Biology and clinical significance of circulating tumor cell subpopulations in lung cancer
Introduction: “liquid biopsy” as blood-based biomarkers
According to the latest global report on cancer instances and mortality, lung cancer accounts for 13% of new cancer diagnoses and 19.4% of cancer-related deaths in both men and women (1). A recent study on cancer mortality rates in Europe predicts that lung cancer deaths in women will be comparable to breast cancer deaths (92,300 vs. 92,600, respectively) for 2017 (2). Despite improvements in drug development and treatments for non-small cell lung carcinoma (NSCLC) patients, five year survival rates remain unacceptably low at ~15%, with many patients presenting with late stage disease and secondary tumors at distant sites (such as the liver, brain, bone and contra lateral lung) at initial diagnosis. Unfortunately, lung cancer patients presenting with distant metastases have significantly reduced progression-free survival (PFS) and overall survival (OS) (3). Recently, a number of actionable genetic driver mutations and translocations have been identified for targeted therapy for NSCLC patients. Among these gene alterations are echinoderm microtubule-associated protein-like 4 (EML4)-anaplastic lymphoma kinase (ALK) fusion, epidermal growth factor receptor (EGFR) activating (exon 21 L585R point mutations and exon 19 deletions) and T790M mutations [confer resistance to first and second generation tyrosine kinase inhibitors (TKIs)], and ROS Proto-Oncogene 1 (ROS1) translocations. Furthermore, there is tremendous excitement about the potential of immunotherapy to treat NSCLC patients. For the first time, a substantial number of patients with advanced NSCLC have benefited greatly from immunotherapies, experiencing durable remissions and prolonged survival (4). Thus in recent years, checkpoint inhibitors have become an important tool for treating advanced NSCLC. The currently approved immunotherapies in NSCLC target PD-1, whereby the PD-1 inhibitors target an immune checkpoint found on T-cells. Cancer cells can use the PD 1 pathway to deactivate T cells and escape from the immune system. However, not everyone benefits from immunotherapy and a good predictive biomarker is still missing and urgently needed.
By identifying and tracking the genetic changes in primary and secondary tumors, patients can be stratified for the most efficient therapeutic regimen on the basis of screening of known biomarkers. Although solid biopsies remain the standard approach for confirming tumor histology and staging, there are limitations to their use. Namely, (I) depending on the tumor location it may not be possible to take a biopsy, (II) serial biopsy or biopsy of metastatic sites is often not feasible and (III) solid biopsies do not fully capture the heterogeneity of the tumor. A less invasive approach, involving a standard blood draw or so-called “liquid-biopsy”, can facilitate multiple sampling both prior to and during treatment without risk to the patient. Importantly, genetic analysis can be carried out on liquid biopsies thus providing a ‘real-time’ snapshot of tumor genotype evolution and the efficacy of treatment response on a patient-to-patient basis.
”Liquid biopsy” is an umbrella term that includes, but is not limited to, molecular analysis of CTCs, circulating tumor DNA (ctDNA), tumor-derived exosomes, and circulating miRNAs. Such blood-based biomarkers may provide an important insight into tumor heterogeneity and the opportunity to investigate the biological mechanisms driving metastatic initiator cells. Therefore the benefit of a liquid biopsy over a solid biopsy is that patient screening can be carried out in a non-invasive manner to better stratify patients for therapeutic interventions and monitor patient response.
To date, a higher number of studies investigating the clinical relevance of liquid biopsies have performed analyses on CTCs or ctDNA. Although the isolation of ctDNA is less challenging and thus easier to implement in a clinical setting, analysis of CTCs offers an opportunity to investigate individual clones on a single-cell level that originate from distant tumor sites in metastatic disease. Furthermore, CTCs represent living cells with the potential of forming metastasis and thus allows for functional analyses. The presence of CTCs in the blood of a cancer patient was first observed as far back as 1869 (5) and in the past 20 years research in the field is rapidly growing to investigate the potential of CTCs in cancer diagnostic and prognostic to the clinic. As evidence of this, for lung cancer alone, there are >160 clinical trials listed on ClinicalTrials.gov which involve CTCs as investigative biomarkers. Furthermore, the successful generation of patient CTC-derived eXplant (CDX) models from SCLC (6) and NSCLC (7) patient CTCs heralded an exciting new phase of research in the field of liquid biopsy. These CDX models offer a dynamic system for better understanding drug resistance mechanisms and testing new drugs and treatment combinations.
Despite such developments, the detection and characterization of NSCLC CTCs from background noise of normal blood cells remains challenging, especially when inter- and intra-patient heterogeneity is taken into consideration. This problem could potentially be mitigated by identifying a broader spectrum of molecular markers that could be utilized for CTC enrichment. In this review we look at a number of protein and genetic markers which may contribute to the biology and clinical significance of various CTC subpopulations in lung cancer.
Biology and detection of CTCs in NSCLC
The clinical utility of CTCs is not confined to their enumeration. CTCs lend themselves well to molecular characterization such as proteomic, genomic (mutation and copy number analyses) or even mRNA and methylation analysis at a single cell level, due to the stability conferred by the whole cell structure of CTCs. Furthermore, the CTC population presents a snapshot of clonal diversity from cells originating from not only primary but also metastatic tumor sites, which cannot be accurately determined by ctDNA analysis. While ctDNA offers the advantage of not requiring a pre-enrichment step, the specific antigens expressed on the surface of circulating tumor cells (CTCs) can also be utilized for enrichment of CTCs from the background of blood cells found in the circulation. The ongoing challenge in the ctDNA field is identifying tumor derived ctDNA from total circulating free DNA (cfDNA) which is also released into the circulation by necrotic and apoptotic healthy cells (8).
The isolation of CTCs also presents a challenge due to the very low numbers of CTCs (usually between 1–100 CTCs/7.5 mL of blood) compared to between 106 and 108 leukocytes. Thus, CTC isolation generally requires two steps: first is the enrichment of these rare cells from background blood cells and a second downstream assay, which further characterizes the enriched CTCs in a sensitive and specific manner (9). Isolation of CTCs from the whole blood can be done using the physical properties of the cell, such as size and density and also migratory properties. There are many devices and assay systems commercially available, some of which are approved as clinical diagnostics that have enabled the identification, enumeration and analysis of CTCs and nucleic acids. The most well-known device is the gold-standard, U.S. Food and Drug Administration (FDA)-cleared CellSearch® system (Janssen Diagnostics, LLC, Raritan, USA) for enumeration and isolation of CTCs from breast, castration-resistant prostate and colorectal cancers (10-12). This system identifies CTCs based on binding to anti-EpCAM ferromagnetic microbeads (enrichment) and cytokeratin (CK) and CD45 expression (detection). To date, no detection threshold has yet been defined for lung cancers for diagnostic use with the CellSearch® system. A growing number of EpCAM-independent labelling technologies detect CTCs based on their physical traits, such as size and plasticity, and negative depletion of leukocytes. Among the commercially available microfluidic platforms, Parsortix (Angle Plc, UK) and, recently, ClearCell1 (Clearbridge BioMedics Pte Ltd, Singapore), are capable of isolating CTCs in an EpCAM-independent manner based on their size and plasticity. Filtration devices, such as isolation based on size of epithelial cells (ISET) (RareCell Diagnostics, France) capture CTCs based on their size (>4 µm), excluding smaller blood cells found in the circulation. Other established methods for isolating CTCs from whole blood samples include density gradients (e.g., Ficoll) and negative depletion of CD45-expressing leukocytes using magnetic-activated cell separation (MACS) beads (Miltenyi Biotec GmbH, Germany) or the RosetteSep kit (StemCell Technologies, Canada). As this review aims to focus predominantly on the various subpopulations of CTCs in lung cancer, we will not be discussing in detail the technologies available for CTC enumeration and detection as they have been comprehensively reviewed in more depth elsewhere (9,13-21).
Heterogeneity and subpopulations of CTCs
Unlike other cancer entities, such as breast and prostate, it remains a challenge to detect CTCs in NSCLC. Interestingly, this is not the case for small-cell lung cancer (SCLC) CTCs which are more numerous and have shown a significant association with PFS and OS (22). When one considers the highly metastatic nature of NSCLC tumors, it is surprising to find so few CTCs in the circulation of NSCLC patients (15). It may be that NSCLC CTCs are present but evade detection due to the limitations of current technologies and protocols, namely the dependence of such approaches on EpCAM labelling (15).
Epithelial to mesenchymal transition and CTCs
For breast, prostate and colorectal cancers, detection of CTCs can be achieved with high sensitivity and specificity using standard epithelial markers, including cell-surface EpCAM and pan Cytokeratin markers (CK8, 18 and 19) (23).
Nevertheless, some CTCs may evade detection due to loss of epithelial marker expression, especially in NSCLC were CTCs are generally difficult to detect. In order to enter the vasculature of the circulatory system and colonize distant organs to form metastases, epithelial tumor cells need to down-regulate their epithelial properties and acquire stem-like features to enhance cell mobility and invasiveness (24). This transition is referred to as an epithelial-mesenchymal transition (EMT) and is often associated with tumor aggressiveness and resistance to therapeutic intervention (25,26). EMT is characterized by the down-regulation of epithelial markers, such as E-cadherin and EpCAM, and by the expression of mesenchymal markers, such as N-Cadherin (also referred to as CDH2 or neural-cadherin) (27) and vimentin, including transcriptional factors Snail, Zeb-1 and TWIST (28,29). In light of this, it may be likely that CTCs in NSCLC patients have more mesenchymal characteristics and thus are overlooked by EpCAM- or epithelial maker-dependent technologies.
It has been observed in NSCLC tumor tissues that an upregulation of N-cadherin and vimentin occurs along the peripheral leading-edge compared to central tumor tissue collected from the same patient (30). From 25 NSCLC patient samples analyzed by immunohistochemical staining, Mahmood et al. showed a significant correlation between the expression of N-cadherin and vimentin and advanced tumor stage. Isolation of CTCs from NSCLC by ISET filtration combined with immunofluorescent imaging of mesenchymal markers (N-cadherin and vimentin) and epithelial markers (EpCAM and pan-Cytokeratin) showed that CTCs can be identified on the basis of mesenchymal marker expression (31,32). Moreover, CTCs, isolated by CD45-depletion, from 43 NSCLC patients showed an association between N-cadherin positivity and significantly shorter PFS (5 vs. 8 months, P=0.03, HR =2.63) (33). In this study, heterogeneous subpopulations of CTCs detected included CTCs that had the following features: N-cadherin+/CK–/CD45–; CK+/N-cadherin−/CD45−; CK+/EpCAM+/CD45– and CK+/N-cadherin+/CD45−. Expression of N-cadherin and vimentin has also been documented in CTCs as having an association with advanced disease in breast and prostate cancer patients (34,35).
In support of the prognostic relevance of mesenchymal markers, expression of vimentin has previously been linked to poor prognosis in NSCLC (36,37). NSCLC CTCs isolated by EpCAM-independent ISET filtration showed positive expression of cell surface vimentin and higher levels of vimentin mRNA in NSCLC CTCs compared to SCLC CTCs (7). Within the same study, tumor tissues from NSCLC CTC-derived CDx models showed stronger immunohistochemical staining of vimentin than observed in SCLC CDx tumors. These findings offer further support to the hypothesis that NSCLC CTCs may be more difficult to detect than SCLC CTCs due to the mesenchymal-like features of the former. However, vimentin is not an exclusive cancer-associated marker but is also expressed by leukocytes. In light of this, vimentin expression alone may not provide a suitable basis for enriching CTCs from surrounding blood cells but, instead, may be better suited to the characterization of CTCs.
EGFR mutations status and CTCs
Approximately 15% of lung adenocarcinoma patients in Europe harbor activating EGFR mutations, with this frequency increasing to ~40% in Asia (38). On this basis, adenocarcinoma patients are routinely tested for EGFR mutations to determine their suitability for targeted therapy. Despite initial good responses to TKIs, the majority of patients relapse due to the occurrence of new resistant clones. The most common resistance mechanism is acquirement of an additional T790M point mutation in the EGFR gene which accounts for ~50% of cases of acquired resistance towards EGFR TKIs (39). The clinical power of assessment of the EGFR mutation status in liquid biopsy has been shown in several studies and today an FDA-approved diagnostic assay [cobas® EGFR Mutation Test v2 (Roche Molecular Systems, Inc., CA, USA)] is commercially available for detection EGFR mutations in ctDNA.
Although detection of EGFR mutations in ctDNA is undoubtedly a powerful tool, a number of publications have shown that ctDNA and CTCs are complementary assays enabling, for example, a more comprehensive screening of the different resistance mechanisms (40,41). Sundaresan et al., investigated 40 patients with EGFR-mutant tumors progressing on EGFR TKI therapy. T790M genotypes were successfully obtained in 30 (75%) tumor biopsies, 28 (70%) CTC samples, and 32 (80%) ctDNA samples. When the results from ctDNA and CTC were combined a positive result was obtained in all patients, with T790M mutations detected in an additional 35% patients in whom the concurrent biopsy was negative or indeterminate (41).
The value of CTCs as a solitary method for detecting EGFR mutations has previously been controversially discussed (42). However, the rapid technological development of reliable single cell assays have substantially increased assay sensitivity and currently enables analysis of mRNA and DNA of single cells (43-45). Accordingly, Liu et al., recently performed a meta-analysis on 170 patients from eight studies with EGFR mutation data on CTCs. A high diagnostic performance of CTCs in detecting EGFR mutations was shown (AUSROC =0.99, 95% CI: 0.98–1.00) (46). Similarly, in a recent single-center study, serial sampling from 120 NSCLC patients demonstrated that activating EGFR mutations (L858R point mutations and exon 19 deletions) detected in CTCs had an overall 94% concordance rate with initial patient-matched solid tumor biopsies (47). For this CTC assay, CTCs were enriched from background blood cells by CD45 depletion and DNA profiling carried out by droplet digital PCR (ddPCR) with a reported specificity and sensitivity of 47% and 64%, respectively. In a pilot study a spiral microfluidic device allowing high throughput, selective picking and isolation of single CTC was used to detect T790M mutations in seven NSCLC patients. This method also showed concordant results with the biopsy sample for six samples (48). The presence of the EGFR point mutation, T790M, in CTCs isolated from 27 NSCLC patients by CTC-chip was shown in another study to coincide with disease progression and reduced response to targeted therapies (17). Moreover, the authors showed a significant concordance (92%) between expression of an activating EGFR mutation in CTCs and patient-matched tumor tissues. Marchetti et al., used the CellSearch CTC analysis platform combined with next-generation sequencing (NGS) for EGFR mutation detection. EGFR mutations were detected in 31 (84%) patients, corresponding to those present in matching tumor tissue (49). Finally, a recent publication by Park et al., used a nanotechnology based CTC detection platform in NSCLC patients (50). Using this highly sensitive device the authors were able to accurately detect each patient’s known mutation on single CTCs. This mRNA based method also showed that NSCLC patient CTCs heterogeneously express MET as another bypass mechanisms mediating resistance to EGFR TKIs.
Despite the still rather challenging technical requirements for sensitive mutation detection on CTCs (45), detection of EGFR mutations on CTCs, instead of difficult to acquire solid biopsies, can clearly facilitate the monitoring a patient’s response to target therapy and identify early signs of drug resistance.
Detection of anaplastic lymphoma kinase and c-ros oncogene 1 rearrangements on CTCs
EML4-ALK fusions are found in 1–5% of NSCLC patients (51) and traditionally these rearrangements are tested in tumor biopsies to determine if a patient is a suitable candidate for targeted therapy against ALK. In recent developments, a growing number of studies have demonstrated that ALK-rearrangements can also be detected in CTCs of NSCLC patients. In an earlier study, CTCs from 18 ALK+ and 14 ALK− NSCLC patients were enriched from whole blood using a combination of ISET filtration for CTC enrichment and the FDA-approved Vysis ALK Break Apart FISH Probe Kit (Abbott Molecular, USA) for detection of ALK-rearrangements (31). Using this approach, ≥4 ALK-rearranged CTCs per 1 mL of blood were found in all ALK+ NSCLC patients (mean value of 11 CTCs per mL) but only one CTC with an ALK-rearrangement was detected in the ALK- NSCLC cohort. The concordance between CTCs and corresponding tumor samples was 99.9% (quantified by the κ coefficient). Interestingly, CTCs from six representative ALK+ patients expressed the mesenchymal markers, vimentin and N-cadherin; however, expression levels of these EMT markers were heterogeneous in tumors from five of the six patients. In a subsequent study, Pailler et al. utilized a semi-automated microscopy approach in combination with ISET to identify ALK rearrangements with filter-adapted-FISH (FA-FISH) revealing that the number of FISH spots per ALK+ NSCLC CTC increased with the use of optimized settings (z-stacks spaced 0.6 µm apart) during scanning (32).
NSCLC CTCs enriched using a spiral microfluidic chip and analyzed with the Vysis ALK Break Apart FISH Probe Kit, 25.4% of CTCs (n=177) were positive for ALK-rearrangements, with ALK-signal also detected in 54% of 200 cells from FFPE samples (52). Using a novel device for CTC-capture, the NanoVelcro Chip, He et al. demonstrated that the ALK-rearranged CTC ratio correlated to pTNM staging in ALK-positive advanced NSCLC patients (53). Within the cohort of 21 ALK+ positive patients, the percentage of ALK-rearranged CTCs ranged from 19–96%, with mean percentages of 64.76±24.03%. A further study established that ALK+ CTCs counts were higher in ALK+ patients (n=14; 3–15 cells per 1.88 mL of blood) compared to ALK- patients (n=12; 0–2 cells per 1.88 mL of blood) (54). ALK positivity in NSCLC CTCs was recapitulated in all tumor tissue biopsies tested from the 14 ALK+ patients. Interestingly, it was also reported that one patient undergoing crizotinib treatment had increasing numbers of ALK-positive CTCs corresponding to partial response and progression, thus demonstrating the potential for ALK+CTC detection as a diagnostic companion. Of further interest, the successful expansion of CTCs from an ALK-positive lung adenocarcinoma patient revealed a drug resistant mutation (L1196M on the ALK gene) and responded to in vitro ceritinib treatment (IC50 of 1,664 nM) with higher efficiency than crizotinib (IC50 of 2,268 nM) (55). These findings highlight the possibility of utilizing ALK+ CTCs for ex vivo drug testing and selection of effective drug treatments for ALK-positive patients.
Despite affecting only 1% of NSCLC cases, patients with ROS1 gene fusions respond well to targeted therapy with crizotinib in clinical pilot trials (56). Pailler et al. isolated CTCs from four crizotinib treated patients with ROS1 rearrangements (57). Using a similar method described in their ALK study (31), ROS1 fusions were detected in CTCs by FA-FISH and revealed heterogeneous levels of ROS1 copy numbers in the CTCs analyzed. Interestingly, patient tumor tissues had lower ROS1 copy numbers compared to levels detected in CTCs of corresponding patients. Moreover, a reduction in ROS1 fusions was observed in the CTCs of two patients who responded to treatment with crizotinib, whereas ROS1 fusion levels remained stable in one patient non-responsive to crizotinib. Although limited by its small size, this study demonstrated the utility of CTCs in tracking patient response through analysis of biomarkers in the blood.
Programmed death ligand-1 expression on CTCs
Another notable CTC biomarker of interest includes programmed death ligand-1 (PD-L1). Advancements in immunotherapy most notably include the approval of programmed death-1 (PD-1) checkpoint inhibitors, nivolumab and pembrolizumab, for second line treatment of NSCLC patients (58,59). Just recently pembrolizumab was approved also as a first line therapy among patients with high PD-L1 expression. Although the prognostic role of PD-L1 expression on primary NSCLC tumor tissue has long been considered controversial, a recent meta-analysis clearly indicated a poor prognosis for patients with PD-L1 overexpression (60). Despite this, only few studies so far have reported the clinical relevance of PD-L1 expression on CTCs and response to immunotherapy.
In a recent study Nicolazzo et al. investigated PD-L1 expression on CTCs in 24 patients receiving nivolumab (61). Despite showing a significance between PD-L1 positivity and poor outcome in late stage treatment (6 months) of NSCLC patients, the authors highlighted a lack of distinction between PD-L1 positivity and outcome at baseline and early treatment (3 months) in this small patient cohort (61). Evidence for PD-L1 expression on circulating metastatic breast cancer cells (HR+ and HER2−) was also reported after analysis of EpCAM+ subpopulations of CTCs using the CellSearch system (62). However, Mazel et al. also observed considerable inter-patient variability in the population of PD-L1+ CTCs.
Recently, it has been demonstrated that PD-L1 expression is not restricted to tumor cells (63). Schehr et al. assessed the contribution of CD11b+ myeloid cells in false positive identification of CTCs and the subsequent impact on interpretation of PD-L1 positivity in CTCs. CTCs from 19 NSCLC patients were enriched from CD45-depleted peripheral blood mononuclear cells (PBMCs) using anti-CD45 MACS beads (Miltenyi) and fluorescently stained for CD45, panCK and the myeloid marker, CD11b+. By means of FACS sorting, they showed that mean expression of PD-L1 in NSCLC patient CTCs reduced substantially when CD11b+ events were removed from the overall CTC population, thus highlighting the need for more rigorous exclusion markers that may otherwise contribute to the false positive identification of PD-L1+ CTCs.
The role of nuclear PD-L1 (nPD-L1) expression was also reported in vimentin positive CTCs of metastatic colorectal and prostate cancer patients (64). Following enrichment by CD45 depletion and subsequent positive selection using a novel cell-surface vimentin (CSV) antibody (37), vimentin+ CTCs from colon cancer patients (n=62) and prostate cancer patients (n=30) were assessed for nPD-L1 expression by immunofluorescent staining. The findings showed a significant negative impact of nPD-L1+ CTCs on OS of colorectal cancer patients but no substantial effect on PFS. The inverse was, however, true for prostate cancer patients, with nPD-L1+ CTCs associated with poor PFS but no effect on OS. While the robustness of PD-L1 as a prognostic biomarker for patient stratification in NSCLC requires further investigation, standardization of PD-L1 detection protocols may go some way in addressing this.
Folate Receptor alpha expression on CTCs
Another molecular target of interest is the cell-surface glycoprotein, folate receptor alpha (FRα) of the FR family (FRα, FRβ, FRγ and FRδ). Responsible for the transport of folate into the cell, FRα is the most studied isoform owing to its expression on malignant cells of epithelial origin versus negligible expression on normal cells. Importantly, with the exception of activated monocytes and CTCs, FR is not expressed on any other cells found in the circulatory system (65). In the context of primary lung cancer tissue, expression of FRα is capable of distinguishing lung adenocarcinomas (higher FRα positivity) from squamous cell carcinomas (lower FRα positivity) in an immunohistological setting (66,67). The clinical relevance of FRα was also demonstrated by an association between FRα positivity and improved OS in patients with resected adenocarcinomas (67,68). Moreover, a monoclonal antibody targeting FRα, farletuzumab, is well tolerated in a clinical setting (69) and has shown promising results, in combination with chemotherapy, in clinical trials for ovarian cancer (70) and NSCLC (71).
A recent study investigated the prevalence of FR+ CTCs in NSCLC using a ligand-target PCR (LT-PCR) approach (71). CTCs were isolated from peripheral blood cells by CD45-depletion and anti-EpCAM magnetic beads were subsequently used to separate CTCs into either an EpCAM+ fraction or EpCAM− fraction. Interestingly, FRα was expressed on both EpCAM+ and EpCAM− CTCs, with EpCAM− cells showed a higher proportion of FRα positive CTCs compared to EpCAM+ cells (72). Furthermore, the presence of FRα on CTCs, using the LT-PCR detection assay, has consistently demonstrated a robust correlation with disease stage and thus may be considered to have potential clinical utility (73,74). In support of this, a recent study quantifying FR+ CTCs in 162 NSCLC patients showed that 77% of the patients had ≥8.70 FR+-CTC units/3 mL blood which significantly decreased following surgical resection (75). Importantly, a clear distinction was reported between individuals with lung cancer (n=197), benign lung disease (n=119) and healthy controls (n=52), thus supporting the potential prognostic relevance of FR+-CTCs in NSCLC. When compared with several established clinical biomarkers, FR+-CTC showed the highest diagnostic efficiency. However, as with all other potential CTC markers, not all CTCs may express FRα. Therefore, it would be advisable to include FR with a broader spectrum of circulating biomarkers in order to capture a higher proportion of the heterogeneous CTC population in NSCLC.
V-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog expression on CTCs
KRAS mutations occur in ~30% of lung adenocarcinomas (76) and although there are conflicting reports on its utility as a prognostic marker, KRAS G12C mutations have been shown to predict lower OS in late stage NSCLC (77,78). Moreover, 9 of 38 (24%) of lung adenocarcinomas refractory to either erlotinib or gefitinib harbored KRAS mutations, further highlighting the potential of KRAS mutational status as a predictive marker for therapeutic response (79). Yet despite numerous trials, a TKI against KRAS has yet to be approved for clinical use. Guibert et al. could detect KRAS mutations in the blood of 82% of patients (n=32) by extracting DNA from ctDNA and CTCs (80). This study used a digital droplet PCR (ddPCR) approach and showed that KRAS-mutated DNA was detected in 78% (n=25/32) of cfDNA samples but that the sensitivity of this assay reduced to 34.3% (n=11/32) in CTC DNA. Challenges in detecting KRAS in CTCs has previously been reported by Freidin et al., who also reported ctDNA-based assays to be more sensitive than CTC-based methods for mutant KRAS detection (81).
Detection of genetic alterations associated with resistance on CTCs
Apart from EGFR T790M mutations, other mechanisms for acquired TKI resistance include: amplification of MET (also known as the hepatocyte growth factor receptor alpha) (82), B-Raf proto-oncogene (BRAF) (83), and phosphatidylinositol 3-kinase, catalytic, alpha polypeptide (PIK3CA) (84), upregulation of Erb-B2 Receptor Tyrosine Kinase 3 (HER3) and AXL Receptor Tyrosine Kinase (Axl) (85).
MET is a target of particular interest as it has been reported to correlate with clinical progression and shorter OS in lung as well as many other cancer patients (86). It is also relevant in terms of liquid biopsy as MET expression in CTC-induced bone xenografts was shown to correlate with MET expression patterns in bone metastasis in breast cancer patients (n=3) (87). In a prospective trial, Yanagita et al. detected MET amplifications in CTCs from 3 of 39 available samples (8%) in erlotinib-treated NSCLC patients with progressive disease (40). No MET amplifications were found in CTCs from paired baseline samples. In a recent study of NSCLC patients (n=256), 72% of ISET- and CellSearch-enriched CTCs expressed MET with 65% of patient-matched FFPE tumor sections also positive for MET expression (93% concordance) (88). Despite a significant association between CTC number and PFS, this study showed no correlation between MET expression and PFS. In a separate study using a semi-quantitative RT-PCR approach to characterize CTCs from 22 NSCLC patients, Hanssen et al. reported only 14.3% of cases as positive for MET (89). In the same study, Hanssen et al. also showed 85.7% positivity for HER3, a member of the epidermal growth factor family, in NSCLC CTCs. This is of interest as, alongside MET, HER3 overexpression is a key player in driving the mechanism of TKI resistance (90).
The frequency of BRAF mutations in NSCLC is relatively low (<2%) with the BRAF V600E mutation accounting for 50% of these and linked to reduced OS (91,92). Despite the low frequency, monitoring this mutation in the blood could identify patients who may benefit from treatment with BRAF inhibitors, such as dabrafenib, which is currently under clinical development. In a small study, Guibert et al. tested DNA extracted from CTCs (isolated by ISET) and cfDNA of 6 patients with lung adenocarcinomas for the BRAF V600E mutation. Using ddPCR, CTCs from only one patient were found positive for BRAF V600E whereas the mutation was detected in the cfDNA of all 6 patients (93). However, this study was not significantly powered to draw substantive conclusions on whether CTCs are inferior to cfDNA for detecting BRAF mutations in NSCLC. Indeed, in a larger study of colorectal cancer CTCs, Kidess-Sigal et al. revealed a considerable concordance of 73.9% for BRAF mutations between 23 CTCs and ctDNA samples (94).
Although relatively rare [~4% of lung cancer cases (95)], the clinical importance of PIK3CA mutations in NSCLC cancer cases is evident from its role in acquired resistance to EGFR TKIs and response to PI3K inhibitors (84,96). To date, there are limited publications which describe the clinical utility of PIK3CA-positive CTCs in NSCLC. Nonetheless, detection of PIK3CA mutations in CTCs from 16 colorectal cancer patients showed a 77.8% correlation with the mutations found in patient-matched tumor tissues using PCR-based Sanger sequencing (94). Using a similar method, PIK3CA mutations in exon 9 and exon 20 were also detected by Gasch et al. in CTCs from HER2-negative breast cancer patients (97). In the only published study, to our knowledge, investigating PIK3CA expression in NSCLC CTCs, Hanssen et al. demonstrated that PIK3CA was expressed in 42.9% of CTCs enriched from 48 NSCLC patients (90). Moreover, detection of PIK3CA+ CTCs corresponded to patients who were chemo-naïve or had progressive disease compared to resected and non-progressive patients with metastases.
Discussion
To date, the potential for CTCs as surrogate markers for the detection of minimal residual disease is limited due to the challenges in detecting CTCs in NSCLC. Until a reliable and robust method for detecting CTCs can be established, setting a CTCs detection threshold, which coincides with diagnostic relevance and prognostic outcome, remains to be determined. Although the frequency of CTCs detected in NSCLC using the CellSearch and EpCAM-dependent approaches remains low, a high CTC count at either baseline or during therapy has been repeatedly shown in a number of studies to negatively impact patient survival rates (42,98). However, clinically relevant results have not only been obtained for CellSearch but for various other CTC assays. Consequently, a comprehensive meta-analysis of CTCs counts from 1,576 NSCLC patients, across 20 studies, reported a significant link between poor PFS and OS and the detection of CTCs in the blood (99). Yet different technologies and detection markers report highly variable CTC counts, also when two assays are performed in paralleled, indicating differences in both sensitivities but also the existence of heterogeneous subpopulations of CTCs in NSCLC.
In this review we introduce a number of alternative biomarkers, which contribute to the heterogeneous subpopulation of circulation tumor cells and which, furthermore, could be utilized to improve the detection and isolation of NSCLC CTCs from the blood circulatory system (Figure 1). A number of novel therapeutic agents are either under clinical development or are already approved for clinical use for targeting the majority of the markers discussed in this review. Here we have illustrated the diverse range of studies investigating not only dysregulation of protein expression but also genetic abnormalities in CTCs which, in many cases, showed a correlation with disease progression, patient outcome or both. For most of these markers larger independent studies are still, however, needed to assess their real clinical relevance as liquid biomarkers.
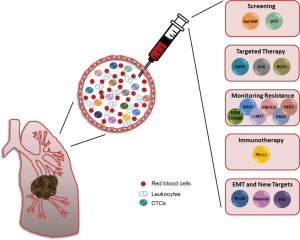
By identifying and characterizing blood-based biomarkers, clinicians could track tumor progression, monitor treatment response and make informed decisions regarding the management of the patient’s disease without the need for an invasive solid biopsy. Keeping in mind the considerable heterogeneity of NSCLC tumors, inclusion of a diverse range of targets for identification of CTCs in NSCLC increases the probability of detection thus providing insight into tumor burden and disease progression. Furthermore, a multi-targeted approach offers a unique opportunity to identify potential metastatic initiator cells and develop novel detection assays.
Conclusions
With the assistance of state-of-the-art technologies and the development of highly sensitive and specific assays, the identification and characterization of the dynamic subpopulations of CTCs in lung cancer, and indeed all cancer types, has the potential to deepen our understanding of the underlying mechanisms driving tumor evolution, metastases and drug resistance. By improving our understanding of these biological mechanisms we can develop novel diagnostic approaches and fundamentally improve therapeutic interventions. Importantly, the non-invasive nature of liquid biopsies allows for sequential sampling, with low risk to the patient, which is conducive to: ‘real-time’ monitoring of therapeutic response, early detection of resistance markers, prognosis and prediction of patient outcome. With such growing evidence in support of the clinical utility of liquid biopsies, the prognostic value of CTCs as surrogate biomarkers of disease progression cannot be underestimated.
Acknowledgements
Funding: The authors are supported by CANCER-ID, an Innovative Medicines Initiative Joint Undertaking under grant agreement n° 115749, resources of which are composed of financial contribution from the European Union’s Seventh Framework Programme (FP7/2007-2013) and EFPIA companies’ in kind contribution.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015;136:E359-86. [Crossref] [PubMed]
- Malvezzi M, Carioli G, Bertuccio P, et al. European cancer mortality predictions for the year 2017, with focus on lung cancer. Ann Oncol 2017;28:1117-23. [Crossref] [PubMed]
- Group NM-aC. Preoperative chemotherapy for non-small-cell lung cancer: a systematic review and meta-analysis of individual participant data. Lancet 2014;383:1561-71. [Crossref] [PubMed]
- Heigener D, Reck M. Exploring the potential of immuno-oncology-based treatment for patients with non-small cell lung cancer. Expert Rev Anticancer Ther 2015;15:69-83. [Crossref] [PubMed]
- Ashworth TR. A case of cancer in which cells similar to those in the tumours were seen in the blood after death. Med J Aust 1869;14:146-7.
- Hodgkinson CL, Morrow CJ, Li Y, et al. Tumorigenicity and genetic profiling of circulating tumor cells in small-cell lung cancer. Nat Med 2014;20:897-903. [Crossref] [PubMed]
- Morrow CJ, Trapani F, Metcalf RL, et al. Tumourigenic non-small-cell lung cancer mesenchymal circulating tumour cells: a clinical case study. Ann Oncol 2016;27:1155-60. [Crossref] [PubMed]
- Pantel K. Blood-Based Analysis of Circulating Cell-Free DNA and Tumor Cells for Early Cancer Detection. PLoS Med 2016;13:e1002205. [Crossref] [PubMed]
- Joosse SA, Gorges TM, Pantel K. Biology, detection, and clinical implications of circulating tumor cells. EMBO Mol Med 2015;7:1-11. [Crossref] [PubMed]
- Cristofanilli M, Budd GT, Ellis MJ, et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med 2004;351:781-91. [Crossref] [PubMed]
- de Bono JS, Scher HI, Montgomery RB, et al. Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin Cancer Res 2008;14:6302-9. [Crossref] [PubMed]
- Cohen SJ, Punt CJ, Iannotti N, et al. Prognostic significance of circulating tumor cells in patients with metastatic colorectal cancer. Ann Oncol 2009;20:1223-9. [Crossref] [PubMed]
- Chudziak J, Burt DJ, Mohan S, et al. Clinical evaluation of a novel microfluidic device for epitope-independent enrichment of circulating tumour cells in patients with small cell lung cancer. Analyst 2016;141:669-78. [Crossref] [PubMed]
- Fusi A, Metcalf R, Krebs M, et al. Clinical utility of circulating tumour cell detection in non-small-cell lung cancer. Curr Treat Options Oncol 2013;14:610-22. [Crossref] [PubMed]
- Hanssen A, Loges S, Pantel K, et al. Detection of Circulating Tumor Cells in Non-Small Cell Lung Cancer. Front Oncol 2015;5:207. [Crossref] [PubMed]
- Hvichia GE, Parveen Z, Wagner C, et al. A novel microfluidic platform for size and deformability based separation and the subsequent molecular characterization of viable circulating tumor cells. Int J Cancer 2016;138:2894-904. [Crossref] [PubMed]
- Maheswaran S, Sequist LV, Nagrath S, et al. Detection of mutations in EGFR in circulating lung-cancer cells. N Engl J Med 2008;359:366-77. [Crossref] [PubMed]
- Nagrath S, Sequist LV, Maheswaran S, et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature 2007;450:1235-9. [Crossref] [PubMed]
- Ozkumur E, Shah AM, Ciciliano JC, et al. Inertial focusing for tumor antigen-dependent and -independent sorting of rare circulating tumor cells. Sci Transl Med 2013;5:179ra47. [Crossref] [PubMed]
- Riethdorf S, Fritsche H, Muller V, et al. Detection of circulating tumor cells in peripheral blood of patients with metastatic breast cancer: a validation study of the CellSearch system. Clin Cancer Res 2007;13:920-8. [Crossref] [PubMed]
- Rolfo C, Castiglia M, Hong D, et al. Liquid biopsies in lung cancer: the new ambrosia of researchers. Biochim Biophys Acta 2014;1846:539-46.
- Huang CH, Wick JA, Sittampalam GS, et al. A multicenter pilot study examining the role of circulating tumor cells as a blood-based tumor marker in patients with extensive small-cell lung cancer. Front Oncol 2014;4:271. [Crossref] [PubMed]
- Pantel K, Alix-Panabieres C. Circulating tumour cells in cancer patients: challenges and perspectives. Trends Mol Med 2010;16:398-406. [Crossref] [PubMed]
- Bednarz-Knoll N, Alix-Panabieres C, Pantel K. Plasticity of disseminating cancer cells in patients with epithelial malignancies. Cancer Metastasis Rev 2012;31:673-87. [Crossref] [PubMed]
- Byers LA, Diao L, Wang J, et al. An epithelial-mesenchymal transition gene signature predicts resistance to EGFR and PI3K inhibitors and identifies Axl as a therapeutic target for overcoming EGFR inhibitor resistance. Clin Cancer Res 2013;19:279-90. [Crossref] [PubMed]
- De Craene B, Berx G. Regulatory networks defining EMT during cancer initiation and progression. Nat Rev Cancer 2013;13:97-110. [Crossref] [PubMed]
- Hazan RB, Qiao R, Keren R, et al. Cadherin switch in tumor progression. Ann N Y Acad Sci 2004;1014:155-63. [Crossref] [PubMed]
- Zeisberg M, Neilson EG. Biomarkers for epithelial-mesenchymal transitions. J Clin Invest 2009;119:1429-37. [Crossref] [PubMed]
- Alix-Panabières C, Mader S, Pantel K. Epithelial-mesenchymal plasticity in circulating tumor cells. J Mol Med (Berl) 2017;95:133-42. [Crossref] [PubMed]
- Mahmood MQ, Ward C, Muller HK, et al. Epithelial mesenchymal transition (EMT) and non-small cell lung cancer (NSCLC): a mutual association with airway disease. Med Oncol 2017;34:45. [Crossref] [PubMed]
- Pailler E, Adam J, Barthelemy A, et al. Detection of circulating tumor cells harboring a unique ALK rearrangement in ALK-positive non-small-cell lung cancer. J Clin Oncol 2013;31:2273-81. [Crossref] [PubMed]
- Pailler E, Oulhen M, Billiot F, et al. Method for semi-automated microscopy of filtration-enriched circulating tumor cells. BMC Cancer 2016;16:477. [Crossref] [PubMed]
- Nel I, Jehn U, Gauler T, et al. Individual profiling of circulating tumor cell composition in patients with non-small cell lung cancer receiving platinum based treatment. Transl Lung Cancer Res 2014;3:100-6. [PubMed]
- Armstrong AJ, Marengo MS, Oltean S, et al. Circulating tumor cells from patients with advanced prostate and breast cancer display both epithelial and mesenchymal markers. Mol Cancer Res 2011;9:997-1007. [Crossref] [PubMed]
- Bock C, Rack B, Huober J, et al. Distinct expression of cytokeratin, N-cadherin and CD133 in circulating tumor cells of metastatic breast cancer patients. Future Oncol 2014;10:1751-65. [Crossref] [PubMed]
- Tadokoro A, Kanaji N, Liu D, et al. Vimentin Regulates Invasiveness and Is a Poor Prognostic Marker in Non-small Cell Lung Cancer. Anticancer Res 2016;36:1545-51. [PubMed]
- Satelli A, Mitra A, Cutrera JJ, et al. Universal marker and detection tool for human sarcoma circulating tumor cells. Cancer Res 2014;74:1645-50. [Crossref] [PubMed]
- Midha A, Dearden S, McCormack R. EGFR mutation incidence in non-small-cell lung cancer of adenocarcinoma histology: a systematic review and global map by ethnicity (mutMapII). Am J Cancer Res 2015;5:2892-911. [PubMed]
- Rolfo C, Giovannetti E, Hong DS, et al. Novel therapeutic strategies for patients with NSCLC that do not respond to treatment with EGFR inhibitors. Cancer Treat Rev 2014;40:990-1004. [Crossref] [PubMed]
- Yanagita M, Redig AJ, Paweletz CP, et al. A Prospective Evaluation of Circulating Tumor Cells and Cell-Free DNA in EGFR-Mutant Non-Small Cell Lung Cancer Patients Treated with Erlotinib on a Phase II Trial. Clin Cancer Res 2016;22:6010-20. [Crossref] [PubMed]
- Sundaresan TK, Sequist LV, Heymach JV, et al. Detection of T790M, the Acquired Resistance EGFR Mutation, by Tumor Biopsy versus Noninvasive Blood-Based Analyses. Clin Cancer Res 2016;22:1103-10. [Crossref] [PubMed]
- Punnoose EA, Atwal S, Liu W, et al. Evaluation of circulating tumor cells and circulating tumor DNA in non-small cell lung cancer: association with clinical endpoints in a phase II clinical trial of pertuzumab and erlotinib. Clin Cancer Res 2012;18:2391-401. [Crossref] [PubMed]
- de Bourcy CF, De Vlaminck I, Kanbar JN, et al. A quantitative comparison of single-cell whole genome amplification methods. PLoS One 2014;9:e105585. [Crossref] [PubMed]
- van Dijk EL, Auger H, Jaszczyszyn Y, et al. Ten years of next-generation sequencing technology. Trends Genet 2014;30:418-26. [Crossref] [PubMed]
- Babayan A, Alawi M, Gormley M, et al. Comparative study of whole genome amplification and next generation sequencing performance of single cancer cells. Oncotarget 2016. [Epub ahead of print].
- Liu Y, Xing Z, Zhan P, et al. Is it feasible to detect epidermal growth factor receptor mutations in circulating tumor cells in nonsmall cell lung cancer?: A meta-analysis. Medicine (Baltimore) 2016;95:e5115. [Crossref] [PubMed]
- He J, Tan W, Ma J. Circulating tumor cells and DNA for real-time EGFR detection and monitoring of non-small-cell lung cancer. Future Oncol 2017;13:787-97. [Crossref] [PubMed]
- Yeo T, Tan SJ, Lim CL, et al. Microfluidic enrichment for the single cell analysis of circulating tumor cells. Sci Rep 2016;6:22076. [Crossref] [PubMed]
- Marchetti A, Del Grammastro M, Felicioni L, et al. Assessment of EGFR mutations in circulating tumor cell preparations from NSCLC patients by next generation sequencing: toward a real-time liquid biopsy for treatment. PLoS One 2014;9:e103883. [Crossref] [PubMed]
- Park SM, Wong DJ, Ooi CC, et al. Molecular profiling of single circulating tumor cells from lung cancer patients. Proc Natl Acad Sci U S A 2016;113:E8379-E86. [Crossref] [PubMed]
- Soda M, Choi YL, Enomoto M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature 2007;448:561-6. [Crossref] [PubMed]
- Khoo BL, Warkiani ME, Tan DS, et al. Clinical validation of an ultra high-throughput spiral microfluidics for the detection and enrichment of viable circulating tumor cells. PLoS One 2014;9:e99409. [Crossref] [PubMed]
- He W, Xu D, Wang Z, et al. Detecting ALK-rearrangement of CTC enriched by nanovelcro chip in advanced NSCLC patients. Oncotarget 2016.
- Tan CL, Lim TH, Lim T, et al. Concordance of anaplastic lymphoma kinase (ALK) gene rearrangements between circulating tumor cells and tumor in non-small cell lung cancer. Oncotarget 2016;7:23251-62. [Crossref] [PubMed]
- Zhang Z, Shiratsuchi H, Palanisamy N, et al. Expanded Circulating Tumor Cells from a Patient with ALK-Positive Lung Cancer Present with EML4-ALK Rearrangement Along with Resistance Mutation and Enable Drug Sensitivity Testing: A Case Study. J Thorac Oncol 2017;12:397-402. [Crossref] [PubMed]
- Gainor JF, Shaw AT. Novel targets in non-small cell lung cancer: ROS1 and RET fusions. Oncologist 2013;18:865-75. [Crossref] [PubMed]
- Pailler E, Auger N, Lindsay CR, et al. High level of chromosomal instability in circulating tumor cells of ROS1-rearranged non-small-cell lung cancer. Ann Oncol 2015;26:1408-15. [Crossref] [PubMed]
- Brahmer J, Reckamp KL, Baas P, et al. Nivolumab versus Docetaxel in Advanced Squamous-Cell Non-Small-Cell Lung Cancer. N Engl J Med 2015;373:123-35. [Crossref] [PubMed]
- Borghaei H, Paz-Ares L, Horn L, et al. Nivolumab versus Docetaxel in Advanced Nonsquamous Non-Small-Cell Lung Cancer. N Engl J Med 2015;373:1627-39. [Crossref] [PubMed]
- Pan ZK, Ye F, Wu X, et al. Clinicopathological and prognostic significance of programmed cell death ligand1 (PD-L1) expression in patients with non-small cell lung cancer: a meta-analysis. J Thorac Dis 2015;7:462-70. [PubMed]
- Nicolazzo C, Raimondi C, Mancini M, et al. Monitoring PD-L1 positive circulating tumor cells in non-small cell lung cancer patients treated with the PD-1 inhibitor Nivolumab. Sci Rep 2016;6:31726. [Crossref] [PubMed]
- Mazel M, Jacot W, Pantel K, et al. Frequent expression of PD-L1 on circulating breast cancer cells. Mol Oncol 2015;9:1773-82. [Crossref] [PubMed]
- Schehr JL, Schultz ZD, Warrick JW, et al. High Specificity in Circulating Tumor Cell Identification Is Required for Accurate Evaluation of Programmed Death-Ligand 1. PLoS One 2016;11:e0159397. [Crossref] [PubMed]
- Satelli A, Batth IS, Brownlee Z, et al. Potential role of nuclear PD-L1 expression in cell-surface vimentin positive circulating tumor cells as a prognostic marker in cancer patients. Sci Rep 2016;6:28910. [Crossref] [PubMed]
- Reddy JA, Haneline LS, Srour EF, et al. Expression and functional characterization of the beta-isoform of the folate receptor on CD34(+) cells. Blood 1999;93:3940-8. [PubMed]
- Nunez MI, Behrens C, Woods DM, et al. High expression of folate receptor alpha in lung cancer correlates with adenocarcinoma histology and EGFR [corrected] mutation. J Thorac Oncol 2012;7:833-40. [Crossref] [PubMed]
- O'Shannessy DJ, Yu G, Smale R, et al. Folate receptor alpha expression in lung cancer: diagnostic and prognostic significance. Oncotarget 2012;3:414-25. [Crossref] [PubMed]
- Iwakiri S, Sonobe M, Nagai S, et al. Expression status of folate receptor alpha is significantly correlated with prognosis in non-small-cell lung cancers. Ann Surg Oncol 2008;15:889-99. [Crossref] [PubMed]
- Kamen BA, Smith AK. Farletuzumab, an anti-folate receptor alpha antibody, does not block binding of folate or anti-folates to receptor nor does it alter the potency of anti-folates in vitro. Cancer Chemother Pharmacol 2012;70:113-20. [Crossref] [PubMed]
- Konner JA, Bell-McGuinn KM, Sabbatini P, et al. Farletuzumab, a humanized monoclonal antibody against folate receptor alpha, in epithelial ovarian cancer: a phase I study. Clin Cancer Res 2010;16:5288-95. [Crossref] [PubMed]
- Bronte G, Lo Vullo F, Pernice G, et al. Farletuzumab for NSCLC: exploiting a well-known metabolic pathway for a new therapeutic strategy. Expert Opin Investig Drugs 2015;24:125-32. [Crossref] [PubMed]
- Lou J, Ben S, Yang G, et al. Quantification of rare circulating tumor cells in non-small cell lung cancer by ligand-targeted PCR. PLoS One 2013;8:e80458. [Crossref] [PubMed]
- Chen X, Zhou F, Li X, et al. Folate Receptor-Positive Circulating Tumor Cell Detected by LT-PCR-Based Method as a Diagnostic Biomarker for Non-Small-Cell Lung Cancer. J Thorac Oncol 2015;10:1163-71. [Crossref] [PubMed]
- Wan JW, Gao MZ, Hu RJ, et al. A preliminary study on the relationship between circulating tumor cells count and clinical features in patients with non-small cell lung cancer. Ann Transl Med 2015;3:352. [PubMed]
- Wang L, Wu C, Qiao L, et al. Clinical Significance of Folate Receptor-positive Circulating Tumor Cells Detected by Ligand-targeted Polymerase Chain Reaction in Lung Cancer. J Cancer 2017;8:104-10. [Crossref] [PubMed]
- Cancer Genome Atlas Research N. Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014;511:543-50. [Crossref] [PubMed]
- Svaton M, Fiala O, Pesek M, et al. The Prognostic Role of KRAS Mutation in Patients with Advanced NSCLC Treated with Second- or Third-line Chemotherapy. Anticancer Res 2016;36:1077-82. [PubMed]
- Wood K, Hensing T, Malik R, et al. Prognostic and Predictive Value in KRAS in Non-Small-Cell Lung Cancer: A Review. JAMA Oncol 2016;2:805-12. [Crossref] [PubMed]
- Pao W, Wang TY, Riely GJ, et al. KRAS mutations and primary resistance of lung adenocarcinomas to gefitinib or erlotinib. PLoS Med 2005;2:e17. [Crossref] [PubMed]
- Guibert N, Pradines A, Farella M, et al. Monitoring KRAS mutations in circulating DNA and tumor cells using digital droplet PCR during treatment of KRAS-mutated lung adenocarcinoma. Lung Cancer 2016;100:1-4. [Crossref] [PubMed]
- Freidin MB, Freydina DV, Leung M, et al. Circulating tumor DNA outperforms circulating tumor cells for KRAS mutation detection in thoracic malignancies. Clin Chem 2015;61:1299-304. [Crossref] [PubMed]
- Engelman JA, Zejnullahu K, Mitsudomi T, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science 2007;316:1039-43. [Crossref] [PubMed]
- de Langen AJ, Smit EF. Therapeutic approach to treating patients with BRAF-mutant lung cancer: latest evidence and clinical implications. Ther Adv Med Oncol 2017;9:46-58. [Crossref] [PubMed]
- Wu SG, Chang YL, Yu CJ, et al. The Role of PIK3CA Mutations among Lung Adenocarcinoma Patients with Primary and Acquired Resistance to EGFR Tyrosine Kinase Inhibition. Sci Rep 2016;6:35249. [Crossref] [PubMed]
- Chong CR, Janne PA. The quest to overcome resistance to EGFR-targeted therapies in cancer. Nat Med 2013;19:1389-400. [Crossref] [PubMed]
- Finocchiaro G, Toschi L, Gianoncelli L, et al. Prognostic and predictive value of MET deregulation in non-small cell lung cancer. Ann Transl Med 2015;3:83. [PubMed]
- Baccelli I, Schneeweiss A, Riethdorf S, et al. Identification of a population of blood circulating tumor cells from breast cancer patients that initiates metastasis in a xenograft assay. Nat Biotechnol 2013;31:539-44. [Crossref] [PubMed]
- Ilie M, Szafer-Glusman E, Hofman V, et al. Expression of MET in circulating tumor cells correlates with expression in tumor tissue from advanced-stage lung cancer patients. Oncotarget 2017;8:26112-21. [PubMed]
- Hanssen A, Wagner J, Gorges TM, et al. Characterization of different CTC subpopulations in non-small cell lung cancer. Sci Rep 2016;6:28010. [Crossref] [PubMed]
- Sergina NV, Rausch M, Wang D, et al. Escape from HER-family tyrosine kinase inhibitor therapy by the kinase-inactive HER3. Nature 2007;445:437-41. [Crossref] [PubMed]
- Davies H, Bignell GR, Cox C, et al. Mutations of the BRAF gene in human cancer. Nature 2002;417:949-54. [Crossref] [PubMed]
- Marchetti A, Felicioni L, Malatesta S, et al. Clinical features and outcome of patients with non-small-cell lung cancer harboring BRAF mutations. J Clin Oncol 2011;29:3574-9. [Crossref] [PubMed]
- Guibert N, Pradines A, Casanova A, et al. Detection and Monitoring of the BRAF Mutation in Circulating Tumor Cells and Circulating Tumor DNA in BRAF-Mutated Lung Adenocarcinoma. J Thorac Oncol 2016;11:e109-12. [Crossref] [PubMed]
- Kidess-Sigal E, Liu HE, Triboulet MM, et al. Enumeration and targeted analysis of KRAS, BRAF and PIK3CA mutations in CTCs captured by a label-free platform: Comparison to ctDNA and tissue in metastatic colorectal cancer. Oncotarget 2016;7:85349-64. [PubMed]
- Kawano O, Sasaki H, Endo K, et al. PIK3CA mutation status in Japanese lung cancer patients. Lung Cancer 2006;54:209-15. [Crossref] [PubMed]
- Engelman JA, Chen L, Tan X, et al. Effective use of PI3K and MEK inhibitors to treat mutant Kras G12D and PIK3CA H1047R murine lung cancers. Nat Med 2008;14:1351-6. [Crossref] [PubMed]
- Gasch C, Oldopp T, Mauermann O, et al. Frequent detection of PIK3CA mutations in single circulating tumor cells of patients suffering from HER2-negative metastatic breast cancer. Mol Oncol 2016;10:1330-43. [Crossref] [PubMed]
- Krebs MG, Sloane R, Priest L, et al. Evaluation and prognostic significance of circulating tumor cells in patients with non-small-cell lung cancer. J Clin Oncol 2011;29:1556-63. [Crossref] [PubMed]
- Wang J, Wang K, Xu J, et al. Prognostic significance of circulating tumor cells in non-small-cell lung cancer patients: a meta-analysis. PLoS One 2013;8:e78070. [Crossref] [PubMed]


