KRAS mutant NSCLC, a new opportunity for the synthetic lethality therapeutic approach
Introduction
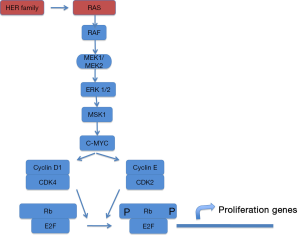
Lung cancer is the leading cause of cancer deaths worldwide and non–small cell lung cancer (NSCLC) accounts for 80% of all lung cancer cases (1). The standard first-line therapy for patients with advanced NSCLC was a platinum-based doublet combination chemotherapy but modest progress has been made with the use of chemotherapy, and additional treatment strategies are needed. So cancer drug development has shifted from cytotoxic, nonspecific chemotherapies to molecularly targeted, rationally designed drugs with greater efficacy and lower toxicities. For this challenge, the best knowledge of cancer biology is required. Nowadays, we are able to identify different genetic changes that allow us to consider NSCLC as a major disease which can be molecularly reclassified into several subsets of diseases (2). RAS gene family members encode small GTPases that activate various signaling pathways involved in proliferation, differentiation and cell survival (Figure 1). RAS proteins function as molecular switches that cycle between a GDP-bound inactive state and GTP-bound active state. Ras proto-oncogenes are the most frequent mutated genes in NSCLC, with mutations detected in about 25% of all tumors, mainly adenocarcinoma subtype (3).
v-Ki-ras2 Kirsten rat sarcoma viral oncogene (K-RAS) accounts for 90% of RAS mutations in lung adenocarcinomas. Most oncogenic forms of RAS impair their intrinsic GTPase activity, preventing GTP hydrolysis.
RAS proteins acquire the potential to transform the cells when an amino acid at position 12, 13, or 61 is replaced as a result of a point mutation in the gene but 97% of K-RAS mutations in NSCLC involve codons 12 or 13 at P-Loop also known as Walker A motif. This domain interacts with the phosphate group of GTP helped by GAP protein. In this regard, mutations at codon 12 avoid K-Ras to be stimulated by GAP protein. As GAP acts as a catalyst to speed up GTPase activity, mutations at that position slow GTP transition to GDP increasing GTP levels. Mutations at codon 61 affect the energy gradient needed to transform substrate (GTP) into product (GDP) because wild-type residue at that position stabilizes the transition state for GTP hydrolysis. So, it is critical to know specific site and biochemical effects when a K-Ras mutation is diagnosed because pharmacological modulation is completely different.
Although KRAS mutations have been widely hypothesized to be related to direct tobacco exposure, they do occur in approximately 15% of lung adenocarcinomas from never-smokers (4). Thus, KRAS tumor status cannot be easily predicted on the basis of smoking history alone. KRAS transversion mutations (G/TorG/C) are more common in former or current smokers and transition mutations (G/A) are more common in patients who never smoked cigarettes.
KRAS mutations have been associated with a poor prognosis such as a lower expectancy for survival (5), reduced benefit from adjuvant chemotherapy, they predict resistance towards EGFR tyrosine kinase inhibitors (6), and obtain less clinical benefits from chemotherapy compared with the general NSCLC population (7).
Treatment of KRAS mutated NSCLC: an unresolved issue
Direct inhibition of KRAS has proven clinically challenging. Although KRAS mutations were identified in lung cancer nearly 30 years ago (8), no successful targeted therapy has been developed and remains an elusive target for cancer therapy (9). So far, there is no yet effective treatment for patients with these types of tumors although we consider that K-RAS is not a unique target but a myriad of targets that combine absence of affinity for a catalyst (GAP) or decreasing affinity for GTP (P-Loop impairing) as well as other biochemical complexities.
Until now, all efforts to inhibit mutant KRAS in NSCLC have failed and few compounds have been assessed by clinical trial. One of the reasons to explain this point is because RAS enzyme kinetics is hard to inhibit due to affinity to substrates, catalyst proteins and sequential conformational changes after first signal that occurs inside this multi-target protein. In fact, the lack of specificity of KAS inhibitors could be related to this biochemical complexity that could be targeted at different levels: membrane attachment, P-Loop and thermodynamic requirements.
Various potent and selective inhibitors of RAS function were developed in the 1990s, with the aim to prevent association of RAS with the inner face of cell membrane (10). First, farnesyl transferase inhibitors avoid a critical post- translational modification in pre-RAS protein blocking isoprenylation. As farnesyl residues are needed to attach K-RAS to membrane it was hypothesized that this sort of inhibitors could inhibit RAS proteins (11). In fact, these inhibitors blocked RAS-dependent oncogenic activity “in vitro” and in preclinical animal models, but unfortunately failed in the clinical practice and showed little clinical efficacy because of a sequential post-translational modification at pre-Ras that compensates first steps of K-RAS maturation (12).
Although effective KRAS inhibitors are not currently available, genetic approaches have identified novel drug targets that are essential for RAS cellular localization and function, raising hope that new inhibitors of specific biochemical functionality of K-RAS will soon be developed.
Rationale for a new treatment strategy for K-RAS mutated NSCLC
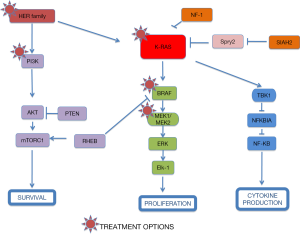
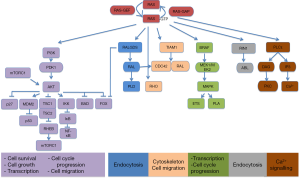
A different approach has been based on testing drugs or combinations of agents that work downstream of activated K-RAS. If you take into account that different KRAS-mutant tumors can activate several signalling pathways, a new treatment strategy for KRAS-mutant NSCLC should be based on the combination of targeted agents that inhibit downstream effectors of K-RAS dependent-tumors according to the “RAS-ome” (Figures 2,3). In this way, a specific knowledge of individual tumor molecular abnormalities that result in oncogene-specific “synthetic lethal” interactions will allow the rationale to combine promising targeted therapies for KRAS-mutated NSCLC.
Targeting HER pathway
Epiregulin (EREG) is ligand of the EGF receptor/EGFR and ERBB4 and is a putative transcriptional target of mutated KRAS dependent signaling that contributes to an aggressive phenotype and could be a promising therapeutic target in oncogenic KRAS-driven NSCLC (13) (Figure 2).
Targeting MEK pathway
Initial efforts focused on proteins downstream K-Ras at the RAS/RAF/mitogen-activated protein kinase (MEK)/extracellular signal-regulated kinase (ERK) signaling pathway. The MAPK pathway converges at the MEK1/MEK2 kinases, for which the only known substrates are the ERK1/ERK2 kinases (Figures 2,3). In fact, MEK inhibition would block ERK signalling irrespective of the upstream stimulus.
MEK1 and MEK2 are dual specificity kinases, RAF- phosphorylated, that phosphorylate the tyrosine and threonine residues on ERK1 and 2, leading to proliferation and migration activation. Mutations in RAS or RAF lead to a sustained oncogenic signal and predict response to MEK inhibition in laboratory models.
Selumetinib (AZD6244, ARRY-142886; AstraZeneca, Alderley Park, Cheshire, UK) is an orally available, potent, selective, non-ATP competitive inhibitor of MEK1/MEK2 kinases (IC50 14 nM for MEK1). Preclinical data from KRAS-mutant NSCLC tumor xenografts showed that selumetinib significantly suppressed tumor growth (14), especially in tumors harboring RAS mutations (15). Initial clinical studies of selumetinib showed target inhibition and tumor responses (16). A phase I trial demonstrating tolerability and preliminary efficacy of selumetinib at 100 mg twice daily (17), identified an acneiform rash as the main dose-limiting toxicity (DLT). However, treatment with selumetinib alone, showed little clinical efficacy in a phase II clinical trial in unselected pre-treated patients with NSCLC when selumetinib was compared with pemetrexed (18).
Results of additional preclinical in-vivo studies have shown that the combination of selumetinib and docetaxel leads to greater tumor-growth inhibition or regression, and apoptosis (19,20). This combination showed a manageable tolerability profile in advanced solid tumors (21) in phase I. With this rationale, a randomised, double-blind, phase II clinical trial combining docetaxel (75 mg/m2 on day 1 of a 21-day cycle) with or without oral selumetinib (75 mg twice daily in a 21-day cycle) in KRAS-mutant NSCLC patients after first-line progression (22). Mature data evidenced a promising trend in overall survival for patients treated at experimental arm (median OS 9.4 vs. 5.2 mo; HR 0.80; 80% CI, 0.56-1.14; one-sided P=0.21). Additionally, median progression-free survival was statistically significant (5.3 vs. 2.1 mo, HR 0.58; 80% CI, 0.42-0.79; one-sided P=0.014), and an impressive response rate around 37% in the combination group compared with 0% in the docetaxel alone group (P<0.0001). In post-hoc analyses, there were also improvements in lung cancer symptoms and all these benefits might be attributable to the cytoreductive effects of the treatment. However, a higher rate of febrile neutropenia (18% vs. 0%), diarrhea, vomiting, stomatitis, and dry skin with selumetinib plus docetaxel were communicated.
Obviously, this is a phase II study and requires further validation in a large phase III clinical trial. Furthermore, the study has potential limitations such as the small sample size and the absence of independent confirmation of progression-free survival and tumor response. Moreover, the control group of the study who received docetaxel alone clearly had poor evolution, lower than expected in previous clinical trials in unselected patients receiving docetaxel at second line setting (23). Furthermore, a new question emerges because poor efficacy of docetaxel in K-RAS mutant NSCLC patients should be investigated. Conversely, the potential synergy of docetaxel and selumetinib remains unclear and additional studies are needed. In-vivo mechanistic drug sequencing studies have shown that administration of selumetinib after docetaxel, rather than before, induced more apoptosis. This finding could have important clinical implications for any dosing schedule of this combination. This contrasts with the majority of previous studies in NSCLC, in which addition of a targeted agent to chemotherapy has not resulted in improved efficacy.
Another important issue is the therapeutic effect of specific KRAS mutations, to define a subpopulation of KRAS-mutant NSCLC in which the combination of selumetinib and docetaxel leads to improved efficacy. Previous studies showed that KRAS mutation subtype seems to be an important predictor of treatment outcome (24).
Wide genomic approaches have evidenced that it is usual for many mutations to co-exist. In this regard, K-RAS mutations in NSCLC patients could be co-expressed with additional sequence alterations. Thus, a recent study done in mice showed that overlapping mutations at p53 or LKB1 affect efficacy of selumetinib plus docetaxel (25) as well as docetaxel alone in tumors that harbors a mutated Kras sequence. For example, combination of selumetinib plus docetaxel provides substantial benefit in K-Rasmt/p53mt lung cancer models. Conversely, mice harboring Krasmt/LKB1mt tumors show primary resistance to this schedule. LKB1 (liver kinase B1) also known as serine threonine kinase 11 (STK11) the defective sequence of which is a cause of Peutz-Jeghers syndrome. Its role is critical in p53-dependent apoptosis, mainly involved at mitochondrion steps. When LKN1 is unable to exert its activity, p53-dependent death is impaired. LKB1 is somatically inactivated in about 30% of NSCLC (26), and the combination of LKB1 loss and KRAS mutation results in a more aggressive phenotype than tumors only harboring KRAS mutations (27). In fact, the decreased activation of ERK phosphorylation in KRAS/LKB1 tumors suggests that the proliferation of these tumors may be driven through other signaling pathways. KRAS/LKB1-mutant tumors have heightened activation of both AKT and SRC. This type of tumors with KRAS mutated and LKB1 inactivated show sensitivity to rapamycin or the MEK inhibitor CI-1040.
Several selumetinib trials are currently enrolling patients, including a phase II study (NCT01229150) in previously treated NSCLC stratified by KRAS status. Mutated KRAS and wild-type KRAS patients are randomized to receive selumetinib and erlotinib or selumetinib alone (28). In addition, the drug is being evaluated with thoracic radiation in one trial (NCT01146756) and in two multi-arm trials (NCT01306045 and NCT01248247) that assign treatment by molecular tumor characteristics.
Other MEK inhibitors have been already tested. Trametinib (GSK 1120212 or JTP-74057) is a reversible, allosteric MEK1/MEK2 inhibitor with an IC50 of 0.7 nM for MEK1, and a high specificity as demonstrated by limited activity against a panel of 180 other kinases. A multi-arm phase I/Ib trial (NCT01192165) is assessing many treatment combinations, specifically with a goal of identifying appropriate regimens for lung and pancreatic cancer treatment. An open-label, randomized phase II trial (NCT01362296) in second-line NSCLC that harbors muta¬tion in KRAS, NRAS, BRAF, or MEK1 is currently recruiting patients.
Dual targeting of MEK with inhibition of other kinases in the same pathway, such as EGFR, or with inhibition of a parallel pathway are also promising directions for ongoing trials.
Targeting PI3K pathway
PI3K is a site of convergence and stem for multiple pathways resulting in complex regulation of signaling and the potential for significant off-target effects, including activation of alternative networks to promote oncogenesis (Figure 3).
NSCLC harbors several molecular alterations involving the PI3K pathway, including PIK3CA amplification and mutation, decrease or loss of phosphate, and tensin homologue (PTEN), AKT mutations, LKB1 loss and KRAS mutation. For all of these features, PI3K pathway is one of the promising approaches to target RAS downstream signaling proteins. Conversely, K-RAS mutations have been predicted to mediate resistance to PI3K inhibitors (29). For this reason, a potential strategy of treatment of KRAS mutant tumors will be focused on dual inhibition of PI3K and MEK/ERK signaling.
MK-2206 is an oral pan-Akt inhibitor that binds Akt in its inactive configuration. MK-2206 has shown preclinical activity in a panel of NSCLC lines, with the greatest activity in a PIK3CA-mutated model (30). Combination therapy with selumetinib demonstrated synergy (31) and is being evaluated clinically (NCT01021748) (32).
Targeting nuclear factor kappa-light-chain enhancer of activated B cells (NF-κB) pathway
KRAS mutated tumors can activate nuclear factor kappa-light-chain enhancer of activated B cells (NF-κB) pathway and produce anti-apoptotic signals, essential for NSCLC survival through cREL and Bcl-xL (33) (Figures 1,3). So, NF-κB signaling and the non-canonical IκB kinase, TBK1, may represent an alternative strategy for targeting KRASmt-driven tumors. These observations suggest a pharmacological alternative for potential treatment of cancers harboring RAS mutations (34).
Neurofibromatosis type 1 pathway
Neurofibromatosis type 1 (NF1) gene regulates cell motility and invasion, and displays high homology with RAS GTPase activating protein (Figure 3). Loss of NF1 produces hyper-activation of RAS signaling in 40% of NSCLC (35). NF1-deficient malignancies and KRas/p53-mutant lung cancer exhibit an aggressive phenotype in murine models. However, agents that enhance proteotoxic stress, including the HSP90 inhibitor IPI-504 showed relevant responses when combined with rapamycin (36). Other HSP90 inhibitors are under evaluation (37). Since the mTOR inhibitor rapamycin has shown potential activity against NF1-associated tumors, it could be a new option of treatment (38).
Wilms tumor gene pathway
The Wilms Tumor gene (WT1) is a tumor suppressor gene that recognizes and binds to the DNA sequence 5'-CGCCCCCGC-3'. Curiously, function may be isoform-specific as isoforms lacking the KTS motif may act as transcription factors and isoforms containing the KTS motif may bind mRNA and play a role in mRNA metabolism or splicing. This biological complexity offers many possibilities for drug development, including those that affect KRASmt driven biology. Recently, a study in both mouse and human cells has shown that the loss of WT1 could activate a senescence program in KRASmt cells (39). If this observation is confirmed, a new approach of treatment will be opened.
GATA2 pathway
The development of RNA interference technology has enabled the possibility of testing biological roles of putative genes in wide-genome scale. In this regard, several screenings assays have been carried out in cell libraries aimed to identify genes the inhibition of which is selectively deleterious to K-RASmt cells (40). Candidate genes were then tested in larger panel of KRAS mutant and wild-type cancer cells. Finally, K-RASmt cancer cell lines were found to be dependent on some genes such as the transcription factor GATA2 (41).
GATA-binding Factor 2 or erythroid transcription factor (GATA2) can be involved in regulation of the proteasome activity, IL-1 and Rho-signaling pathways. Recently, it has been observed that loss of GATA2 reduced the viability of NSCLC cells harboring RAS mutations, whereas wild-type cells were unaffected (42). Although GATA2 itself is likely undruggable, combined suppression of GATA2-regulated pathways with clinically approved inhibitors caused marked tumor clearance. Pharmacological inhibition of GATA2-mediated pathways with bortezomib and fasudil results in dramatic tumor inhibition (43). These observations present a new treatment option to KRAS mutant NSCLC.
Seven in absentia homolog 2 pathway
The human homolog of Drosophila seven-in-absentia--SIAH-1 and SIAH-2 are ubiquitin E3 ligases and driving ubiquitin-mediated degradation of conserved downstream components of the RAS pathway that are required for mammalian RAS signal transduction (Figure 3). In this regard, SIAH-2 regulates the tumor growth by degradation of SPRY2 and subsequent activation of the RAS-ERK pathway. Since SIAH-2 can be involved in different NSCLC, SIAH-2 may be a viable target for novel anti-RAS and anticancer agents aimed at inhibiting EGFR and/or RAS-mediated tumorigenesis (44).
RNA-binding motif 5 pathway
RBM5 (RNA-binding motif protein 5, also named H37/LUCA-15) gene is a component of the spliceosome. A complex (also known as the prespliceosome) that regulates the alternative splicing of a number of mRNAs. It has demonstrated tumor suppressor activity (45). RBM5 can inhibit the growth of lung cancer cells and induce apoptosis both in vitro and in vivo (46). RBM5 is downregulated by the constitutively activated RAS mutant protein, RAS (G12V), in rat embryonic fibroblast cells, which indicates a correlation between the RAS pathways and RBM5 activity (47). Further evaluation of interrelationships between RBM5 expression and KRAS gene must be carried out to open a novel therapeutic approach.
IL-8 pathway
Interleukin-8 (IL-8; CXCL8) is a cytokine of the CXC chemokine family that is involved in neutrophil recruitment and activation. In addition, IL-8 is an angiogenic growth factor that is overexpressed in different cancers, including NSCLC (48). Lung adenocarcinoma and muco-epidermoid carcinoma cells produce substantial amounts of IL-8, and express both CXCR1 and CXCR2 IL-8 receptors. Activating mutations of KRAS upregulate IL-8 expression in NSCLC and IL-8 can play a role in cell growth and migration in oncogenic KRAS-driven NSCLC (49).
Twist-related protein 1 pathway
Twist1 acts as a transcriptional regulation as a heterodimer with E proteins. Interestingly, Twist1 regulates gene expression differentially, depending on dimer composition: homodimers induce expression of FGFR2 and POSTN while heterodimers repress FGFR2 and POSTN expression and induce THBS1 expression. Additionally, it has been suggested to play an important role during tumor progression. For example, transgenic mouse models have shown that Twist1 cooperates with KRAS (G12D) to markedly accelerate lung tumorigenesis by abrogating cellular senescence programs and promoting the progression from benign adenomas to adenocarcinomas. Moreover, the suppression of Twist1 to physiological levels is enough to cause KRAS mutant lung tumors to undergo senescence losing their neoplastic features (50). The suppression of TWIST1 in human tumors may be an effective example of pro-senescence therapy.
Conclusions
Traditionally, treatment decisions for patients with lung cancer have historically been based on tumor histology and TNM stage. One promising treatment strategy involves the further subdivision of NSCLC into clinically relevant molecular subsets, according to a classification schema based on specific so-called driver mutations.
Although mutational activation of the KRAS pathway is the most frequent genetic event in NSCLC, it remains an elusive target for cancer therapy. In fact, it has been considered an “undruggable” genetic alteration.
A key goal in cancer research is the discovery of new drug targets that will selectively impair the viability of tumoral cells such as KRAS mutant NSCLC. Therefore, a specific knowledge of individual tumor molecular abnormalities that result in oncogene-specific “synthetic lethal” interactions will allow the rationale to combine promising targeted therapies for KRAS-mutated NSCLC. Recently, a MEK inhibitor, selumetinib, has shown interesting efficacy when combined with docetaxel in patients with KRAS-mutant tumors. Several pathways may provide attractive approaches to develop new treatments in KRAS-mutated NSCLC.
Acknowledgements
Disclosure: The authors declare no conflict of interest.
References
- Jemal A, Bray F, Center MM, et al. Global cancer statistics. CA Cancer J Clin 2011;61:69-90.
- Herbst RS, Heymach JV, Lippman SM. Lung cancer. N Engl J Med 2008;359:1367-80.
- Mitsudomi T, Viallet J, Mulshine JL, et al. Mutations of ras genes distinguish a subset of non-small-cell lung cancer cell lines from small-cell lung cancer cell lines. Oncogene 1991;6:1353-62.
- Riely GJ, Kris MG, Rosenbaum D, et al. Frequency and distinctive spectrum of KRAS mutations in never smokers with lung adenocarcinoma. Clin Cancer Res 2008;14:5731-4.
- Mascaux C, Iannino N, Martin B, et al. The role of RAS oncogene in survival of patients with lung cancer: a systematic review of the literature with meta-analysis. Br J Cancer 2005;92:131-9.
- Mitsudomi T, Yatabe Y. Mutations of the epidermal growth factor receptor gene and related genes as determinants of epidermal growth factor receptor tyrosine kinase inhibitors sensitivity in lung cancer. Cancer Sci 2007;98:1817-24.
- Eberhard DA, Johnson BE, Amler LC, et al. Mutations in the epidermal growth factor receptor and in KRAS are predictive and prognostic indicators in patients with non-small-cell lung cancer treated with chemotherapy alone and in combination with erlotinib. J Clin Oncol 2005;23:5900-9.
- Santos E, Martin-Zanca D, Reddy EP, et al. Malignant activation of a K-ras oncogene in lung carcinoma but not in normal tissue of the same patient. Science 1984;223:661-4.
- Ahearn IM, Haigis K, Bar-Sagi D, et al. Regulating the regulator: post-translational modification of RAS. Nat Rev Mol Cell Biol 2011;13:39-51.
- Sousa SF, Fernandes PA, Ramos MJ. Farnesyltransferase inhibitors: a detailed chemical view on an elusive biological problem. Curr Med Chem 2008;15:1478-92.
- Blumenschein G, Ludwig C, Thomas G, et al. O-082 A randomized phase III trial comparing ionafarnib/carboplatin/paclitaxel versus carboplatin/paclitaxel (CP) in chemotherapy-naive patients with advanced or metastatic non-small cell lung cancer (NSCLC). Lung Cancer 2005;49:S30.
- Whyte DB, Kirschmeier P, Hockenberry TN, et al.K- and N-Ras are geranylgeranylated in cells treated with farnesyl protein transferase inhibitors. J Biol Chem 1997;272:14459-64.
- Sunaga N, Kaira K, Imai H, et al. Oncogenic KRAS-induced epiregulin overexpression contributes to aggressive phenotype and is a promising therapeutic target in non-small-cell lung cancer. Oncogene 2012. [Epub ahead of print].
- Dry JR, Pavey S, Pratilas CA, et al. Transcriptional pathway signatures predict MEK addiction and response to selumetinib (AZD6244). Cancer Res 2010;70:2264-73.
- Garon EB, Finn RS, Hosmer W, et al. Identification of common predictive markers of in vitro response to the Mek inhibitor selumetinib (AZD6244; ARRY-142886) in human breast cancer and non-small cell lung cancer cell lines. Mol Cancer Ther 2010;9:1985-94.
- Banerji U, Camidge DR, Verheul HM, et al. The first-in-human study of the hydrogen sulfate (Hyd-sulfate) capsule of the MEK1/2 inhibitor AZD6244 (ARRY-142886): a phase I open-label multicenter trial in patients with advanced cancer. Clin Cancer Res 2010;16:1613-23.
- Adjei AA, Cohen RB, Franklin W, et al. Phase I pharmacokinetic and pharmacodynamic study of the oral, small-molecule mitogen-activated protein kinase kinase 1/2 inhibitor AZD6244 (ARRY-142886) in patients with advanced cancers. J Clin Oncol 2008;26:2139-46.
- Hainsworth JD, Cebotaru CL, Kanarev V, et al. A phase II, open-label, randomized study to assess the efficacy and safety of AZD6244 (ARRY-142886) versus pemetrexed in patients with non-small cell lung cancer who have failed one or two prior chemotherapeutic regimens. J Thorac Oncol 2010;5:1630-6.
- Holt SV, Logié A, Odedra R, et al. The MEK1/2 inhibitor, selumetinib (AZD6244; ARRY-142886), enhances anti-tumour efficacy when combined with conventional chemotherapeutic agents in human tumour xenograft models. Br J Cancer 2012;106:858-66.
- Davies BR, Logie A, McKay JS, et al. AZD6244 (ARRY-142886), a potent inhibitor of mitogen-activated protein kinase/extracellular signal-regulated kinase kinase 1/2 kinases: mechanism of action in vivo, pharmacokinetic/pharmacodynamic relationship, and potential for combination in preclinical models. Mol Cancer Ther 2007;6:2209-19.
- Kim K, Infante J, Cohen R, et al. A phase I dose-escalation study of selumetinib in combination with docetaxel in patients with advanced solid tumors. Mol Cancer Ther 2011;10:B225.
- Jänne PA, Shaw AT, Pereira JR, et al. Selumetinib plus docetaxel for KRAS-mutant advanced non-small-cell lung cancer: a randomised, multicentre, placebo-controlled, phase 2 study. Lancet Oncol 2013;14:38-47.
- Douillard JY, Shepherd FA, Hirsh V, et al. Molecular predictors of outcome with gefitinib and docetaxel in previously treated non-small-cell lung cancer: data from the randomized phase III INTEREST trial. J Clin Oncol 2010;28:744-52.
- Ihle NT, Byers LA, Kim ES, et al. Effect of KRAS oncogene substitutions on protein behavior: implications for signaling and clinical outcome. J Natl Cancer Inst 2012;104:228-39.
- Chen Z, Cheng K, Walton Z, et al. A murine lung cancer co-clinical trial identifies genetic modifiers of therapeutic response. Nature 2012;483:613-7.
- Ding L, Getz G, Wheeler DA, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature 2008;455:1069-75.
- Mahoney CL, Choudhury B, Davies H, et al. LKB1/KRAS mutant lung cancers constitute a genetic subset of NSCLC with increased sensitivity to MAPK and mTOR signalling inhibition. Br J Cancer 2009;100:370-5.
- Goldman JW, Garon EB. Targeting MEK for the treatment of non-small-cell lung cancer. J Thorac Oncol 2012;7:S377-8.
- Ihle NT, Lemos R Jr, Wipf P, et al. Mutations in the phosphatidylinositol-3-kinase pathway predict for antitumor activity of the inhibitor PX-866 whereas oncogenic Ras is a dominant predictor for resistance. Cancer Res 2009;69:143-50.
- Hirai H, Sootome H, Nakatsuru Y, et al. MK-2206, an allosteric Akt inhibitor, enhances antitumor efficacy by standard chemotherapeutic agents or molecular targeted drugs in vitro and in vivo. Mol Cancer Ther 2010;9:1956-67.
- Engelman JA, Chen L, Tan X, et al. Effective use of PI3K and MEK inhibitors to treat mutant Kras G12D and PIK3CA H1047R murine lung cancers. Nat Med 2008;14:1351-6.
- Meng J, Dai B, Fang B, et al. Combination treatment with MEK and AKT inhibitors is more effective than each drug alone in human non-small cell lung cancer in vitro and in vivo. PLoS One 2010;5:e14124.
- Meylan E, Dooley AL, Feldser DM, et al. Requirement for NF-kappaB signalling in a mouse model of lung adenocarcinoma. Nature 2009;462:104-7.
- Barbie DA, Tamayo P, Boehm JS, et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature 2009;462:108-12.
- Roberts PJ, Stinchcombe TE, Der CJ, et al. Personalized medicine in non-small-cell lung cancer: is KRAS a useful marker in selecting patients for epidermal growth factor receptor-targeted therapy? J Clin Oncol 2010;28:4769-77.
- De Raedt T, Walton Z, Yecies JL, et al. Exploiting cancer cell vulnerabilities to develop a combination therapy for ras-driven tumors. Cancer Cell 2011;20:400-13.
- Acquaviva J, Smith DL, Sang J, et al. Targeting KRAS-mutant non-small cell lung cancer with the Hsp90 inhibitor ganetespib. Mol Cancer Ther 2012;11:2633-43.
- Johannessen CM, Johnson BW, Williams SM, et al. TORC1 is essential for NF1-associated malignancies. Curr Biol 2008;18:56-62.
- Vicent S, Chen R, Sayles LC, et al. Wilms tumor 1 (WT1) regulates KRAS-driven oncogenesis and senescence in mouse and human models. J Clin Invest 2010;120:3940-52.
- Luo J, Emanuele MJ, Li D, et al. A genome-wide RNAi screen identifies multiple synthetic lethal interactions with the Ras oncogene. Cell 2009;137:835-48.
- Steckel M, Molina-Arcas M, Weigelt B, et al. Determination of synthetic lethal interactions in KRAS oncogene-dependent cancer cells reveals novel therapeutic targeting strategies. Cell Res 2012;22:1227-45.
- Kumar MS, Hancock DC, Molina-Arcas M, et al. The GATA2 transcriptional network is requisite for RAS oncogene-driven non-small cell lung cancer. Cell 2012;149:642-55.
- Barbacid M. Opening a new GATAway for treating KRAS-driven lung tumors. Cancer Cell 2012;21:598-600.
- Ahmed AU, Schmidt RL, Park CH, et al. Effect of disrupting seven-in-absentia homolog 2 function on lung cancer cell growth. J Natl Cancer Inst 2008;100:1606-29.
- Oh JJ, Razfar A, Delgado I, et al. 3p21.3 tumor suppressor gene H37/Luca15/RBM5 inhibits growth of human lung cancer cells through cell cycle arrest and apoptosis. Cancer Res 2006;66:3419-27.
- Shao C, Zhao L, Wang K, et al. The tumor suppressor gene RBM5 inhibits lung adenocarcinoma cell growth and induces apoptosis. World J Surg Oncol 2012;10:160.
- Edamatsu H, Kaziro Y, Itoh H. LUCA15, a putative tumour suppressor gene encoding an RNA-binding nuclear protein, is down-regulated in ras-transformed Rat-1 cells. Genes Cells 2000;5:849-58.
- Luppi F, Longo AM, de Boer WI, et al. Interleukin-8 stimulates cell proliferation in non-small cell lung cancer through epidermal growth factor receptor transactivation. Lung Cancer 2007;56:25-33.
- Sunaga N, Imai H, Shimizu K, et al. Oncogenic KRAS-induced interleukin-8 overexpression promotes cell growth and migration and contributes to aggressive phenotypes of non-small cell lung cancer. Int J Cancer 2012;130:1733-44.
- Tran PT, Shroff EH, Burns TF, et al. Twist1 suppresses senescence programs and thereby accelerates and maintains mutant Kras-induced lung tumorigenesis. PLoS Genet 2012;8:e1002650