EGFR molecular testing in African-American non-small cell lung cancer patients - a review of discrepant data
Substantial discrepancy has recently emerged regarding the true frequency of activating epidermal growth factor receptor (EGFR) mutations in African American (AA) NSCLC subpopulations. Three recent reports appear to contradict earlier, and generally consistent, findings of a low mutation frequency in AA patients. The first round of sequencing studies by our group and others (2005 to 2009, Table 1) indicated that the frequency of activating EGFR mutations was significantly lower in African Americans (AA), ranging from 2-3% (4 of 160 cases combined), compared to a White cohort (1-5). However, in 2011 two large studies (Table 1: Reinersman, Cote) observed a much higher frequency of activating EGFR mutations in AA NSCLC cohorts, ranging from 12-19% (31 of 188 cases combined), without significant differences compared to White NSCLC cohorts. A third 2011 study (Table 1: Harada) reported an even higher EGFR mutation frequency of 31% for AA NSCLC, though this is based on a limited cohort of only sixteen cases (6-8).
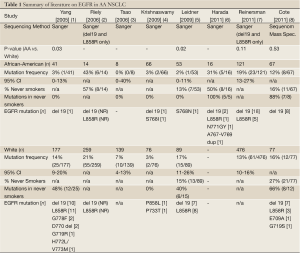
Full Table
What could account for these discordant results? One explanation may lie in the heterogenous study cohorts. While not all prior studies have provided data on smoking status - and this is somewhat surprising given the disease at hand - comparison of the studies which do report this information, demonstrates that the proportion of never-smokers in the AA NSCLC cohorts varies widely from 13% to 57% (see Table 1). We would argue that the lower end of this range (13% Leidner) is more informative to community practice, and is more congruent with previously reported smoking rates in a large AA NSCLC cohort (7% in a series of 1,288 patients) (9); than the high proportion of never-smokers in recent studies indicating a higher frequency of EGFR mutation for AA NSCLC (57% Cote, 50% Harada). This skew may be due to archival specimen sourcing from referral-based tertiary/quaternary centers, and also points to the likely explanation for discrepancy - Simpson’s paradox - a statistical phenomenon in which a correlation observed between heterogeneous groups in the aggregate, is reversed when groups are disaggregated (10-12).
As an illustration of Simpson’s paradox, we can disaggregate smoking status and EGFR mutation findings based on results from the only two studies which published sufficient data to allow for this analysis (see Table 2, Leidner, Cote). Notably, these two studies had similar sized cohorts of AA NSCLC cases, but arrived at divergent conclusions regarding EGFR mutation frequency. Table 2 presents disaggregated results according to ever/never smoking status for each study. In both studies, the ever-smoker group represents the large majority of the AA NSCLC cohort, (87% Leidner 46/53 and 84% Cote 56/67), as would be expected. Restricting analysis to AA ever-smokers, demonstrates good agreement between studies at 2% EGFR mutation frequency (1/46 Leidner and 1/56 Cote). In contrast, when the small AA never-smoker groups are compared, the real driver of divergence comes sharply into view, with a 64% discrepancy in EGFR mutation frequency between studies (0/7 Leidner 0% and 7/11 Cote 64%). Strikingly, 7 of the 8 total EGFR mutations identified by Cote et al., were detected in the very limited AA never-smoker subgroup (n=11).
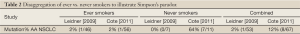
Full Table
The discrepancy between the two studies above, which disappears for ever-smokers when data are disaggregated, illustrates of Simpson’s paradox - a statistical phenomenon with implications both in the theoretical context and in practical application. In the famous Berkeley court case, alleged bias favoring males over females was claimed, based on analysis of admissions data in the aggregate. The alleged bias was subsequently shown to be driven by consistent and disproportionately higher rates of female applicants to more competitive departments, which came into clear view only after disaggregated analysis at the departmental level (13). This is also the likely explanation for apparent discrepancy between EGFR tissue profiling studies in AA NSCLC. In this case, disaggregation of the data according to smoking status (for the two studies where this is possible) demonstrates that the apparent divergence is driven entirely by results from very small never-smoker subgroups.
Additional discrepancies between the various studies which should be mentioned, include differences in the specific EGFR mutations being reported and the varying sensitivity of detection for the sequencing methods employed. Two activating EGFR mutations are routinely tested in clinical practice: a short oligonucleotide deletion in exon 19 (del 19) and a non-synonymous point mutation in exon 21 leading amino acid substitution at residue 858 (L858R). Together, these two mutations (del 19 and L858R) account for up to 90% of identified EGFR mutations. Several, much rarer mutations have been reported, including prior reports in the AA NSCLC tissue profiling literature (Table 1). Because these rare mutations are not routinely tested in clinical practice, the actual sensitivity they may, or may not, confer to EGFR targeted therapy is not known. In fact, there is evidence to suggest that some EGFR mutations may actually confer resistance. In the recent study by Harada et al., five EGFR mutations were observed in a cohort of 16 AA NSCLC cases, and notably, all were observed in AA never-smokers (6). Two of these five mutations represented rare insertions in exon 20 (N771GY and 767A-769V dup). Subsequent laboratory modeling using YFP-tagged MCF-7 cells expressing these mutations actually showed increased resistance to erlotinib in vitro, which may open an intriguing line of future investigation surrounding mechanisms of proclivity to rare variant EGFR mutations in specific population groups.
A further source of potential discrepancy between studies bears mention - the use of different sequencing technologies, which could influence the scope and threshold of mutations being detected. While most prior studies relied on standard Sanger sequencing for mutational profiling, the study by Cote et al. used a higher sensitivity platform (Sequenom mass spectrometry) which can detect mutation in as few as 5-10% of tumor cells (14). Whether response to EGFR TKI in a tumor consisting of >90% wild-type EGFR cells is clinically meaningful, remains to be determined, but may ultimately reveal that higher sensitivity is not a sine qua non of clinical benefit.
A final consideration must be given to patient self-reporting for determination of race which may lead to selection bias. Objective measure of genetic admixture is now theoretically possible, for example using ancestry SNP genotyping panels (15). The complex interaction of race and genetics in somatic tumorigenesis is not mechanistically well characterized or clinically interpretable at the present time. However, as hinted at by clustering of rare variant EGFR mutations in NSCLC, this may be an area ripe for future investigations as genomic advances proceed apace.
In the end, tissue-based EGFR mutational analysis is only a surrogate for what is actually of primary clinical interest: a reasonable prediction of treatment efficacy. In order to further assess the treatment effects of EGFR TKI’s, we reviewed treatment response in an unselected AA NSCLC patient population treated in the community setting (Cleveland, OH). If indeed activating EGFR mutations are significantly rarer among AA NSCLC patients, a significantly lower rate of objective response would be expected in comparison to a White, North American counterpart where previous objective response rates of roughly 10% have been observed (12.3% Perez-Soler et al. 2004 and 8.9% Shepherd et al. 2005) (16,17). Self-identified AA patients with advanced NSCLC treated with empiric EGFR TKI (either erlotinib or gefitinib) prior to widespread mutation testing, were evaluated for radiographic response by RECIST criteria (18). Patient characteristics are summarized in Table 3.
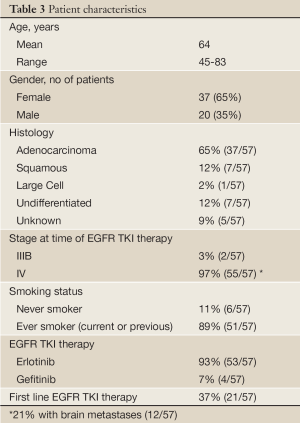
Full Table
We observed a 5% rate of response to EGFR TKI among unselected AA NSCLC patients treated in the community setting prior to the advent of routine EGFR mutation screening, using objective RECIST criteria and chart review of 57 cases. While this result did not reach significance (P=0.223), when compared against a 10% response rate as reported in large North American trials of unselected primarily White NSCLC patients (16,17), it represents a trend toward reduced rate of response. This trend is more in line with the results of early tissue profiling studies, including our own, which pointed to a lower frequency of activating EGFR mutations in AA vs. White NSCLC, and is consistent with current clinical practice of limiting EGFR TKI to patients with activating EGFR mutations by tissue analysis, regardless of race. As EGFR mutations are more frequent in never-smokers, a true evaluation of mutational frequency by race should be studied in a never-smoker cohort to help clarify the relationship between race and EGFR mutational status.
Acknowledgements
Funding: R. Leidner, NCI 5K12 CA076917-12.
Disclosure: B. Clifford, None; P. Fu, None; N. Pennell: Teva, Oncogenex; B. Halmos: Eli-Lilly, Pfizer, Oncothyreon, Daiichi and OSI; R. Leidner, None.
References
- Yang SH, Mechanic LE, Yang P, et al. Mutations in the tyrosine kinase domain of the epidermal growth factor receptor in non-small cell lung cancer. Clin Cancer Res 2005;11:2106-10. [PubMed]
- Riely GJ, Pao W, Pham D, et al. Clinical course of patients with non-small cell lung cancer and epidermal growth factor receptor exon 19 and exon 21 mutations treated with gefitinib or erlotinib. Clin Cancer Res 2006;12:839-44. [PubMed]
- Tsao AS, Tang XM, Sabloff B, et al. Clinicopathologic characteristics of the EGFR gene mutation in non-small cell lung cancer. J Thorac Oncol 2006;1:231-9. [PubMed]
- Krishnaswamy S, Kanteti R, Duke-Cohan JS, et al. Ethnic differences and functional analysis of MET mutations in lung cancer. Clin Cancer Res 2009;15:5714-23. [PubMed]
- Leidner RS, Fu P, Clifford B, et al. Genetic abnormalities of the EGFR pathway in African American Patients with non-small-cell lung cancer. J Clin Oncol 2009;27:5620-6. [PubMed]
- Harada T, Lopez-Chavez A, Xi L, et al. Characterization of epidermal growth factor receptor mutations in non-small-cell lung cancer patients of African-American ancestry. Oncogene 2011;30:1744-52. [PubMed]
- Reinersman JM, Johnson ML, Riely GJ, et al. Frequency of EGFR and KRAS mutations in lung adenocarcinomas in African Americans. J Thorac Oncol 2011;6:28-31. [PubMed]
- Cote ML, Haddad R, Edwards DJ, et al. Frequency and type of epidermal growth factor receptor mutations in African Americans with non-small cell lung cancer. J Thorac Oncol 2011;6:627-30. [PubMed]
- 9. Schwartz AG, Swanson GM. Lung carcinoma in African Americans and whites. A population-based study in metropolitan Detroit, Michigan. Cancer 1997;79:45-52. [PubMed]
- Simpson EH. The Interpretation of Interaction in Contingency Tables. J Roy Stat Soc B 1951;13:238-41.
- Mittal Y. Homogeneity of subpopulations and Simpson’s paradox. J Am Stat Assoc 1991;86:167-72.
- Fu P, Panneerselvam A, Clifford B, et al. Simpson’s paradox - aggregating and partitioning populations in health disparities of lung cancer patients. Stat Methods Med Res 2012. [Epub ahead of print]. [PubMed]
- Bickel PJ, Hammel EA, O’connell JW. Sex bias in graduate admissions: data from berkeley. Science 1975;187:398-404. [PubMed]
- MacConaill LE, Campbell CD, Kehoe SM, et al. Profiling critical cancer gene mutations in clinical tumor samples. PLoS One 2009;4:e7887. [PubMed]
- Via M, Ziv E, Burchard EG. Recent advances of genetic ancestry testing in biomedical research and direct to consumer testing. Clin Genet 2009;76:225-35. [PubMed]
- Pérez-Soler R, Chachoua A, Hammond LA, et al. Determinants of tumor response and survival with erlotinib in patients with non--small-cell lung cancer. J Clin Oncol 2004;22:3238-47. [PubMed]
- Shepherd FA, Rodrigues Pereira J, Ciuleanu T, et al. Erlotinib in previously treated non-small-cell lung cancer. N Engl J Med 2005;353:123-32. [PubMed]
- Eisenhauer EA, Therasse P, Bogaerts J, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer 2009;45:228-47. [PubMed]


