Circulating tumor cells and CDX models as a tool for preclinical drug development
Introduction
Lung cancers are the most common tumor type worldwide, with 1.82 million cases in 2012 and 1.6 million deaths (19.4% of all tumor-related deaths) (1). Lung cancers can be subdivided into two main cytological subgroups: small cell lung cancer (SCLC) and non-small cell lung cancer (NSCLC) that is further subdivided into: adenocarcinoma, squamous cell carcinoma and large cell carcinoma (2). Early stage NSCLC and some cases of limited stage SCLC undergo surgery with curative intent (3-6). Systemic chemotherapy is the first-line treatment for advanced stage disease. SCLC patients are treated with a combination of a platinum-based agent and a topoisomerase inhibitor, whilst NSCLC patients usually receive a platinum-based agent in combination with pemetrexed or gemcitabine, unless specific targeted therapies are indicated (4,6). Significant progresses in the past few years with targeted therapies for NSCLC include gefitinib (EGFR tyrosine-kinase inhibitor) and crizotinib (ALK, ROS1, MET tyrosine kinase inhibitor) approved for the treatment of EGFR mutant and ALK positive patients, respectively (4,7). More and more targeted agents are under evaluation to inhibit specific driver mutations and deal with mechanisms of resistance (7). Examples are alectinib and osimertinib that target crizotinib refractory ALK-rearranged and EGFR T790M mutated NSCLC patients, respectively (8,9). In contrast, the lack of actionable driver mutations in SCLC led to disappointing results with novel therapeutics (10). Although, in a Phase I trial on recurrent SCLC patients, responses have been seen with a DLL3-antibody-drug conjugate (NCT01901653). Overall, despite these considerable efforts to improve clinical care of lung cancer patients, the median survival rates remain very low, with 5-year survival rates <7% and <17% in SCLC and NSCLC, respectively (11,12).
There are several reasons underlying the lack of success in improving clinical care of lung cancer patients. Most challenging is the late detection of disease coupled with the high mutation rate that drives intra- and inter-tumor heterogeneity in advanced lung cancers (13). As mentioned before, surgery is restricted only to patients with early stage disease and biopsy can be performed for patients with advanced stage diseases, however not always without risk to the patient. Moreover, biopsies are small and may not capture intratumor-heterogeneity sufficiently confounding a personalised medicine approach (14-16). Ongoing efforts are focused on improvement of minimally-invasive biomarkers, such as imaging and liquid biopsies, to aid early diagnosis and identify patients who would benefit from curative surgery, to stratify the patients and to predict or monitor response to therapies. A further obstacle to progress in the management of lung cancer patients is the availability of preclinical models that faithfully mimic the patient’s tumor to study biology, test novel therapies and characterize mechanisms of drug resistance.
In this review, we summarise and evaluate the existing preclinical models of lung cancers, comprising established cell lines, genetically engineered mouse models (GEMMs) and patient derived xenografts (PDXs). We also highlight the recent advances in the enrichment of patients’ circulating tumor cells (CTCs) used as a liquid biopsy to generate lung cancer CTC-derived preclinical models.
Pre-clinical models of lung cancer
Lung cancer cell lines were among the first to be generated (17). SCLC cells were first successfully cultured in 1971 (18) and at the present time approximately 300–400 cell lines have been established from SCLC and NSCLC (17). Since most SCLC patients are diagnosed with metastatic disease, curative surgery is rarely performed and consequently tumor tissue for clinical studies is rarely obtainable. As a result, most of our knowledge about the pathogenesis and biology of SCLC is derived from studies on SCLC cell lines (19,20). Most SCLC cell lines were derived from metastatic sites such as bone marrow aspirates and pleural effusions (21). On the contrary, most NSCLC cell lines were established from primary tumors with relatively low culture success rates compared with SCLC (21). Despite challenges in the culture of NSCLC cell lines, various important discoveries were achieved (22,23).
Regardless of the knowledge gained from studies on SCLC and NSCLC cell lines, this has not always translated to improvements in the clinic (24). There is some contention regarding the degree of drift that lung cancer cell lines undergo in prolonged culture (17). Whilst NSCLC cell lines are reported to be representative of the parental tumors and seem to maintain these characteristics over longer culture periods (25) and SCLC cell lines maintain neuroendocrine differentiation after establishment (26), SCLC cell lines established from patient-derived xenografts (PDX), after months in culture, exhibit significantly different gene expression profiles compared with PDX transcriptomes that faithfully matched tumor biopsies (27). Moreover, cancer cell lines grow in plastic where they lack the tissue structure and the stromal cells which have an essential role in tumor growth, angiogenesis, and metastasis (17).
GEMMs provide a complementary approach for lung cancer research, notably to study the biology and development of lung cancers in vivo (28). In 2003, Meuwissen et al. established a SCLC GEMM based on conditional alleles of TP53 and RB1, the two most frequently mutated tumor suppressor genes (29). Homozygous deletion of TP53 and RB1 in pulmonary neuroendocrine cells resulted in tumors mimicking the histopathology of human SCLC that have been used to study metastatic dissemination (29). This GEMM was used to examine the importance of other potential candidate tumor suppressors and oncogenes in SCLC (30); for instance, knocking out P130, a member of the Rb family on this background resulted in accelerated tumor growth, confirming P130 as a tumor suppressor in SCLC (31). Similar studies established PTEN and NOTCH as important tumor suppressors (32,33). GEMMs have also been used to investigate the origin of SCLC; Sutherland et al. demonstrated that targeting TP53 and RB1 in neuroendocrine cells as opposed to other cell types within the healthy lung using a calcitonin gene-related peptide (CGRP) virus most likely resulted in the development of SCLC (34).
The majority of GEMMs for NSCLC were generated for the adenocarcinoma subtype by expressing conditional alleles of KrasG12D or EgfrL858R thereby mimicking tumor development in approximately 40% of patients (35,36). These GEMMs helped to identify pathways involved in the development and maintenance of NSCLC, notably components of the MAPK pathway (37,38). Similar to GEMMs for SCLC, mouse models for NSCLC proved useful to perform cell-of-origin studies showing that different cell types can lead to the development of NSCLC (28). The first GEMM for lung squamous cell carcinoma in which conditional alleles of SOX2, PTEN, and CDKN2AB were deleted was described in 2016 (39).
Whilst GEMMs are undoubtedly elegant research tools that bring insights to tumor biology, they have had limited utility for pharmacology studies. In contrast to human tumors that harbour a high mutation load stemming from exposure to carcinogens in tobacco smoke, GEMMs do not exhibit this genetic complexity (30). GEMMs are biased based on the few genetic alterations introduced and, also, they take lot of time and money to be developed (40). Although SCLC GEMMs fail to recapitulate the spectrum of patient responses to platinum-based therapies, some NSCLC GEMMs have proven useful in assessing therapeutic efficacy of both targeted and cytotoxic agents (41).
PDXs are generated by implantation of tumor biopsies derived from SCLC (27) and NSCLC (42) patients into immune-deficient mice. PDXs better reflect the tumor-stroma interaction present in the primary tumor, despite the stroma derived from the host. Next generation sequencing (NGS) studies of SCLC PDX models proved useful in the dissection of the molecular landscape of this disease, where patient biopsies are often small and necrotic limiting research applications (33,43). Similarly, NSCLC PDX models have proven utility for testing novel therapeutics and to further understand disease biology (44). However, the limitation for PDX modelling of lung cancer resides in the challenges in obtaining sufficient quality and quantity of tumor biopsies as curative surgery is only performed for early stage disease (14-16,45). Further caveats are that PDX models are generated in immunodeficient mice and therefore inappropriate for studies of immunotherapeutics as the murine stromal interactions with the PDX are different from the human microenvironment (46). Immunodeficient mouse models closer mimicking human stromal components or engrafted with human immune system are currently being developed (47,48) and whilst expensive, could provide new opportunities to generate PDXs in a potentially more translatable model system.
To overcome the problem of tumor tissue availability to generate PDXs, we developed a new approach with a readily accessed clinical sample: a 10ml blood draw to generate CTC derived explants (CDXs) (49), as it will be discussed in the next sections.
CTCs as liquid biopsies
A readily accessible sample from the peripheral blood of cancer patients can provide tumor-derived resources, such as CTCs and circulating tumor DNA (ctDNA). These ‘liquid-biopsies’ have the potential to allow non-invasive molecular description of a patient’s tumor and monitoring of tumor growth and response to therapies (50). The utility of ctDNA in cancer patient management has already been extensively reviewed elsewhere (51-54). CTCs released from the primary tumor that survive in the bloodstream have the potential to seed at secondary sites to form metastases (55). Though technically more challenging than analysis of ctDNA, CTCs, with continuing improvements in detection and enrichment techniques (56,57), offer a broader repertoire of potential biomarkers (nucleic acids and proteins) and the opportunity to grow disseminating tumor cells in vitro and in vivo. Multiple studies have demonstrated that enumeration of EpCAM+ CTCs are prognostic in several cancer types including NSCLC and SCLC (58-62). The development of next generation sequencing platforms to dissect the genomic and transcriptomic profiles of single CTCs has provided a new window into the molecular landscape of cancers. Whole-exome sequencing of CTCs collected from prostate cancer patients identified 90% and 73% of mutations in early and advanced disease respectively, that matched single nucleotide variants found in the primary tumor and lymph node metastasis (63). Miyamoto et al. isolated an average of 6 prostate cancer CTCs per patient and performing single cell RNAseq profiling, they showed activation of the non-canonical Wnt signalling in patients progressing on anti-androgen receptor therapy (64). RNAseq, CNVs and mutational profiling were also performed on single CTCs isolated from the blood of melanoma, breast, and colorectal cancer patients (65-68). Immunomagnetic enrichment of melanoma-associated chondroitin sulphate proteoglycan+ CTCs and single cell comparative genomic hybridization showed chromosomal changes and aberrations typical of melanoma (65). With a dielectrophoresis-based device, Fabbri and co-workers were able to isolate single CTCs from colorectal cancer patients to perform whole genome amplification and targeted mutational sequencing (68). A similar study was done by Gasch et al, exploiting an EpCAM-dependent enrichment of CTCs from colorectal cancer patients (67). Both studies on colorectal cancer CTCs, showed intra- and inter-patient heterogeneity of genetic alterations in EGFR, KRAS and PIK3CA suggesting that molecular characterization of single CTCs is a useful tool to characterize the overall complexity of the tumor and to identify emerging drug resistant clones. Heterogeneity among individual CTCs was also observed in breast cancer patients where the transcriptomic profile of single CTCs showed expression of genes involved in EMT, metastasis and the AKT/mTOR pathway, suggesting potential therapeutic candidates for these patients (66). Molecular characterisation of CTCs from lung cancer patients has also been reported. Yeo and co-workers optimized a microfluidic device to isolate single cell CTCs from the blood sample of NSCLC patients and showed 100% concordance between EGFR mutations detected in CTCs and the primary tumor (69). Two recent studies performed single cell profiling of CTCs from both SCLC and NSCLC (70,71). In the first study, Carter et al. identified a CTC copy number alteration-based classifier to discriminate between chemorefractory and chemosensitive SCLC patient (70). In the other one, a panel of genes expressed in EpCAM+ CTCs derived from NSCLC patients showed to be predictive of progression free survival and highlights the relevance of the NOTCH1 pathway in these advanced stage NSCLC patients (71).
These studies highlight the potential for molecular characterization of single CTCs as clinical tools to support precision medicine, but so far, the direct clinical benefits for the patients have been limited. Moreover, whilst there have been significant increases to the overall survival of subgroups of NSCLC patients treated with EGFR and ALK targeted inhibitors, drug resistance and disease relapse are common and the effectiveness of precision medicine in its current format has been called into question (72,73). In part, this criticism stems from our relatively simple methods for predicting drug sensitivity based on genomic information only. Patients are generally selected only based on the presence or absence of a specific driver mutation, without taking into account the context in which the mutation is emerging. An outstanding example is the BRAF inhibitor vemurafenib. This targeted therapy recognizes constitutive active mutant BRAF V600 and showed impressive response in melanoma patients (74). Initial attempts to utilize vemurafenib in BRAF mutant colorectal cancer were unsuccessful, and it is now understood that both genomic background and cellular context can govern drug sensitivity (75). To help bridge the gap between our ability to detect mutations and to translate this knowledge into a clinically efficacious treatment, here we focus on the preclinical value of CTCs to perform functional downstream analysis, demonstrating how they can improve our understanding of lung cancer biology and how CTCs can be exploited to identify novel therapeutic options for those patients.
CDX models
PDX mouse models are the most useful models to identify the best treatment for the patient (76,77). Several groups have tried to optimize the protocol for lung cancer tissue sampling and processing to improve the success rate of lung cancer PDXs generation (44,78-80). However, as mentioned before, lung cancer PDX development remains challenging with limited tumor availability. The majority of NSCLC PDXs are derived from either pleural effusion or a small biopsy of the primary tumor collected during surgery of primary diagnosed NSCLC patients (44,78). SCLC patients, instead, rarely undergo surgery and biopsies are usually obtained via procedures that use radiologically guided approaches (e.g., percutaneously, bronchoscopically) and recently, ultrasound-guided transbronchial needle aspirates (EBUS-TBNA) of lymph nodes (14,79,80). In particular, EBUS-TBNA is considered safe with risk of complications in only 1% of cases (81). However, risks like mediastinitis, pericarditis and death have been reported (82,83). Open lung biopsy is a higher risk procedure but allow to biopsy larger pieces of tissue (14) and is only performed during surgery when there is a clinical benefit for the patient.
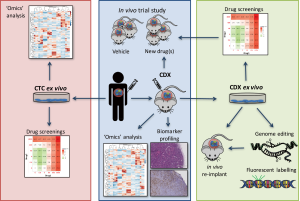
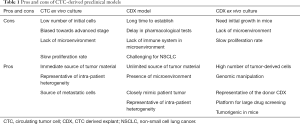
Conversely, CTCs can be easily collected from a blood withdrawal. It is non-invasive for the patient and samples can be collected independently of disease stage. Hodgkinson et al. demonstrated, for the first time, that CTCs enriched from the blood of SCLC patient are tumorigenic in immunocompromised mice, thus generating CTC-derived explants (CDXs) (49). The molecular profiles of CDXs demonstrated broad similarity with the primary tumor and matched single CTCs. Most importantly, from a pharmacological perspective, the response of CDXs to standard chemotherapies mirrored the donor patient’s response to the same treatment (49). Therefore, CDXs can be complementary to tumor biopsies and PDXs and can be a source of tumor material for research purposes (Figure 1). CDXs offer an opportunity to generate models for those patients that cannot undergo surgery or an alternative invasive procedure. Moreover, CDXs can be derived from CTCs collected at different time points during patient’s follow up, allowing the generation of paired models that recapitulate the patient’s tumor evolution (Table 1).
Full table
Unlike PDXs, CDXs are derived from a subset of tumor cells that have already acquired invasive behaviour and are disseminating in the bloodstream. Additionally, in metastatic patients, the degree of tumor heterogeneity captured by CTCs is likely to be higher than obtained by a single small biopsy. As a precedent, Heitzer and co-workers demonstrated the presence of alterations uniquely present in the CTCs of colorectal cancer patients. They confirmed that these alterations were not sequencing artefacts by ultra-deep sequencing of primary tumor and metastases and reveal that 85% of CTC ‘private mutations’ were also present at a minor subclonal level in the primary tumor or metastases (mutation frequencies 0.02–0.42) (84).
Despite the advantages of CDX models and the relatively high ‘take rate’ in SCLC, their generation for NSCLC patients has proven far more challenging. The prevalence of CTCs in SCLC is much higher than observed for NSCLC using the CellSearch platform that captures and enriches the EpCAM+ CTC subpopulation, as evidenced by the tenfold higher prognostic cut off of 50 CTCs vs. 5 CTCs respectively (61,62). With a median EpCAM+ CTC count of only four, it seems likely that the relative lack of CTCs in NSCLC contributes to difficulties in generating a CDX (85). However, the first and only NSCLC CDX model described to date (86), was generated from a patient whose parallel EpCAM+ CTC count was zero, and whose CTCs, enumerated by filtration were predominantly mesenchymal, consistent with epithelial to mesenchymal transition. A number of variables, including CTC enrichment and implantation methodologies, will likely require optimization to improve the ‘take rate’ of NSCLC CDX. We and others have recently reported that sampling blood from the draining pulmonary vein yields higher CTC numbers from stage I–III NSCLC patients immediately prior to tumor resection (1–3,093 vs. 0–4 CTCs in the peripheral blood) (87,88). Although the reason for this difference is not yet known, it is likely that the unfavourable environment in the bloodstream and CTC filtration via capillary beds could account for reduced CTC counts in the periphery. Alternatively, epithelial to mesenchymal transition may results in both loss of EpCAM expression and provide a survival advantage, thus allowing only the most aggressive CTCs to survive transit to the periphery.
CDXs maintain the histopathological characteristics of the donor tumor and growth dynamics with passage, therefore providing a renewable source of patient tumor material that can be exploited for multiple research purposes. SCLC CDXs can be generated from both chemosensitive and chemorefractory patients, especially for advanced stage disease. Moreover, as mentioned before, blood samples can be collected at several time points during the course of patients’ disease, allowing for the relatively easy generation of evolutionarily related longitudinal models. Such matched pairs of CDX allow comparison of tumor biology pre-treatment (baseline) and upon disease progression (relapse) providing unique models to interrogate intrinsic and acquired mechanisms of chemo-resistance. Preclinical models of rapidly progressing SCLC have hitherto been scarce. Given the substantial genomic heterogeneity in SCLC, a large panel of CDXs will now be required to provide a tractable discovery platform to identify new druggable targets and pathways via comprehensive multi-omic analyses.
Compared with PDX and CDX models developed in other cancer types (89), sadly the time frame for the generation of SCLC CDXs often exceeds the lifespan of the donor patient making the ‘one mouse, one patient’ paradigm incompatible for SCLC (90). Nevertheless with a large panel of CDX models, pharmacological studies to test novel therapeutics can be performed with parallel biomarker development; promising results can be translated to the clinic using pharmacodynamic and predictive CTC based biomarkers and CTC number as a surrogate of response.
Clearly, as CDX and PDX models are established in immunodeficient mice they cannot provide a panacea for therapy testing, and, to date, preclinical evaluation of immunotherapies relies on syngeneic and GEMMs (91,92). However, humanized mouse models are starting to enter the research field. These mice are normal immunocompromised mice in which a human immune system has been engrafted (48). The generation of CDX and PDX in these new models, may improve the possibility to predict the response to specific therapies, particularly the one linked to the immune system.
CTCs ex vivo culture
Several major obstacles in translating lung cancer CDX research remain, they cannot be generated from every patient, they take several months to establish, and in vivo pharmacology is expensive and time consuming. One way forward to improve the efficiency of mouse model generation, perform molecular analysis, and examine drug efficacy in a shorter time frame is the generation of CTC ex vivo cultures (Figure 1). There are several technologies that facilitate single-cell analysis, however, very few maintain the viability of the cells to perform downstream functional assays (56,93). In the past few years, several groups have tried to expand CTCs in culture from different cancer types. Yu et al. were able to derive CTC cell lines from off-treatment or progressing breast cancer patients (94). They used a microfluidic platform, CTC-iChip, to deplete normal blood cells and enrich un-manipulated CTCs. The selected CTCs were viable and grew for more than 6 months. They showed molecular and phenotypic similarities with uncultured primary CTCs and the derived donor tumor, validating their tumor origin. Moreover, they optimized a drug screening platform to screen small numbers of cells with high reproducibility. In this way, CTC culture can help predicting the response to specific drug combinations, and tailor the treatment of the patients accordingly. Similarly, Cayrefourcq et al. derived CTC cell lines from a metastatic colorectal cancer (CRC) patient that maintained some of the characteristics of the donor tumor, especially regarding aggressiveness and metastatic potential (95). One CTC cell line was also obtained from a prostate cancer patient and cultured as organoid in a 3D system (96) and two CTC cell lines were derived from extensive stage SCLC patients and kept in culture for more than 4 months (97).
These emerging studies show the potential of CTC ex vivo cultures as promising tool to assist delivery of personalised cancer therapy (98). For example, a recent study demonstrated the feasibility of CTC ex vivo expansion from metastatic CRC patients and subsequent drug screening in a very short time frame (less than a month) (99). However, some problems in CTC cultures are still prominent: the success rate is still low and generally biased towards advanced stage disease, the number of CTCs collected from a single blood sample is usually small, limiting the number of manipulations that could be done in the short term and longer term expansion may be often associated with phenotypic drift in culture (27) (Table 1).
CDX-derived ex vivo culture
The establishment of short term cell cultures generated from dissociated CDX tumors offers a useful intermediate between the CDX and the CTC ex vivo culture. In our group, we established a short-term ex vivo culture of cells derived from SCLC CDXs (Lallo et al., manuscript under review). These cells maintain similar transcriptomic and immunohistochemical profiles of the corresponding CDX and donor patient over several weeks and mimic the chemotherapy responses of the donor patient and their CDX model. Compared with direct culture of CTCs sampled from the patient where the number of CTCs is maximally 100 to 1,000 for NSCLC and SCLC respectively, the number of cells derived from a CDX is in the range of 15–30 million cells per tumor (Table 1). This approach where CDX derived cultures can be genetically manipulated now offers a rapid and tractable system to study SCLC biology, function test hypotheses and conduct drug screening of compound libraries. Upfront selection of promising therapeutics via short term CDX culture screens can then refine in vivo testing reducing the number of animals used for pharmacological studies in accordance with the 3Rs’ principals (100) to accelerate the identification of promising drug candidates (Figure 1).
Future perspectives and challenges
The identification of CTCs in the bloodstream of cancer patients has opened new opportunities in the study of these diseases. CTC enumeration can have an immediate clinical utility as independent prognostic biomarkers and in both SCLC and NSCLC they have been suggested to be predictive of chemotherapy response (62,85). On the other hand, single-cell CTC profiling can give insight into tumor heterogeneity both at the mutational and gene-expression levels (66-68,84,101). Therefore, the opportunity to study viable CTCs will allow functional validation of correlative hypotheses generated through single-cell molecular profiling. CTC ex vivo cultures are promising, but improvements in their establishment and subsequent in vitro expansion are required for routine use. In this regard, CDX models present an unprecedented opportunity to study the biology and mechanisms of chemoresistance, especially in SCLC, which could ultimately lead to the discovery of novel synthetic lethality and targeted therapy approaches. One key advantage of CDXs is the ability to generate longitudinal models made during disease evolution, allowing analysis of transcriptional and genomic alterations occurring prior to or after disease relapse. At the moment, because of the aggressive nature of the disease and the time needed to establish CDXs, it is challenging to generate longitudinal models in real time. Tail vein injection or orthotopic implantation of CTCs into the lung could hasten tumor growth by providing a more natural microenvironment, as observed in other cancer models (102). Optimization of direct culturing of CTCs is another way to facilitate the development of real time longitudinal models for lung cancer.
The unprecedent opportunity to have both refractory and progression (relapse) lung cancer models can help to understand the differences between mechanisms of acquired and innate therapy resistance. Moreover, re-challenging chemosensitive CDXs with the standard chemotherapy regimen until the drugs are no longer effective could lead to the generation of new models of resistance, as has already been shown in PDX models (6). In this regard, it will be particularly interesting to genomically characterize and compare these laboratory-derived relapse models with the patient-derived relapse CDX and assess their potential to recapitulate progression of the tumor.
Finally, we recently demonstrated that CDX-derived cells can be expanded ex vivo and re-injected subcutaneously in immunocompromised mice, where they form tumors with the same characteristic of the original CDX (Lallo et al., manuscript under review). That means that several manipulations, such as CRISPR genome editing (103), or fluorescent labelling of single clones (104) can be performed ex vivo and subsequently tested in vivo. These systems will contribute to the identification of candidate genes that mediate chemoresistance, metastases, cell-cooperativity and tumor evolution.
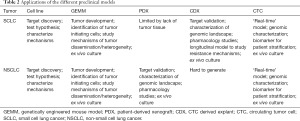
In summary, we think that all preclinical models mentioned in the text, including cell lines, GEMMs, PDXs, CDXs and CTCs, can be useful tools to help understand lung cancer biology. Considering the advantages and disadvantages of each model (Table 2) can help to choose the proper system to answer specific scientific questions. A comprehensive overview of the various preclinical models has been covered elsewhere (27,105), however it is worth noting that these models are complementary. CDXs may be easier to generate in patients where obtaining biopsies is challenging, such as extensive stage SCLC, whereas PDXs should be considered when tissue is more available, such as NSCLC and limited stage SCLC patients. Future studies on CTCs particularly with refined marker independent enrichment and isolation workflows that capture CTC heterogeneity with increased sensitivity are poised to extend our understanding of disseminating lung cancers (at earlier or late stage) with future utility as liquid biomarkers to aid treatment management. The use of CTCs in lung cancer research offers a plethora of opportunities, from single cell CTC profiling, through the generation of CDX models to the genetic manipulation of CDX ex vivo cultures. CTC cultures and/or CDXs provide an invaluable opportunity to identify novel biomarkers, to test several therapeutic options and discover new candidate targets for SCLC and NSCLC, both diseases for which new treatment strategies are urgently needed (Figure 1).
Full table
Acknowledgements
None.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015;136:E359-86. [Crossref] [PubMed]
- Davidson MR, Gazdar AF, Clarke BE. The pivotal role of pathology in the management of lung cancer. J Thorac Dis 2013;5 Suppl 5:S463-78. [PubMed]
- Reck M, Heigener DF, Mok T, et al. Management of non-small-cell lung cancer: recent developments. Lancet 2013;382:709-19. [Crossref] [PubMed]
- Goldstraw P, Ball D, Jett JR, et al. Non-small-cell lung cancer. Lancet 2011;378:1727-40. [Crossref] [PubMed]
- Schneider BJ, Saxena A, Downey RJ. Surgery for early-stage small cell lung cancer. J Natl Compr Canc Netw 2011;9:1132-9. [Crossref] [PubMed]
- van Meerbeeck JP, Fennell DA, De Ruysscher DK. Small-cell lung cancer. Lancet 2011;378:1741-55. [Crossref] [PubMed]
- Chan BA, Hughes BG. Targeted therapy for non-small cell lung cancer: current standards and the promise of the future. Transl Lung Cancer Res 2015;4:36-54. [PubMed]
- Greig SL. Osimertinib: first global approval. Drugs 2016;76:263-73. [Crossref] [PubMed]
- Ou SH, Ahn JS, De Petris L, et al. Alectinib in crizotinib-refractory ALK-rearranged non-small-cell lung cancer: a phase II global study. J Clin Oncol 2016;34:661-8. [Crossref] [PubMed]
- Arcaro A. Targeted therapies for small cell lung cancer: where do we stand? Crit Rev Oncol Hematol 2015;95:154-64. [Crossref] [PubMed]
- National Cancer Institute. Scientific framework for small cell lung cancer (SCLC). Available online: http://deainfo.nci.nih.gov/advisory/ctac/workgroup/SCLC/SCLC Congressional Response.pdf. 2014 Updated 2014.
- Cetin K, Ettinger DS, Hei YJ, et al. Survival by histologic subtype in stage IV nonsmall cell lung cancer based on data from the surveillance, epidemiology and end results program. Clin Epidemiol 2011;3:139-48. [Crossref] [PubMed]
- Alexandrov LB, Nik-Zainal S, Wedge DC, et al. Signatures of mutational processes in human cancer. Nature 2013;500:415-21. [Crossref] [PubMed]
- Manhire A, Charig M, Clelland C, et al. Guidelines for radiologically guided lung biopsy. Thorax 2003;58:920-36. [Crossref] [PubMed]
- Scott WJ, Howington J, Feigenberg S, et al. Treatment of non-small cell lung cancer stage I and stage II: ACCP evidence-based clinical practice guidelines (2nd edition). Chest 2007;132:234S-42S.
- Koletsis EN, Prokakis C, Karanikolas M, et al. Current role of surgery in small cell lung carcinoma. J Cardiothorac Surg 2009;4:30. [Crossref] [PubMed]
- Gazdar AF, Gao B, Minna JD. Lung cancer cell lines: useless artifacts or invaluable tools for medical science? Lung Cancer 2010;68:309-18. [Crossref] [PubMed]
- Oboshi S, Tsugawa S, Seido T, et al. A new floating cell line derived from human pulmonary carcinoma of oat cell type. Gan 1971;62:505-14. [PubMed]
- Pleasance ED, Stephens PJ, O'Meara S, et al. A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature 2010;463:184-90. [Crossref] [PubMed]
- Watkins DN, Berman DM, Burkholder SG, et al. Hedgehog signalling within airway epithelial progenitors and in small-cell lung cancer. Nature 2003;422:313-7. [Crossref] [PubMed]
- Gazdar AF, Minna JD. NCI series of cell lines: an historical perspective. J Cell Biochem Suppl 1996;24:1-11. [Crossref] [PubMed]
- Johnson RL, Huang W, Jadhav A, et al. A quantitative high-throughput screen identifies potential epigenetic modulators of gene expression. Anal Biochem 2008;375:237-48. [Crossref] [PubMed]
- Virmani AK, Fong KM, Kodagoda D, et al. Allelotyping demonstrates common and distinct patterns of chromosomal loss in human lung cancer types. Genes Chromosomes Cancer 1998;21:308-19. [Crossref] [PubMed]
- Joshi M, Ayoola A, Belani CP. Small-cell lung cancer: an update on targeted therapies. Adv Exp Med Biol 2013;779:385-404. [Crossref] [PubMed]
- Wistuba II, Bryant D, Behrens C, et al. Comparison of features of human lung cancer cell lines and their corresponding tumors. Clin Cancer Res 1999;5:991-1000. [PubMed]
- Gazdar AF, Carney DN, Russell EK, et al. Establishment of continuous, clonable cultures of small-cell carcinoma of lung which have amine precursor uptake and decarboxylation cell properties. Cancer Res 1980;40:3502-7. [PubMed]
- Daniel VC, Marchionni L, Hierman JS, et al. A primary xenograft model of small-cell lung cancer reveals irreversible changes in gene expression imposed by culture in vitro. Cancer Res 2009;69:3364-73. [Crossref] [PubMed]
- Kwon MC, Berns A. Mouse models for lung cancer. Mol Oncol 2013;7:165-77. [Crossref] [PubMed]
- Meuwissen R, Linn SC, Linnoila RI, et al. Induction of small cell lung cancer by somatic inactivation of both Trp53 and Rb1 in a conditional mouse model. Cancer Cell 2003;4:181-9. [Crossref] [PubMed]
- Semenova EA, Nagel R, Berns A. Origins, genetic landscape, and emerging therapies of small cell lung cancer. Genes Dev 2015;29:1447-62. [Crossref] [PubMed]
- Schaffer BE, Park KS, Yiu G, et al. Loss of p130 accelerates tumor development in a mouse model for human small-cell lung carcinoma. Cancer Res 2010;70:3877-83. [Crossref] [PubMed]
- Cui M, Augert A, Rongione M, et al. PTEN is a potent suppressor of small cell lung cancer. Mol Cancer Res 2014;12:654-9. [Crossref] [PubMed]
- George J, Lim JS, Jang SJ, et al. Comprehensive genomic profiles of small cell lung cancer. Nature 2015;524:47-53. [Crossref] [PubMed]
- Sutherland KD, Proost N, Brouns I, et al. Cell of origin of small cell lung cancer: inactivation of Trp53 and Rb1 in distinct cell types of adult mouse lung. Cancer Cell 2011;19:754-64. [Crossref] [PubMed]
- Jackson EL, Willis N, Mercer K, et al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev 2001;15:3243-8. [Crossref] [PubMed]
- Ji H, Li D, Chen L, et al. The impact of human EGFR kinase domain mutations on lung tumorigenesis and in vivo sensitivity to EGFR-targeted therapies. Cancer Cell 2006;9:485-95. [Crossref] [PubMed]
- Shaw AT, Meissner A, Dowdle JA, et al. Sprouty-2 regulates oncogenic K-ras in lung development and tumorigenesis. Genes Dev 2007;21:694-707. [Crossref] [PubMed]
- Blasco RB, Francoz S, Santamaría D, et al. c-Raf, but not B-Raf, is essential for development of K-Ras oncogene-driven non-small cell lung carcinoma. Cancer Cell 2011;19:652-63. [Crossref] [PubMed]
- Ferone G, Song JY, Sutherland KD, et al. SOX2 is the determining oncogenic switch in promoting lung squamous cell carcinoma from different cells of origin. Cancer Cell 2016;30:519-32. [Crossref] [PubMed]
- Richmond A, Su Y. Mouse xenograft models vs GEM models for human cancer therapeutics. Dis Model Mech 2008;1:78-82. [Crossref] [PubMed]
- Hayes SA, Hudson AL, Clarke SJ, et al. From mice to men: GEMMs as trial patients for new NSCLC therapies. Semin Cell Dev Biol 2014;27:118-27. [Crossref] [PubMed]
- Maheswaran S, Sequist LV, Nagrath S, et al. Detection of mutations in EGFR in circulating lung-cancer cells. N Engl J Med 2008;359:366-77. [Crossref] [PubMed]
- Oltersdorf T, Elmore SW, Shoemaker AR, et al. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature 2005;435:677-81. [Crossref] [PubMed]
- Zhang XC, Zhang J, Li M, et al. Establishment of patient-derived non-small cell lung cancer xenograft models with genetic aberrations within EGFR, KRAS and FGFR1: useful tools for preclinical studies of targeted therapies. J Transl Med 2013;11:168. [Crossref] [PubMed]
- Davenport RD. Diagnostic value of crush artifact in cytologic specimens. Occurrence in small cell carcinoma of the lung. Acta Cytol 1990;34:502-4. [PubMed]
- Jeffers M, Rong S, Vande Woude GF. Hepatocyte growth factor/scatter factor-Met signaling in tumorigenicity and invasion/metastasis. J Mol Med (Berl) 1996;74:505-13. [Crossref] [PubMed]
- Francone TD, Landmann RG, Chen CT, et al. Novel xenograft model expressing human hepatocyte growth factor shows ligand-dependent growth of c-Met-expressing tumors. Mol Cancer Ther 2007;6:1460-6. [Crossref] [PubMed]
- Ito R, Takahashi T, Katano I, et al. Current advances in humanized mouse models. Cell Mol Immunol 2012;9:208-14. [Crossref] [PubMed]
- Hodgkinson CL, Morrow CJ, Li Y, et al. Tumorigenicity and genetic profiling of circulating tumor cells in small-cell lung cancer. Nat Med 2014;20:897-903. [Crossref] [PubMed]
- Heitzer E, Auer M, Ulz P, et al. Circulating tumor cells and DNA as liquid biopsies. Genome Med 2013;5:73. [Crossref] [PubMed]
- Siravegna G, Bardelli A. Genotyping cell-free tumor DNA in the blood to detect residual disease and drug resistance. Genome Biol 2014;15:449. [Crossref] [PubMed]
- Diaz LA Jr, Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol 2014;32:579-86. [Crossref] [PubMed]
- Crowley E, Di Nicolantonio F, Loupakis F, et al. Liquid biopsy: monitoring cancer-genetics in the blood. Nat Rev Clin Oncol 2013;10:472-84. [Crossref] [PubMed]
- Bettegowda C, Sausen M, Leary RJ, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med 2014;6:224ra24. [Crossref] [PubMed]
- Pantel K, Speicher MR. The biology of circulating tumor cells. Oncogene 2016;35:1216-24. [Crossref] [PubMed]
- Alix-Panabières C, Pantel K. Challenges in circulating tumour cell research. Nat Rev Cancer 2014;14:623-31. [Crossref] [PubMed]
- Krebs MG, Metcalf RL, Carter L, et al. Molecular analysis of circulating tumour cells-biology and biomarkers. Nat Rev Clin Oncol 2014;11:129-44. [Crossref] [PubMed]
- Scher HI, Jia X, de Bono JS, et al. Circulating tumour cells as prognostic markers in progressive, castration-resistant prostate cancer: a reanalysis of IMMC38 trial data. Lancet Oncol 2009;10:233-9. [Crossref] [PubMed]
- Aggarwal C, Meropol NJ, Punt CJ, et al. Relationship among circulating tumor cells, CEA and overall survival in patients with metastatic colorectal cancer. Ann Oncol 2013;24:420-8. [Crossref] [PubMed]
- Cristofanilli M, Budd GT, Ellis MJ, et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med 2004;351:781-91. [Crossref] [PubMed]
- Krebs MG, Sloane R, Priest L, et al. Evaluation and prognostic significance of circulating tumor cells in patients with non-small-cell lung cancer. J Clin Oncol 2011;29:1556-63. [Crossref] [PubMed]
- Hou JM, Krebs MG, Lancashire L, et al. Clinical significance and molecular characteristics of circulating tumor cells and circulating tumor microemboli in patients with small-cell lung cancer. J Clin Oncol 2012;30:525-32. [Crossref] [PubMed]
- Lohr JG, Adalsteinsson VA, Cibulskis K, et al. Whole-exome sequencing of circulating tumor cells provides a window into metastatic prostate cancer. Nat Biotechnol 2014;32:479-84. [Crossref] [PubMed]
- Miyamoto DT, Zheng Y, Wittner BS, et al. RNA-Seq of single prostate CTCs implicates noncanonical Wnt signaling in antiandrogen resistance. Science 2015;349:1351-6. [Crossref] [PubMed]
- Ulmer A, Schmidt-Kittler O, Fischer J, et al. Immunomagnetic enrichment, genomic characterization, and prognostic impact of circulating melanoma cells. Clin Cancer Res 2004;10:531-7. [Crossref] [PubMed]
- Powell AA, Talasaz AH, Zhang H, et al. Single cell profiling of circulating tumor cells: transcriptional heterogeneity and diversity from breast cancer cell lines. PLoS One 2012;7:e33788. [Crossref] [PubMed]
- Gasch C, Bauernhofer T, Pichler M, et al. Heterogeneity of epidermal growth factor receptor status and mutations of KRAS/PIK3CA in circulating tumor cells of patients with colorectal cancer. Clin Chem 2013;59:252-60. [Crossref] [PubMed]
- Fabbri F, Carloni S, Zoli W, et al. Detection and recovery of circulating colon cancer cells using a dielectrophoresis-based device: KRAS mutation status in pure CTCs. Cancer Lett 2013;335:225-31. [Crossref] [PubMed]
- Yeo T, Tan SJ, Lim CL, et al. Microfluidic enrichment for the single cell analysis of circulating tumor cells. Sci Rep 2016;6:22076. [Crossref] [PubMed]
- Carter L, Rothwell DG, Mesquita B, et al. Molecular analysis of circulating tumor cells identifies distinct copy-number profiles in patients with chemosensitive and chemorefractory small-cell lung cancer. Nat Med 2017;23:114-9. [Crossref] [PubMed]
- Mariscal J, Alonso-Nocelo M, Muinelo-Romay L, et al. Molecular profiling of circulating tumour cells identifies notch1 as a principal regulator in advanced non-small cell lung cancer. Sci Rep 2016;6:37820. [Crossref] [PubMed]
- Prasad V. Perspective: the precision-oncology illusion. Nature 2016;537:S63. [Crossref] [PubMed]
- Le Tourneau C, Delord JP, Goncalves A, et al. Molecularly targeted therapy based on tumour molecular profiling versus conventional therapy for advanced cancer (SHIVA): a multicentre, open-label, proof-of-concept, randomised, controlled phase 2 trial. Lancet Oncol 2015;16:1324-34. [Crossref] [PubMed]
- Chapman PB, Hauschild A, Robert C, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med 2011;364:2507-16. [Crossref] [PubMed]
- Kopetz S, Desai J, Chan E, et al. PLX4032 in metastatic colorectal cancer patients with mutant BRAF tumors. J Clin Oncol 2010;28:abstr 3534.
- Calles A, Rubio-Viqueira B, Hidalgo M. Primary human non-small cell lung and pancreatic tumorgraft models--utility and applications in drug discovery and tumor biology. Curr Protoc Pharmacol 2013;Chapter 14:Unit 14.26.
- Hidalgo M, Amant F, Biankin AV, et al. Patient-derived xenograft models: an emerging platform for translational cancer research. Cancer Discov 2014;4:998-1013. [Crossref] [PubMed]
- Ilie M, Nunes M, Blot L, et al. Setting up a wide panel of patient-derived tumor xenografts of non-small cell lung cancer by improving the preanalytical steps. Cancer Med 2015;4:201-11. [Crossref] [PubMed]
- Anderson WC, Boyd MB, Aguilar J, et al. Initiation and characterization of small cell lung cancer patient-derived xenografts from ultrasound-guided transbronchial needle aspirates. PLoS One 2015;10:e0125255. [Crossref] [PubMed]
- Leong TL, Marini KD, Rossello FJ, et al. Genomic characterisation of small cell lung cancer patient-derived xenografts generated from endobronchial ultrasound-guided transbronchial needle aspiration specimens. PLoS One 2014;9:e106862. [Crossref] [PubMed]
- Kinsey CM, Arenberg DA. Endobronchial ultrasound-guided transbronchial needle aspiration for non-small cell lung cancer staging. Am J Respir Crit Care Med 2014;189:640-9. [Crossref] [PubMed]
- Eapen GA, Shah AM, Lei X, et al. Complications, consequences, and practice patterns of endobronchial ultrasound-guided transbronchial needle aspiration: results of the AQuIRE registry. Chest 2013;143:1044-53. [Crossref] [PubMed]
- Asano F, Aoe M, Ohsaki Y, et al. Deaths and complications associated with respiratory endoscopy: a survey by the Japan Society for Respiratory Endoscopy in 2010. Respirology 2012;17:478-85. [Crossref] [PubMed]
- Heitzer E, Auer M, Gasch C, et al. Complex tumor genomes inferred from single circulating tumor cells by array-CGH and next-generation sequencing. Cancer Res 2013;73:2965-75. [Crossref] [PubMed]
- Krebs MG, Hou JM, Sloane R, et al. Analysis of circulating tumor cells in patients with non-small cell lung cancer using epithelial marker-dependent and -independent approaches. J Thorac Oncol 2012;7:306-15. [Crossref] [PubMed]
- Morrow CJ, Trapani F, Metcalf RL, et al. Tumourigenic non-small-cell lung cancer mesenchymal circulating tumour cells: a clinical case study. Ann Oncol 2016;27:1155-60. [Crossref] [PubMed]
- Crosbie PA, Shah R, Krysiak P, et al. Circulating tumor cells detected in the tumor-draining pulmonary vein are associated with disease recurrence after surgical resection of NSCLC. J Thorac Oncol 2016;11:1793-7. [Crossref] [PubMed]
- Reddy RM, Murlidhar V, Zhao L, et al. Pulmonary venous blood sampling significantly increases the yield of circulating tumor cells in early-stage lung cancer. J Thorac Cardiovasc Surg 2016;151:852-8. [Crossref] [PubMed]
- Girotti MR, Gremel G, Lee R, et al. Application of sequencing, liquid biopsies, and patient-derived xenografts for personalized medicine in melanoma. Cancer Discov 2016;6:286-99. [Crossref] [PubMed]
- Malaney P, Nicosia SV, Dave V. One mouse, one patient paradigm: New avatars of personalized cancer therapy. Cancer Lett 2014;344:1-12. [Crossref] [PubMed]
- Mosely SI, Prime JE, Sainson RC, et al. Rational selection of syngeneic preclinical tumor models for immunotherapeutic drug discovery. Cancer Immunol Res 2017;5:29-41. [Crossref] [PubMed]
- Moynihan KD, Opel CF, Szeto GL, et al. Eradication of large established tumors in mice by combination immunotherapy that engages innate and adaptive immune responses. Nat Med 2016;22:1402-10. [Crossref] [PubMed]
- Yu M, Stott S, Toner M, et al. Circulating tumor cells: approaches to isolation and characterization. J Cell Biol 2011;192:373-82. [Crossref] [PubMed]
- Yu M, Bardia A, Aceto N, et al. Cancer therapy. Ex vivo culture of circulating breast tumor cells for individualized testing of drug susceptibility. Science 2014;345:216-20. [Crossref] [PubMed]
- Cayrefourcq L, Mazard T, Joosse S, et al. Establishment and characterization of a cell line from human circulating colon cancer cells. Cancer Res 2015;75:892-901. [Crossref] [PubMed]
- Gao D, Vela I, Sboner A, et al. Organoid cultures derived from patients with advanced prostate cancer. Cell 2014;159:176-87. [Crossref] [PubMed]
- Hamilton G, Burghuber O, Zeillinger R. Circulating tumor cells in small cell lung cancer: ex vivo expansion. Lung 2015;193:451-2. [Crossref] [PubMed]
- Maheswaran S, Haber DA. Ex vivo culture of CTCs: an emerging resource to guide cancer therapy. Cancer Res 2015;75:2411-5. [Crossref] [PubMed]
- Grillet F, Bayet E, Villeronce O, et al. Circulating tumour cells from patients with colorectal cancer have cancer stem cell hallmarks in ex vivo culture. Gut 2016. [Epub ahead of print]. [Crossref] [PubMed]
- "What are the 3Rs?". National centre for replacement, refinement and reduction of animals in research. Available online: http://www.nc3rs.org.uk/the-3rs. August 14, 2013
- Ramsköld D, Luo S, Wang YC, et al. Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat Biotechnol 2012;30:777-82. [Crossref] [PubMed]
- Budd GT, Cristofanilli M, Ellis MJ, et al. Circulating tumor cells versus imaging--predicting overall survival in metastatic breast cancer. Clin Cancer Res 2006;12:6403-9. [Crossref] [PubMed]
- Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol 2014;32:347-55. [Crossref] [PubMed]
- Kreso A, O'Brien CA, van Galen P, et al. Variable clonal repopulation dynamics influence chemotherapy response in colorectal cancer. Science 2013;339:543-8. [Crossref] [PubMed]
- Voortman J, Lee JH, Killian JK, et al. Array comparative genomic hybridization-based characterization of genetic alterations in pulmonary neuroendocrine tumors. Proc Natl Acad Sci U S A 2010;107:13040-5. [Crossref] [PubMed]


