Drug screening to target nuclear orphan receptor NR4A2 for cancer therapeutics
Introduction
Cancer remains a major healthcare issue worldwide. According to world health organization, approximately 8.2 million people annually succumb to the disease (1). While many factors contribute to cancer development and mortality, it is clear that more effective anticancer drugs are needed. Progress in understanding cancer biology has led to discovery of molecular alterations that can be pharmacologically targeted by small molecule inhibitors. For instance, somatic activating mutations in epidermal growth factor gene (EGFR) sensitize lung cancer cells to tyrosine kinase inhibitors (TKI) such as erlotinib (2,3). These genetic changes occur relatively infrequently, and efficacy of the targeted therapies is almost always temporally (4-6). So, improved outcomes for precision therapies will require strategies to overcome drug resistance as well as discovery and validation of new, targetable oncogenic pathways for development of new therapies.
Our team has worked on molecular characterization of mucoepidermoid carcinoma (MEC), which commonly occurs in human salivary gland as well as upper respiratory tract (7). A common chromosomal translocation t(11:19) reported in the literature led to our discovery of a unique fusion gene, cAMP responsive element binding (CREB)-regulated transcription coactivator 1 (CRTC1)-Mastermind-like protein 2 (MAML2), in most human MEC tumor samples and cell line (7,8). We and others also found that this fusion gene up-regulates CREB protein signaling and CREB responsive genes such as Amphiregulin (AREG) and NR4A2 (9-11). Our further research determined that CRTC1-MAML2+ MEC tumors and non-small cell lung cancer (NSCLC) with loss of liver kinase B (LKB1) gene, which commonly occurs in NSCLC, are sensitive to gene silencing of NR4A2 in vitro (12).
NR4A2, also known as Nurr1, belongs to a subfamily of the orphan nuclear receptor NR4As (13). These receptors transcriptionally regulate cell proliferation, apoptosis, inflammation, neuronal development, and carcinogenesis (13,14). Several studies suggest an oncogenic role of NR4A2, because its expression was associated with poor patient outcome, invasive phenotype, and resistance to systemic therapy (15-18). These studies include gene silencing of NR4A2 in preclinical models (17). Currently, there is no clinically available agent that is known to target NR4As for the treatment of human cancer.
We conducted high throughput drug screening with a luciferase-based in vitro assay to identify agents to suppress NR4A2 expression which were validated later in our preclinical assays. Compound libraries included Food and Drug Administration (FDA)-approved drugs, which could potentially be quickly repurposed, as well as a diverse collection of drug-like scaffolds. This study may lead to development of NR4A2 inhibitors and also help researchers explore inhibitors of other orphan receptor genes.
Methods
Chemicals, screening compounds, and vectors
Chemicals used for in vitro studies were purchased as follows: G418 Sulfate (Corning, NY, USA); H-89 dihydrochloride hydrate, S3I-201, Sigma-Aldrich (St. Louis, MO, USA); afatinib, rapamycin, sorafenib, dasatinib, LC Laboratories (Woburn, MA, USA); gefitinib, dabrafenib, AZD9291, CO1686, OSI-420, bosutinib, vemurafenib, SelleckChem (Houston, TX, USA); KU171309, Life Chemicals Inc. (Niagara-on-the-Lake, Canada). Reagents for transfection (Lipofectamine 2000) were purchased from Invitrogen (Carlsbad, CA, USA). Dual Luciferase Reporter Assay System, and Steady Glo Luciferase Assay System kits were obtained from Promega (Maddison, MI, USA).
Compounds used for high throughput screening were obtained from Life Chemicals and FDA library. Firefly luciferase vectors (pGL4.26, positive screen; pGL4.29, negative screen) were purchased from Promega (Maddison, MI, USA). GFP vectors (pCMV6-AC-GFP) and plasmid containing open reading frame of human NR4A2 was purchased from OriGene Technologies (Rockville, MD, USA).
Cell lines, growth assay, and establishment of sublines
NCI-H3118, H2087, H2126 were kindly gifted by Dr. Frederic Kaye (University of Florida). All of them were originally derived from human individuals with MEC (H3118) and NSCLC (H2087 and H2126) at National cancer Institute, and have been maintained by standard tissue culture techniques. UM-SCC-81B, a human squamous cell carcinoma cell line, was kindly donated by Dr. Thomas Carey at University of Michigan via material transfer agreement. All cells were cultured in RMPI1640 medium with 10% fetal bovine serum and grown at 5% CO2. All cells were also tested for mycoplasma by Lonza Mycoalert Assay when we received cells.
Unless otherwise noted, in vitro growth assay was performed with 96-well plates, and cells were placed at a density of 500–1,000 cells/well on the day prior to drug exposure which lasted for 48 hours. Thereafter amount of live cells were determined by CellTiterGlo assay (Promega, Maddison, MI, USA) according to manufactures guideline. A triplicate was made for each treatment condition (i.e., time and concentration), and a median and 95% confidence interval were determined based on at least three independent experiments.
A total of five repeated nucleotide sequences of NuRE [5-GAT CGT GAT ATT TAC CTC CAA ATG CCA-3 (19)] were cloned into pGL4.26 luciferase vector by standard molecular biology technique. H3118 cells were then transfected with the pGL4.26 containing 5xNuRE sequence or pGL4.29 which contains CRE using Lipofectamine 2000®, according to manufacturer’s recommendation. Transient transfection was performed, repeated five times, and relative luciferase activity was determined by normalization with Renilla luciferase using Dual-Luciferase Reporter system® (Promega) according to the manufactures guideline. Stably transfected H3118 cell clones were selected by G418, and then further expanded till we confirmed stable expression of the luciferase gene by Steady Glo Luciferase Assay System® (Promega).
H2087 cells stably expressing GFP-NR4A2 or GFP alone were established similarly. GFP tagged pCMV-AC-GFP vectors with or without NR4A2 gene were transfected into NR4A2 negative H2087 cells. Positive clones for each vector construct were selected by G418, and then expanded for cell growth assay.
High throughput screening
The primary screening was performed using a stably transfected H3118 cell line expressing luciferase reporter under the control of 5xNuRE regulatory elements. To identify compounds modulating luciferase expression, 10,000 cells were plated in in each well of 384 well microplates to screen a total of 20,427 compounds. Labcyte Echo 555 was used to acoustically transfer the 5,384 FDA/Bioactive compounds (5 µM) and Life Chemical diversity set (15,040 compounds: 10 µM) to the assay plates. After 48 h of incubation, One-Glo luciferase detection reagent (Promega, Madison, MI, USA) was added to the cells and luminescence was quantified using Perkin Elmer Enspire. An average Z’ score of 0.73±0.05 was obtained across all assay plates screened. The primary hits were identified as compounds that inhibited luminescence to ≥40%, which was a cut-off defined by plate median plus three standard deviations.
Reconfirmation of primary hits
The 316 FDA/bioactive compounds and 254 Life Chemical diversity compounds were cherry-picked from library stocks and their activity reconfirmed in a concentration response assay. The two cell lines H3116.4.26. NuRE as well as H3116.4.29 cell lines were treated with 20, 10, 5 and 2.5 µM FDA/Bioactive compounds and at 40, 20, 20, 5 and 2.5 µM of Life Chemical compounds. At 48 h post exposure, the luminescence was quantified using One Glo luciferase assay kit (Promega). Along with luminescence inhibition, the effect of compounds on cytotoxicity of the two cell lines was also measured in parallel using the Cell Titer Glo assay (Promega).
Immunoblotting
Western blot analyses were performed as previously reported. A total of 18–50 µg protein derived from cell lysate was prepared using standard method, then loaded onto SDS-PAGE gels. Subsequently electrophoresis, transfer to PVDF membrane, and blotting with primary and secondary antibodies followed. Blotting signals were detected by ChemiDocTM MP Imaging System. Primary and secondary antibodies were purchased from as follows: NR4A2 (1:1000), R & D Systems (Minneapolis, MN, USA); EGFR (1:1000), p-EGFR (1:1000), ERK (1:1000), p-ERK (1:1000), Cell Signaling Technology (Danvers, MA, USA); GAPDH (1:1000), Santa Cruz (Dallas, TX, USA); anti-mouse IgG, HRP-linked Antibody (1:5000), Cell Signaling Technology (Danvers, MA, USA), and Goat anti-Rabbit IgG (H + L), HRP Conjugate (1:5000), Thermo Fisher Scientific (Waltham, MA, USA). Blotted membranes were washed, and then incubated with SuperSignalTM West Femto (Thermo Fisher Scientific, Waltham, MA, USA) at room temperature for 5 min prior to image scanning.
RT-PCR
mRNA was extracted from tissue culture cells using RNeasy kit (Qiagen, Germantown, MD, USA). Thereafter cDNA was made using SuperScript® III First-Strand Synthesis SuperMix (Thermo Fisher Scientific). A total of 30 cycles of PCR reaction with the cycle conditions: 98.0 °C for 30 s, followed by 30 cycles of 98.0 °C for 10 s, 59.0 °C for 25 s and 72.0 °C for 25 s was performed by thermal cycler, Veriti® 96-Well Fast Thermal Cycler. PCR primers for human NR4A2 and GAPDH are as follows: NR4A2(Forward 5’-CGACATTTCTGCCTTCTCC-3’, Reverse 5’-GGTAAAGTGTCCAGGAAAAG-3’), GAPDH (Forward 5’-GCCACATCGCTCAGAACCA-3’, Reverse 5’-CCAGCATCGCCCCACTTGAT-3’). Samples were then loaded onto 1–2% agarose gel containing ethidium bromide for visualization.
Statistical analyses
Statistical significance was determined using two-sided Student’s t-test or Mann-Whitney U test (Wilcoxon rank-sum test) as appropriate. P<0.05 was considered to be significant.
Statement of ethics
Ethics approval was not required for this study because no human or animal was involved.
Results
High throughput drug screening for NR4A2 inhibitors
In order to identify candidate agents targeting NR4A2 signaling, we undertook a cell-based high throughput screening using isogenic human cancer cells. Transient transfection of NCI-H3118, a human MEC cell line constitutively expressing endogenous NR4A2, with pGL4.26 luciferase vector containing NR4A2 binding sites (19), exhibited high level luciferase expression as compared to H2087 cell line (Figure 1A). A positive H3118 clone with stable luciferase activity was used to screen a total of 20,427 compounds including 5,387 FDA approved agents and known bioactives and 15,040 compounds from Life Chemical diversity scaffold Library. A total of 316 and 254 compounds, respectively, were shown to suppress luciferase activity by more than 40%, which was a cut-off defined by plate median plus three standard deviations. Secondary screening was conducted with an isogenic H3118 cells expressing cAMP responsive element (CRE)-luciferase (pGL4.29). This was used to eliminate false positive compounds that can suppress luciferase activity, leaving 23 FDA and 43 Life Chemical compounds as potential NR4A2 inhibitors (Figure 1B, Tables 1,2). These positive hits have differential activity to suppress luciferase activity driven by endogenous NR4A2, and minimal effect on CRE-luciferase. Of note, several positive hits among FDA approved agents were known inhibitors for EGFR and Src. Examples of these positive hits are shown in Figure 1C.
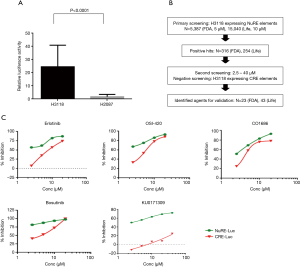

Full table

Full table
In vitro validation studies for positive hits
Several compounds were validated for in vitro sensitivity according to NR4A2 expression status. As we have previously reported, H3118 and H2126 cells showed high NR4A2 expression at both protein and mRNA levels, whereas H2087 and UM-SCC-81B cells showed very low to undetectable expression (Figure 2A,B) (11). RT-PCR analysis showed that UM-SCC-81B cells had no CRTC1-MAML2 translocation (data not shown). These findings are consistent with our previous study demonstrating that cells with either CRTC1-MAML2 translocation (H3118) or loss of LKB1 (H2126) had relatively high NR4A2 level as compared to wild type cells (H2087).
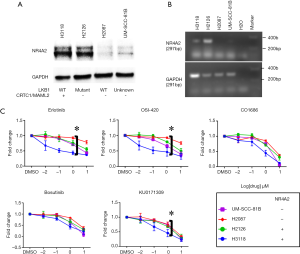
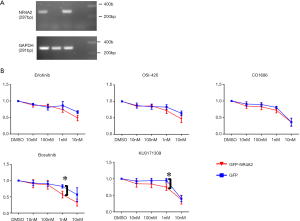
In vitro growth inhibitory activities of the several hits were tested for these four human cancer cell lines. EGFR inhibitors (erlotinib, OSI-420, CO-1686), a Src inhibitor (bosutinib), and a compound from Life Chemical library with unknown mechanism of action (KU0171309) had superior growth inhibitory activity for H3118 as compared to other three cell lines (Figure 2C). An isogenic H2087 cell line stably expressing GFP-NR4A2 gene (H2087-GFP-NR4A2) was more sensitive to these compounds than H2087-GFP cell (Figure 3).
Mechanism of action of NR4A2 inhibitors
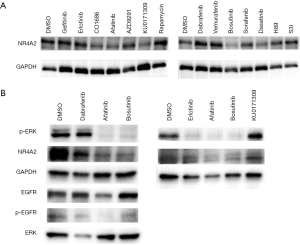
To further determine mechanism of action of the potential NR4A2 inhibitors, H3118 cells were treated with EGFR/Src inhibitors and KU0171309. EGFR inhibitors (Erlotinib, Afatinib) and Src inhibitors (Bosutinib, Dasatinib) suppressed both NR4A2 and phospho-ERK, whereas KU171309 suppress NR4A2 without affecting p-ERK (Figure 4). Compounds targeting downstream of EGFR such as BRAF (dabrafenib, vemurafenib), mTOR/S6K (rapamycin, H89), STAT3 (S3I), MEK (trametinib) did not significantly affect NR4A2 level (Figure 4 and data not shown). Although BRAF inhibitors (Dabrafenib and Vemurafenib) suppressed pERK, they did not consistently suppress NR4A2 expression or cell growth (Figure 4 and data not shown).
Discussion
Orphan nuclear receptor family members share several biological features across species. They act as transcriptional factors to regulate downstream genes, and lack endogenous ligands despite common ligand-binding structures (13). This family includes subfamilies such as NR0Bs, NR1Ds, NR2Es, NR2Fs, NR4As, and NR6As. NR4A2 is a member of NR4A subfamily with significant similarity in ligand-binding domains to NR4A2 and NR4A3. Recent studies demonstrated that NR4A2 is involved in a number of biologic processes to regulate homeostasis. It plays a key role in neuronal development, inflammation, and carcinogenesis (13,14). Unlike its homologues NR4A1 and NR4A3 where their concomitant loss lead to acute leukemia in murine models (20,21), studies on NR4A2 mostly demonstrated its role as an oncogene. For instance, its forced expression leads to resistance to chemotherapy-induced apoptosis in several human cancer types (18,22). Overexpression of NR4A2 and its association with poor patient outcome have been reported for several human cancer types (15,16,18,22). Given lack of inhibitors for any transcription factors in cancer therapy, development of NR4A2 inhibitors will likely encourage researchers to explore other transcription factors as therapeutic targets.
Our previous research indicated that NR4A2 is one of the promising targets for subsets of MEC and NSCLC tumors (12). We found that CRTC1-MAML2 transgene in MEC tumor has activated CREB signaling, whereas LKB1 null NSCLC cells have under-phosphorylated CRTC1 which leads to its nuclear localization to induce CREB signaling (9,12). NR4A2 was found to be a key molecule to sustain their cell growth in vitro.
Our high throughput screening procedure determined that 23 FDA approved/bioactive and 43 Life Chemical compounds suppressed luciferase activity driven by endogenous NR4A2. Subsequent study demonstrated several EGFR inhibitors, Src inhibitors, and a compound KU0171309 had differential activity for NR4A2+ cells. EGFR and Src inhibitors appear to inhibit NR4A2 expression via blocking ERK signaling, which is known to regulate NR4A2 in cancer cells (23). KU0171309, however, inhibits NR4A2 through a different—possibly direct but currently unknown—mechanism. Inhibitors of downstream targets of EGFR/Src such as BRAF, MEK, STAT3, mTOR did not suppress NR4A2 level (Figures 4,5). In particular, BRAF inhibitors (Dabrafenib and Vemurafenib) were unable to suppress NR4A2 level and growth of H3118 cells despite depletion of p-ERK. Therefore ERK signaling does not seem to be the sole mechanisms to suppress NR4A2 level, and other collateral pathways related to EGFR/Src may have a role in NR4A2 regulation. For instance, EGFR and Src inhibitors may suppress PI3K and STAT3 along with ERK. NR4A2 is known to be regulated by PI3K-Akt signaling whose simultaneous suppression with ERK may significantly impact NR4A2 regulation (24).
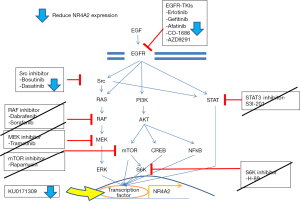
NR4A2 expression can also be regulated by prostaglandins, NFκB, Wnt/beta-catenin and other signals. As a transcription factor, NR4A2 regulates many downstream signal pathways such as osteopontin, angiogenesis, and maturation of regulatory T cells (13,14). Therefore further investigation regarding the mechanism of action of KU171309 may need to consider possible effects on these pathways, in addition to possible direct effects on NR4A2.
Recent studies demonstrated that EGFR-TKIs have selective activity for cells with somatic mutation in EGFR kinase domain that primarily drives cell growth (2,3). These driver/activating mutations in EGFR gene are now used in clinic to select patients who benefit from EGFR-TKIs. As a result of these observations, indications approved by FDA for such TKIs are currently restricted to patients with activating mutations. However, there are several lines of preclinical and clinical evidence to suggest the use of EGFR-TKIs for EGFR wild-type cancers. We and others previously reported that mucoepidermoid cell lines with CRTC1-MAML2 translocation had relatively high sensitivity to EGFR-TKIs in vitro (9,11). Wu et al. revealed that CRTC1-MAML2 chimera gene regulates EGFR ligand AREG which in turn leads to addiction to EGFR signaling, and that EGFR inhibitors were effective for in vitro and in vivo models (11). Whang et al. demonstrated that EGFR wild-type/LKB1 null NSCLC cells are relatively sensitive to EGFR-TKIs due to impaired energy metabolism and mitochondrial dysfunction, although they do not have activating EGFR mutation (25). Moreover, phase III clinical trials showed survival advantage for EGFR-TKIs over control in stage IV/recurrent NSCLC where a vast majority of patient population seemed EGFR wild-type (26,27). Besides activating EGFR mutations, our present study suggests that certain cell types (e.g., CRTC1-MAML2, LKB1 null) might possibly show collateral sensitivity to EGFR-TKIs due to suppression of pERK and downstream effects on NR4A2. This hypothesis awaits validation in future studies in vitro and in vivo.
To the best of our knowledge, this is the first study to identify agents targeting NR4A2 in human cancer cells. Most positive hits except KU0171309 suppressed NR4A2 level via depletion of p-ERK. Future studies must include validations regarding homologue selectivity (lack of effect on NR4A1 and NR4A3), determination of mechanism of action for KU0171309, and in vivo studies. These future investigations should yield insights that may facilitate academic and industry development of inhibitors targeting NR4A2, and possibly additional orphan nuclear receptors.
Acknowledgements
The authors thank Marilyn Nielsen, Kelly Vandiver, and Melinda Broward for administrative assistance. This work was partly supported by University of Kansas Cancer Center start-up fund (T Komiya).
Footnote
Conflicts of Interest: RP Perez is currently employed by Bristol-Myers Squibb; the other authors have no conflicts of interest to declare.
Ethical Statement: Ethics approval was not required for this study because no human or animal was involved.
References
- World Health Organization. Cancer statistics. Available online: http://www.who.int/cancer/en/
- Paez JG, Jänne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 2004;304:1497-500. [Crossref] [PubMed]
- Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med 2004;350:2129-39. [Crossref] [PubMed]
- Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med 2009;361:947-57. [Crossref] [PubMed]
- Yang JC, Wu YL, Schuler M, et al. Afatinib versus cisplatin-based chemotherapy for EGFR mutation-positive lung adenocarcinoma (LUX-Lung 3 and LUX-Lung 6): analysis of overall survival data from two randomised, phase 3 trials. Lancet Oncol 2015;16:141-51. [Crossref] [PubMed]
- Shaw AT, Kim DW, Nakagawa K, et al. Crizotinib versus chemotherapy in advanced ALK-positive lung cancer. N Engl J Med 2013;368:2385-94. [Crossref] [PubMed]
- Tonon G, Modi S, Wu L, et al. t(11;19)(q21;p13) translocation in mucoepidermoid carcinoma creates a novel fusion product that disrupts a Notch signaling pathway. Nat Genet 2003;33:208-13. [Crossref] [PubMed]
- Komiya T, Park Y, Modi S, et al. Sustained expression of Mect1-Maml2 is essential for tumor cell growth in salivary gland cancers carrying the t(11;19) translocation. Oncogene 2006;25:6128-32. [Crossref] [PubMed]
- Coxon A, Rozenblum E, Park YS, et al. Mect1-Maml2 fusion oncogene linked to the aberrant activation of cyclic AMP/CREB regulated genes. Cancer Res 2005;65:7137-44. [Crossref] [PubMed]
- Wu Z, Huang X, Feng Y, et al. Transducer of regulated CREB-binding proteins (TORCs) induce PGC-1alpha transcription and mitochondrial biogenesis in muscle cells. Proc Natl Acad Sci U S A 2006;103:14379-84. [Crossref] [PubMed]
- Chen Z, Chen J, Gu Y, et al. Aberrantly activated AREG-EGFR signaling is required for the growth and survival of CRTC1-MAML2 fusion-positive mucoepidermoid carcinoma cells. Oncogene 2014;33:3869-77. [Crossref] [PubMed]
- Komiya T, Coxon A, Park Y, et al. Enhanced activity of the CREB co-activator Crtc1 in LKB1 null lung cancer. Oncogene 2010;29:1672-80. [Crossref] [PubMed]
- Safe S, Jin UH, Morpurgo B, et al. Nuclear receptor 4A (NR4A) family - orphans no more. J Steroid Biochem Mol Biol 2016;157:48-60. [Crossref] [PubMed]
- Safe S, Jin UH, Hedrick E, et al. Minireview: role of orphan nuclear receptors in cancer and potential as drug targets. Mol Endocrinol 2014;28:157-72. [Crossref] [PubMed]
- Guo J, Zu G, Zhou T, et al. Clinicopathological significance of orphan nuclear receptor Nurr1 expression in gastric cancer. Clin Transl Oncol 2015;17:788-94. [Crossref] [PubMed]
- Wang J, Yang J, Zou Y, et al. Orphan nuclear receptor nurr1 as a potential novel marker for progression in human prostate cancer. Asian Pac J Cancer Prev 2013;14:2023-8. [Crossref] [PubMed]
- Inamoto T, Papineni S, Chintharlapalli S, et al. 1,1-Bis(3′-indolyl)-1-(p-chlorophenyl)methane activates the orphan nuclear receptor Nurr1 and inhibits bladder cancer growth. Mol Cancer Ther 2008;7:3825-33. [Crossref] [PubMed]
- Han Y, Cai H, Ma L, et al. Nuclear orphan receptor NR4A2 confers chemoresistance and predicts unfavorable prognosis of colorectal carcinoma patients who received postoperative chemotherapy Eur J Cancer 2013;49:3420-30. [Crossref] [PubMed]
- Maira M, Martens C, Philips A, et al. Heterodimerization between members of the Nur subfamily of orphan nuclear receptors as a novel mechanism for gene activation. Mol Cell Biol 1999;19:7549-57. [Crossref] [PubMed]
- Mullican SE, Zhang S, Konopleva M, et al. Abrogation of nuclear receptors Nr4a3 and Nr4a1 leads to development of acute myeloid leukemia Nat Med 2007;13:730-5. [Crossref] [PubMed]
- Ramirez-Herrick AM, Mullican SE, Sheehan AM, et al. Reduced NR4A gene dosage leads to mixed myelodysplastic/myeloproliferative neoplasms in mice. Blood 2011;117:2681-90. [Crossref] [PubMed]
- Han Y, Cai H, Ma L, et al. Expression of Orphan Nuclear Receptor NR4A2 in Gastric Cancer Cells Confers Chemoresistance and Predicts an Unfavorable Postoperative Survival of Gastric Cancer Patients With Chemotherapy. Cancer 2013;119:3436-45. [Crossref] [PubMed]
- Smith AG, Lim W, Pearen M, et al. Regulation of NR4A nuclear receptor expression by oncogenic BRAF in melanoma cells. Pigment Cell Melanoma Res 2011;24:551-63. [Crossref] [PubMed]
- Mahajan S, Saini A, Chandra V, et al. Nuclear receptor Nr4a2 promotes alternative polarization of macrophages and confers protection in sepsis. J Biol Chem 2015;290:18304-14. [Crossref] [PubMed]
- Whang YM, Park SI, Trenary IA, et al. LKB1 deficiency enhances sensitivity to energetic stress induced by erlotinib treatment in non-small-cell lung cancer (NSCLC) cells. Oncogene 2016;35:856-66. [Crossref] [PubMed]
- Soria JC, Felip E, Cobo M, et al. Afatinib versus erlotinib as second-line treatment of patients with advanced squamous cell carcinoma of the lung (LUX-Lung 8): an open-label randomised controlled phase 3 trial. Lancet Oncol 2015;16:897-907. [Crossref] [PubMed]
- Cappuzzo F, Ciuleanu T, Stelmakh L, et al. Erlotinib as maintenance treatment in advanced non-small-cell lung cancer: a multicentre, randomised, placebo-controlled phase 3 study. Lancet Oncol 2010;11:521-9. [Crossref] [PubMed]


