawson SJ. Circulating tumor cells and circulating tumor DNA for precision medicine: dream or reality? Ann Oncol 2014;25:2304-13. [Crossref] [PubMed]
- Sirera R, Gil M, Blasco A, et al. Retrospective analysis of the prognostic role of p16 protein inactivation in plasma in patients with locally advanced non-small cell lung cancer. Lung Cancer 2008;61:104-8. [Crossref] [PubMed]
- Bordi P, Del Re M, Danesi R, et al. Circulating DNA in diagnosis and monitoring EGFR gene mutations in advanced non-small cell lung cancer. Transl Lung Cancer Res 2015;4:584-97. [PubMed]
- Rosell R, Moran T, Queralt C, et al. Screening for epidermal growth factor receptor mutations in lung cancer. N Engl J Med 2009;361:958-67. [Crossref] [PubMed]
- Rosell R, Molina MA, Serrano MJ. EGFR mutations in circulating tumour DNA. Lancet Oncol 2012;13:971-3. [Crossref] [PubMed]
- Kim HR, Lee SY, Hyun DS, et al. Detection of EGFR mutations in circulating free DNA by PNA-mediated PCR clamping. J Exp Clin Cancer Res 2013;32:50. [Crossref] [PubMed]
- Douillard J-Y, Ostoros G, Cobo M, et al. Gefitinib treatment in EGFR mutated caucasian NSCLC: circulating-free tumor DNA as a surrogate for determination of EGFR status. J Thorac Oncol 2014;9:1345-53. [Crossref] [PubMed]
- Oxnard GR, Paweletz CP, Kuang Y, et al. Noninvasive detection of response and resistance in egfrmutant lung cancer using quantitative next-generation genotyping of cell-free plasma DNA. Clin Cancer Res 2014;20:1698-705. [Crossref] [PubMed]
- Diehl F, Li M, He Y, et al. BEAMing: single-molecule PCR on microparticles in water-in-oil emulsions. Nat Methods 2006;3:551-9. [Crossref] [PubMed]
- Taniguchi K, Uchida J, Nishino K, et al. Quantitative detection of EGFR mutations in circulating tumor DNA derived from lung adenocarcinomas. Clin Cancer Res 2011;17:7808-15. [Crossref] [PubMed]
- Murtaza M, Dawson S-J, Tsui DWY, et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature 2013;497:108-12. [Crossref] [PubMed]
- Couraud S, Vaca-Paniagua F, Villar S, et al. Noninvasive diagnosis of actionable mutations by deep sequencing of circulating free DNA in lung cancer from never-smokers: a proof-of-concept study from BioCAST/IFCT-1002. Clin Cancer Res 2014;20:4613-24. [Crossref] [PubMed]
- Martinez P, McGranahan N, Birkbak NJ, et al. Computational optimisation of targeted DNA sequencing for cancer detection. Sci Rep 2013;3:3309. [Crossref] [PubMed]
- Kinde I, Wu J, Papadopoulos N, et al. Detection and quantification of rare mutations with massively parallel sequencing. Proc Natl Acad Sci U S A 2011;108:9530-5. [Crossref] [PubMed]
- Lebofsky R, Decraene C, Bernard V, et al. Circulating tumor DNA as a non-invasive substitute to metastasis biopsy for tumor genotyping and personalized medicine in a prospective trial across all tumor types. Mol Oncol 2015;9:783-90. [Crossref] [PubMed]
- Newman AM, Lovejoy AF, Klass DM, et al. Integrated digital error suppression for improved detection of circulating tumor DNA. Nat Biotechnol 2016;34:547-55. [Crossref] [PubMed]
- Goto K, Ichinose Y, Ohe Y, et al. Epidermal growth factor receptor mutation status in circulating free DNA in serum: from IPASS, a phase III study of gefitinib or carboplatin/paclitaxel in non-small cell lung cancer. J Thorac Oncol 2012;7:115-21. [Crossref] [PubMed]
- Huang Z, Wang Z, Bai H, et al. The detection of EGFR mutation status in plasma is reproducible and can dynamically predict the efficacy of EGFR-TKI. Thoracic Cancer 2012;3:334-40. [Crossref]
- Zhao X, Han RB, Zhao J, et al. Comparison of epidermal growth factor receptor mutation statuses in tissue and plasma in stage I-IV non-small cell lung cancer patients. Respiration 2013;85:119-25. [Crossref] [PubMed]
- Kim ST, Sung JS, Jo UH, et al. Can mutations of EGFR and KRAS in serum be predictive and prognostic markers in patients with advanced non-small cell lung cancer (NSCLC)? Med Oncol 2013;30:328. [Crossref] [PubMed]
- Wang S, Han X, Hu X, et al. Clinical significance of pretreatment plasma biomarkers in advanced non-small cell lung cancer patients. Clin Chim Acta 2014;430:63-70. [Crossref] [PubMed]
- Weber B, Meldgaard P, Hager H, et al. Detection of EGFR mutations in plasma and biopsies from non-small cell lung cancer patients by allele-specific PCR assays. BMC Cancer 2014;14:294. [Crossref] [PubMed]
- Karachaliou N, Mayo-de las Casas C, Queralt C, et al. Association of EGFR L858R Mutation in Circulating Free DNA With Survival in the EURTAC Trial. JAMA Oncol 2015;1:149-57. [Crossref] [PubMed]
- Thress KS, Brant R, Carr TH, et al. EGFR mutation detection in ctDNA from NSCLC patient plasma: A cross-platform comparison of leading technologies to support the clinical development of AZD9291. Lung Cancer 2015;90:509-15. [Crossref] [PubMed]
- Wei Z, Shah N, Deng C, et al. Circulating DNA addresses cancer monitoring in non small cell lung cancer patients for detection and capturing the dynamic changes of the disease. Springerplus 2016;5:531. [Crossref] [PubMed]
- Zheng D, Ye X, Zhang MZ, et al. Plasma EGFR T790M ctDNA status is associated with clinical outcome in advanced NSCLC patients with acquired EGFR-TKI resistance. Sci Rep 2016;6:20913. [Crossref] [PubMed]
- Sozzi G, Conte D, Leon M, et al. Quantification of free circulating DNA as a diagnostic marker in lung cancer. J Clin Oncol 2003;21:3902-8. [Crossref] [PubMed]
- Fournié GJ, Courtin JP, Laval F, et al. Plasma DNA as a marker of cancerous cell death. Investigations in patients suffering from lung cancer and in nude mice bearing human tumours. Cancer Lett 1995;91:221-7. [Crossref] [PubMed]
- Li BT, Drilon A, Johnson ML, et al. A prospective study of total plasma cell-free DNA as a predictive biomarker for response to systemic therapy in patients with advanced non-small-cell lung cancersdagger. Ann Oncol 2016;27:154-9. [Crossref] [PubMed]
- Ludovini V, Pistola L, Gregorc V, et al. Plasma DNA, microsatellite alterations, and p53 tumor mutations are associated with disease-free survival in radically resected non-small cell lung cancer patients: a study of the perugia multidisciplinary team for thoracic oncology. J Thorac Oncol 2008;3:365-73. [Crossref] [PubMed]
- Gautschi O, Huegli B, Ziegler A, et al. Origin and prognostic value of circulating KRAS mutations in lung cancer patients. Cancer Lett 2007;254:265-73. [Crossref] [PubMed]
- Camps C, Jantus-Lewintre E, Cabrera A, et al. The identification of KRAS mutations at codon 12 in plasma DNA is not a prognostic factor in advanced non-small cell lung cancer patients. Lung Cancer 2011;72:365-9. [Crossref] [PubMed]
- Wang S, An T, Wang J, et al. Potential clinical significance of a plasma-based KRAS mutation analysis in patients with advanced non-small cell lung cancer. Clin Cancer Res 2010;16:1324-30. [Crossref] [PubMed]
- Mok T, Wu Y-L, Lee JS, et al. Detection and Dynamic Changes of EGFR Mutations from Circulating Tumor DNA as a Predictor of Survival Outcomes in NSCLC Patients Treated with First-line Intercalated Erlotinib and Chemotherapy. Clin Cancer Res 2015;21:3196-203. [Crossref] [PubMed]
- Douillard J-Y, Ostoros G, Cobo M, et al. First-line gefitinib in Caucasian EGFR mutation-positive NSCLC patients: a phase-IV, open-label, single-arm study. Br J Cancer 2014;110:55-62. [Crossref] [PubMed]
- He W, Xu D, Wang Z, et al. Detecting ALK-rearrangement of CTC enriched by nanovelcro chip in advanced NSCLC patients. Oncotarget 2016. [Epub ahead of print]. [PubMed]
- Marchetti A, Palma JF, Felicioni L, et al. Early Prediction of Response to Tyrosine Kinase Inhibitors by Quantification of EGFR Mutations in Plasma of NSCLC Patients. J Thorac Oncol 2015;10:1437-43. [Crossref] [PubMed]
- Marcq M, Vallée A, Bizieux A, et al. Detection of EGFR Mutations in the Plasma of Patients with Lung Adenocarcinoma for Real-Time Monitoring of Therapeutic Response to Tyrosine Kinase Inhibitors? J Thorac Oncol 2014;9:e49-50. [Crossref] [PubMed]
- Yang X, Zhuo M, Ye X, et al. Quantification of mutant alleles in circulating tumor DNA can predict survival in lung cancer. Oncotarget 2016;7:20810-24. [PubMed]
- Sorensen BS, Wu L, Wei W, et al. Monitoring of epidermal growth factor receptor tyrosine kinase inhibitor-sensitizing and resistance mutations in the plasma DNA of patients with advanced non-small cell lung cancer during treatment with erlotinib. Cancer 2014;120:3896-901. [Crossref] [PubMed]
- Zhang Z, Ramnath N, Nagrath S. Current Status of CTCs as Liquid Biopsy in Lung Cancer and Future Directions. Front Oncol 2015;5:209. [Crossref] [PubMed]


