Immunohistochemistry for predictive biomarkers in non-small cell lung cancer
Introduction
In the old paradigm of lung cancer treatment, patients with advanced non-small cell lung cancer (NSCLC), irrespective of histologic subtypes, were typically treated with chemotherapeutic agents, in particular platinum doublets (1). The paradigm of NSCLC treatment started shifting at the beginning of this century, and the discovery of epidermal growth factor receptor (EGFR) mutations in NSCLC paved the way to molecularly targeted therapy and development of predictive biomarkers (2-5). Predictive biomarkers are defined as markers for which the results are essential for therapeutic decision-making, and for treatment with an EGFR tyrosine kinase inhibitor (TKI), the presence of sensitizing EGFR mutations serves as a predictive biomarker. The role of predictive biomarker assays for NSCLC was established in 2011 when the US Food and Drug Administration (FDA) approved both drug and its companion diagnostic test [crizotinib and break-apart fluorescence in situ hybridization (FISH) for anaplastic lymphoma kinase (ALK)] for treatment of patients with advanced ALK-rearranged NSCLC (6,7). Crizotinib, a small molecule TKI of c-MET, ALK and proto-oncogene tyrosine-protein kinase ROS (ROS1), was also approved for treatment of advanced ROS1-rearranged NSCLC in 2016 (8). Thus, break-apart FISH has become the reference standard to detect ALK and ROS1 rearrangements (9,10). However, given the low incidence of ALK (5%) and ROS1 (1–2%) rearrangements in NSCLC (11), expensive FISH assays may not be cost efficient. Thus, immunohistochemistry (IHC) with a sensitive antibody clone targeting ALK or ROS1 protein has been developed as a predictive biomarker assay (12,13).
More recently, the blockade of immune checkpoints to reinstitute host antitumor immunity has been investigated extensively in the field of lung cancer, and a few anti programmed cell death protein 1 (PD-1)/programmed death-ligand 1 (PD-L1) agents have been approved by the US FDA for treatments of advanced NSCLC as the first line or second or more line therapy. In the clinical trials of anti PD-1/PD-L1 agents, PD-L1 IHC assays have been used for predictive biomarker testing, and positive results indicate the presence of an immunomodulatory molecule that can be impacted by the PD-1/PD-L1 blockade (14-20).
In this review, IHC to detect ALK and ROS1 rearrangements and other molecular targets as well as PD-L1 expression will be discussed. Of note, it is important to differentiate the therapeutic decision-making role of IHC for predictive biomarkers from IHC performed for diagnostic purposes, which plays a diagnostically supportive or decisive role and can be critical in distinguishing NSCLC subtypes (21).
IHC for molecular targets
Testing for ALK rearrangements
ALK rearrangements in lung cancer consist mostly of echinoderm microtubule-associated protein-like 4 (EML4)—ALK translocations (22). First described by a Japanese group led by Dr. Mano in 2007, the EML4-ALK fusion results from a small inversion within the short arm of chromosome 2 leading to expression of a chimeric tyrosin kinase. The chimeric protein has been shown to possess potent oncogenic activity in vitro and in vivo (23). ALK rearrangements have been found in approximately 5% (1–15%) of patients with NSCLC (24).
There are several methods that have been used to detect ALK rearrangements, namely, FISH, IHC, multiplex real time polymerase chain reaction (RT-PCR) and next-generation sequencing (NGS). Of those, multiplex RT-PCR can identify all known ALK rearrangements in a single experiment, and the presence of fusion transcripts as detected by RT-PCR provides direct evidence of chromosomal rearrangements. It requires, however, high-quality RNA, which is difficult to extract from formalin-fixed paraffin-embedded (FFPE) samples. Furthermore, RT-PCR can only detect fusion transcripts with known fusion partners (25). NGS is very efficient to detect rearrangements not only of ALK but also of multiple other genes in a single FFPE sample, although the sensitivity of NGS to detect the gene rearrangements varies among the platforms. Hybrid capture-based NGS can detect most genomic breakpoints, which may be located in introns, while targeted DNA-based NGS methods can detect gene rearrangements only when their breakpoints are adequately covered (22). In order to improve the sensitivity of NGS, anchored multiplex PCR (AMP) has recently been introduced. AMP, a rapid target enrichment method for NGS, is compatible with low nucleic acid input from FFPE specimens, and is effective in detecting gene rearrangements without prior knowledge of the fusion partners (26). Unfortunately, however, these molecular techniques are not available in many routine pathology laboratories, and their turn-around-time is typically 2–3 weeks that may be too long for patients with rapidly progressive, advanced lung cancer to wait. ALK FISH is currently considered as the universally accepted reference standard, and is approved by the US FDA as a companion diagnostic kit for crizotinib (7). However, the close proximity of EML4 and ALK genes on the short arm of chromosome 2 and other technical or biological conditions may rarely produce equivocal or erroneous results, even when FISH testing is performed with careful preparation and interpretation strictly adherent to the guidelines (22). Furthermore, ALK FISH assays do not seem to be cost efficient in detecting one ALK positive case out of 20 patients with NSCLC. Conversely, IHC is a routine methodology in most pathology laboratories to detect a protein of interest, and appears to be a cost efficient method for reflex testing.
IHC for ALK
Initially, several different IHC assays were used for detecting ALK protein expression secondary to ALK rearrangements in the lung. In the published studies, the type or source of antibodies, and antigen retrieval, antibody detection, and amplification techniques varied substantially (22). ALK protein is not expressed in normal lung parenchyma, but is usually expressed secondary to gene rearrangements and the promoter of the fusion partner likely drives the expression. EML4, the most common fusion partner for ALK in lung cancer, encodes a protein that is expressed at a low level in normal lung (27). Of the four most commonly used antibody clones, the clone ALK1 is not sensitive enough to detect the low-level expression of ALK protein secondary to gene rearrangements (Table 1) (12,28,29). Furthermore, increasing the sensitivity with an enhanced detection system will likely result in the greatest amount of background staining with ALK1 (12,45), thus the clone is not preferred as a screening tool. Conversely, a novel monoclonal anti-ALK antibody 1A4 (Origene, Rockville, MD, USA) appears to have high sensitivity compared to the other clones and was considered as a promising candidate for ALK rearrangement screening in lung cancer (44); however, it has been shown that the clone 1A4 antibody suffers from lower specificity (70%) (Table 1) (43). Thus, tumors that are positive for ALK with 1A4 IHC require confirmation by another technology such as FISH or NGS. The performance of the other two clones, 5A4 and D5F3, in detecting ALK rearrangements appear to be equally good. As laboratory developed tests, the clone 5A4, in particular that produced by Novocastra (currently Leica Biosystems), carries very high sensitivity and specificity (92.9–100% and 98.1–100%, respectively) equivalent to, if not better than, those of the clone D5F3 (Cell Signalling Technology, Billerica, MA, USA) (75.9–100% and 95.0–100%, respectively) (Table 1) (12,25,30-43). Conklin and colleagues conducted head-to-head comparison of different ALK IHC assays and confirmed that the clones D5F3 and 5A4 (Novocastra, Newcastle, United Kingdom) with the ADVANCE system (Dako, Carpenteria, CA, USA) outperformed ALK1 based assays (45). These results are also supported by the recent analysis of pooled data on the diagnostic operating characteristics in 12 studies (3,754 NSCLC specimens) that had evaluated ALK IHC with the 4-tiered scoring systems (46). The sensitivities of D5F3 and 5A4 antibodies were much higher than that of ALK1. With both D5F3 and 5A4 IHC assays, binary IHC scoring with 3+ as the cut-off for positivity, the ALK IHC-positive and negative categories corresponded to ALK FISH-positive and negative cases, respectively. The nearly 100% concordance in these IHC categories supports the use of IHC as a screening method to identify ALK-rearranged NSCLC. However, when tumors exhibit 1+ or 2+ intensity, the results need to be confirmed with ALK FISH or NGS (46). Of note, the reproducibility of ALK IHC results among different laboratories and pathologists is high for any validated protocols (47-50).
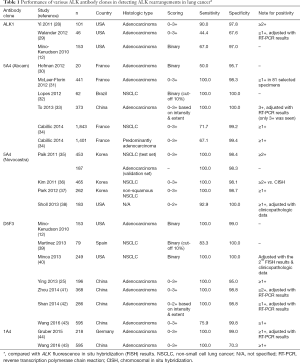
Full table
Now, an immunoassay [Ventana ALK (D5F3) CDx Assay, Ventana Medical Systems, Tucson, AZ, USA] with the clone D5F3 coupled to the automated immunostaining platform BenchMark XT has been developed and approved by the US FDA as a companion diagnostic kit for crizotinib (51). The assay includes the amplification step with the OptiView Amplification Kit which is intended to reduce or eliminate equivocal results by increasing the signal difference between the specific immunoreaction from the non-specific background staining (that may result in false positive interpretation), thus enabling binary scoring (49). Similarly, with tyramide enhancement that works for both the D5F3 and 5A4 clones, the difference in epitope concentration between negative and strongly positive staining intensities is reduced to the extent that scoring is either negative or positive (22,49,52).
The strong IHC amplification systems used in ALK IHC assays, however, may be associated with various artefacts leading to false-positive results. Positive ALK IHC typically shows strong granular cytoplasmic staining, while weaker cytoplasmic staining may be seen in alveolar macrophages, neural elements, glandular epithelium, extracellular mucin, and areas of necrotic tumor as well as ALK-negative NSCLC (22,49). In addition, some neuroendocrine carcinomas reportedly exhibit positive reactions despite the absence of ALK rearrangements (27,53). Conversely, the signet ring cell morphology that is frequently seen in ALK-rearranged adenocarcinomas could be a source of false-negative staining. A thin membranous positive pattern on ALK IHC may be masked by an intracellular mucin vacuole (54-56). Further, the paranuclear dot-like pattern reportedly associated with the KIF5B-ALK rearrangement may be considered as an artefact, leading to a false-negative result (57). It is very important to understand these possible pitfalls when evaluating ALK IHC, in particular as a standalone test.
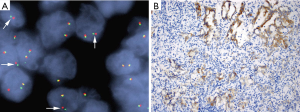
Multiple studies have reported discordant results between ALK FISH and IHC, both IHC (D5F3/5A4)+/FISH− and IHC−/FISH+ results, in NSCLC (25,31,33,37,38,40-42,47,50,52,58-68). The discordant results could be attributed to false negative or positive interpretation of FISH results, false negative or positive interpretation of IHC or yet undetermined mechanisms. False-negative interpretation of FISH results could be due to the paucity of tumor cells in the sample, the presence of reactive normal cells interpreted as malignant cells (38), or the physical close proximity of ALK and EML4 genes on the short arm of chromosome 2 (Figure 1) or complex rearrangements involving the ALK gene leading to narrow splits (28). Atypical FISH patterns including solitary green signals with split 5’ centromeric probe could be mistakenly interpreted as positive (38,48). Amplifications of the ALK gene may be associated with ALK protein expression (typically 1+ or 2+ staining) in some cases (69-71). False-positive interpretation of ALK IHC results may be attributed to non-specific high background (27), while suboptimal tissue preservation and fixation with variation of ALK protein expression among specimens could lead to false-negative results (50,59). Importantly, ALK FISH positivity ranging between 10% and 20% is prone to false negative or positive results, thus it should be interpreted with caution (66). It is worth mentioning a recent study by Pekar-Zlotin and colleagues that applied NGS as a gold standard for ambiguous cases. They identified six cases with FISH/IHC discrepant results (five IHC+/FISH− and one IHC−/FISH+) in 51 lung adenocarcinomas. NGS confirmed the IHC results in five of the six discrepant cases (except for one with weak IHC+/FISH− results), leading to a sensitivity and specificity of 42.9% and 97.7% for FISH and 100% and 97.7% for IHC compared to an NGS-based approach. Based on the results, they concluded that FISH-based testing may miss a significant number of patients who are eligible for ALK inhibition, and suggested an NGS-based approach in cases with inconclusive IHC staining (64).
ALK IHC assays, already recommended by organizations in Europe, Asia and the United States, are validated and standardized and have been implemented in daily practice as a cost-effective screening tool or a standalone test for detecting ALK rearrangements in NSCLC. When the laboratory conducts an ALK IHC assay as a laboratory developed test (LDT), an optimal algorithm for ALK testing (positive or equivocal results confirmed by FISH or NGS) should be designed. It is also important for laboratories to participate regularly in external quality assessment programs to maintain the reliability of assays.
Testing for ROS1 rearrangements
ROS1 is located on chromosome 6q22 and encodes a receptor tyrosine kinase of the insulin receptor family that shares 77% of amino acid sequences of the ATP-binding site of the tyrosine kinase domain with ALK (72). ROS1 fusions were identified as potential driver events in a cell line (HCC78; SLC34A2-ROS1) and an NSCLC patient sample (CD74-ROS1) in 2007 (73). Subsequently, multiple gene rearrangements involving ROS1 have been reported, but two-thirds of those are distributed over three genes: CD74, EZR, and SLC34A2, each of which has two or more fusion patterns (74). The reported incidence of these fusion proteins in NSCLC is generally low and ranges from 1% to 2% (75). ROS1 may be detected by a variety of techniques, including FISH, RT-PCR, NGS and IHC. Currently, a companion diagnostic kit for crizotinib has not been specified by the US FDA, thus any tests that are validated in individual laboratories could be used to detect ROS1 rearrangements in NSCLC, but the majority use FISH and/or more recently NGS (22). Pros and cons of each assay in detecting rearrangements of ROS1 are similar to those of ALK. Given the presence of multiple fusion partners and easier interpretation of break-apart signals due to interchromosomal rearrangements in the majority of ROS1-rearranged NSCLC, FISH has been used in many pathology laboratories (76). However, FISH may not be cost efficient in detecting one ROS1-rearraneged case in 50–100 patients with NSCLC, and ROS1 IHC may serve as an effective screening tool in this context.
IHC for ROS1
Most studies on ROS1 IHC have used the D4D6 rabbit monoclonal antibody (Cell Signaling Technology, Danvers, MA, USA) applied at dilutions ranging from 1:50 to 1:250 with various antigen retrieval methods and amplification and detection systems in automated instruments or with manual testing (22).
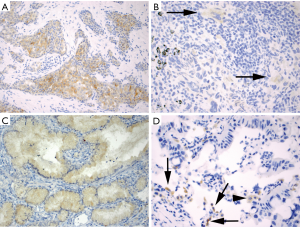
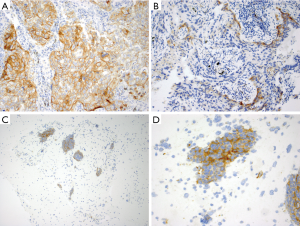
ROS1 overexpression in ROS1-rearranged lung cancers is typically cytoplasmic, but staining patterns vary and may be dependent upon the fusion partner. For instance, a granular cytoplasmic staining with focal or diffuse globular aggregates of protein has been associated with the CD74-ROS1 fusion (77), while membranous staining has been observed in tumors with the EZR-ROS1 fusion (77,78). ROS1-rearranged tumors almost always exhibit diffuse protein expression that is typically homogeneous in a staining pattern but staining can vary in intensify from weak to strong in a tumor (Figure 2A). Detection of ROS1 protein expression in ROS1-rearranged tumors with signet ring cells can be challenging since the cytoplasm may be replaced by non-reactive mucin (77), as described in the ALK-rearranged counterpart (27). ROS1 expression may be seen in tumors without ROS1-rearrangements, most often in a focal or patchy, weak pattern (Figure 2B), but in some cases it can be diffuse and/or strong (79-81). In one study, ROS1 expression was present in 80% of adenocarcinomas with mucinous morphology (invasive mucinous adenocarcinomas) that were negative for ROS1 rearrangements (Figure 2C) (77). Thus, ROS1 expression in this tumor subtype should be interpreted with caution. Similarly, ROS1 expression is occasionally present in non-neoplastic hyperplastic type II pneumocytes and in alveolar macrophages (Figure 2D) (82). In most cases, the expression in these cells is weak-to-moderate (1+ to 2+ in intensity) (22,76), while strong granular cytoplasmic staining of osteoclast-type giant cells has been reported in the setting of a bone metastasis (82).
The performance of ROS1 IHC detecting ROS1 rearrangements is not as good as that of ALK IHC, given the only modest specificities reported in some studies (the reported specificities range from 68% to 100%), while the sensitivity is nearly 100% (Table 2) (78,81-90). Therefore, all IHC positive results need to be confirmed by FISH or another testing method before consideration of treatment with a ROS1 inhibitor. Reported discrepancies between FISH and IHC may also reflect false-negative or false-positive results by FISH (77). Given the easier interpretation of break-apart signals with ROS1 FISH the contribution of such a factor should be limited, however.
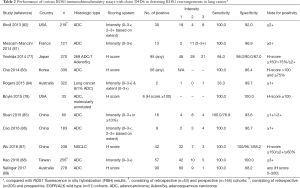
Full table
Based on our experience and others, diffuse and homogeneous protein expression and/or high H-score may be used in selecting cases for the confirmatory testing. For instance, some studies have suggested that application of H-score cutoffs of 100 or 150 can maximize the sensitivity and specificity of ROS1 IHC (77,78). Thus, experienced pathologists may be able to distinguish tumors harboring ROS1 rearrangements from those with false-positive IHC results based on the distinctive patterns of ROS1 expression. Although individual laboratories may opt to perform confirmatory testing on only those samples with diffuse and homogeneous expression patterns, a confirmatory analysis on all samples examined with IHC is recommended to evaluate the performance of ROS1 IHC at least at the beginning of the IHC implementation. In France, a national pathology expert panel recommends a screening with ROS1 IHC and confirmatory FISH for IHC positive or equivalent cases to detect ROS1-rearranged NSCLC (81).
PD-1/PD-L1 blockade in NSCLC
High profile clinical trials have demonstrated impressive anti-tumor activity of anti PD-1 and PD-L1 agents in NSCLC (19,91,92), and significant improvements in overall survival (OS) of previously treated, advanced NSCLC patients compared to single-agent docetaxel (16,18,20,93). Subsequently, the US FDA approved nivolumab, pembrolizumab and atezolizumab for NSCLC patients with disease progression on or after platinum-based chemotherapy (94). Clinical trials with two other agents, durvalumab and avelumab, have also shown promising results (14,15). Furthermore, pembrolizumab was approved by the FDA as a 1st line treatment for advanced NSCLC patients in October, 2016 (94), based on the results of the clinical trial, KEYNOTE-024, that showed significantly improved objective response rate, progression free survival (PFS), and OS when advanced NSCLC patients, whose tumors harbored PD-L1 expression by IHC in 50% or greater of the tumor cells, were treated with pembrolizumab compared to platinum-based chemotherapy in the 1st line setting (95). In these clinical trials, PD-L1 expression in the tumor by the specific IHC assay served as a predictive biomarker. Importantly, five different PD-L1 IHC assays were developed for the five PD-1/PD-L1 agents (Table 3) (96,97), and some of the assays have been approved by FDA as either companion (a requirement for drug eligibility) or complimentary (only for guidance) diagnostic kits along with the corresponding anti PD-1/PD-L1 agents.

Full table
Now, anti PD-1/PD-L1 agents are incorporated in the paradigm of treatment for advanced NSCLC patients. Given the availability of pembrolizumab as a first line therapy, the National Comprehensive Cancer Network (NCCN) guidelines recommend that all advanced NSCLC samples be tested with a PD-L1 IHC assay in a reflex manner (98).
PD-L1 IHC testing
As briefly mentioned, five different PD-L1 IHC assays (the Dako 28-8, Dako 22C3, Ventana SP142, Ventana SP263 and Dako 73-10) have been developed and validated as a predictive biomarker in the clinical trials (Table 3) (96,97). Pathology laboratories need at least one affordable, validated test, but it is not practical for them to conduct several specific assays for one protein from a financial and regulatory perspective, and given the limited availability of tumor tissue for testing and the number of tissue-based diagnostic tests required in the management of an advanced NSCLC patient. However, selecting one assay among several available is challenging. While each of the five IHC assays recognizes PD-L1 protein, each antibody clone appears to be specific for a different epitope of the PD-L1 protein and may not have the same binding affinity for its epitope. In addition, different detection systems, with or without amplification, are used in different assays, thus the performance of these five assays may be different. Scoring systems to determine “positive” results are also quite different between the five assays, since these systems were determined based on a predictive value and clinical data obtained during the therapeutic/diagnostic test co-development process of individual anti PD-1/PD-L1 agents. For instance, complete circumferential or partial linear membranous staining of tumor cells irrespective of intensity is considered positive for the DAKO 22C3, 28-8 and 73-10 assays (Figure 3A,B), while any membranous and/or cytoplasmic expression of tumor cells is considered positive for the Ventana SP263 assay. Ventana SP142 assay is unique in that not only tumor cell expression, but also immune cell expression, are taken into account. Furthermore, each assay has a specific % of positive tumor cells as a cut-off, and the percent may be different depending on the 1st line vs. 2nd or more line of treatment. Importantly, the outcomes of patients, when using these drugs in a cohort selected for “positive” PD-L1 expression, have only been evaluated in trials using the specific drug-assay combinations (Table 3).
In order to see whether we can use only one of these assays or any other laboratory developed tests (LDTs) to select patients for all available anti PD-1/PD-L1 agents, several studies, although only from the technical/analytical perspective, have compared the performance of clinical trial PD-L1 IHC assays (99-101). The International Association for the Study of Lung Cancer (IASLC) and the American Association for Cancer Research (AACR), in collaboration with pharmaceutical companies and diagnostics venders, have evaluated the technical similarities and differences of the Dako 28-8, Dako 22C3, Ventana SP263, and Ventana SP142 assays (101). The initial phase I part of this study was to test the feasibility on a small cohort of 38 NSCLC resections (not treated with anti PD-1/PD-L1 agents), each stained using all four assays and scored by three trained pathologists. Of those, the Dako 22C3, Dako 28-8, and Ventana SP263 assays demonstrated the similar membranous staining on tumor cells, while the Ventana SP142 assay consistently revealed smaller numbers of positive tumor cells. IASLC is now conducting a larger phase II study consisting of different sample types (resection, biopsy and cell block) stained using 5 clinical trial assays (including the Dako 73-10 assay) evaluated by more than 20 pulmonary pathologists. The German ring study assessed interobserver concordance and PD-L1 IHC staining patterns in 15 NSCLC resection specimens using the four PD-L1 IHC assays (the Dako 28-8, Dako 22C3, Ventana SP263, and Ventana SP142 assays), and showed that the tumor cell score could be reproducible, with no differences in interobserver concordance among the tested assays (100). The scoring of immune cells, however, yielded low concordance rates indicating that immune cell scoring might require specific standardization. In addition, they found that staining patterns/intensities might be different among the four assays. While the 28-8 and 22C3 assay stained similar proportions of tumor cells in the majority of cases, the SP142 assay stained fewer tumor cells and SP263 stained more tumor cells compared to the 28-8 and 22C3 assays in some cases. Another recent study, in which 493 NSCLC tissue samples were stained using three assays (the Dako 28-8, Dako 22C3 and Ventana SP263 assays) and scored by a single trained pathologist, has reported the similar patterns of tumor membranous staining with high (>90%) overall percentage agreement between the assays at multiple expression cut-offs (including 1%, 10%, 25% and 50%) (99).
The results of these studies raise the possibility that the Dako 22C3, Dako 22-8 and Ventane SP263 assays can be used interchangeably to identify patients most likely to respond to anti PD-1/PD-L1 agents (nivolumab, pembrolizumab and durvalumab) provided that the appropriate scoring system is used for the corresponding agent (99). Unfortunately, however, the Ventana SP142 assay that consistently shows fewer numbers of positive tumor cells and includes the immune cell component for scoring does not appear to be interchangeable with the other assays. What about the performance of LDTs that could be developed with either clinical trial antibody clones or non-clinical trial clones? A recent study by Neuman and colleagues has reported successful implementation of 22C3 IHC on the Ventana Benchmark XT platform with two of the Ventana’s detection systems after a rigorous optimization process (102). Among multiple non-clinical trial PD-L1 antibody clones, the clone E1L3N (CST) has been optimized with various IHC platforms and detection systems, and have been used in multiple clinical studies (100,103-113). Importantly, a prospective, multi-institutional study sponsored by NCCN and Bristol-Myers Squibb has shown the possible utility of E1L3N IHC on the Leica Bond platform (114). The study, in which 13 pathologists scored 90 surgically resected NSCLCs stained using the Dako 22-8, Dako 22C3, Ventana SP142 and E1L3N/Leica assays have shown analytical equivalency of the Dako 28-8 and E1L3N/Leica assays using the average pathologist scores across all 90 cases. While the Dako 22C3 assays revealed significantly less expression than the other two antibodies (only when averaging the readings of 13 pathologists), there was no difference in sensitivity and specificity equivalents between the Dako 22-8, Dako 22C3 and E1L3N/Leica assays using a “real world” assessment (agreement by individual pathologists). The Ventana SP 142, however, again revealed significantly less expression by large amount with every method of assessment (114). The results of these studies have brought optimism that harmonization between assays, including LDTs, may be possible. Of somber note, the recent French study comparing performance of clinical trial assays and various combination of LDTs has reported only half of the evaluated LDTs demonstrating sufficient concordance with the reference assays for tumor cell scores (115). The study consisted of 41 resected NSCLCs stained with clones 28-8, 22C3, SP263, SP142 and E1L3N in 7 centers (3 with Dako Link 48, 2 with Ventana Benchmark Ultra and 2 with Leica Bond III; 8 clinical trial assays and 27 LDTs) scored by 7 trained pathologists, and found that only 14 of the 27 LDTs achieved >0.75 weighted kappa coefficient compared to the reference assay (115). Thus, not all LDTs will be interchangeable with the clinical trial assays.
As for interobserver concordance on PD-L1 tumor cells scoring, in the German Ring study with highly selected cases, the overall percent agreement at the 1% threshold ranged for all 4 assays from 90.4% to 97.2%, and at the 50%, from 91.5% to 94.8% (100). An Australian study by Cooper and colleagues using the Dako PD-L1 22C3 assay on a highly selected set of cases showed the overall percent agreement at the 1% threshold of 84.2%, and at the 50% threshold of 81.9% (116). Rehman and colleagues examined the reproducibility of 5 pathologists on a selected set of cases stained with the Ventana SP142 assay and showed an intraclass correlation coefficient of 94% agreement among the pathologists for the assessment of PD-L1 in tumor cells, but only 27% agreement on stromal/immune cell PD-L1 expression. The subjective interpretation of the guidelines for scoring stromal cells may have contributed to the fair agreement on PD-L1 expression on stromal/immune cells (117).
IHC for other predictive biomarkers
IHC may be used to detect other predictive biomarkers in NSCLC. Of those, EGFR mutation specific antibodies recognize the protein conformation change due to the mutation, but do not bind to the wild type EGFR protein. There are two types of EGFR mutation specific antibodies—the one specific for a 15 bp deletion in exon 19 and the other for a L858R point mutation in exon 21. Currently, these are not recommended for predictive testing, since the sensitivity for detecting the corresponding mutations is modest, while the specificity is high (118). However, EGFR mutation specific IHC may be useful when available tissue samples are insufficient for molecular assays due to scarcity of tumor cells and/or fixation with decalcification or heavy metal solutions.
In Europe, EMA approved monoclonal anti-EGFR drug, necitumumab, for patients with advanced-stage squamous NSCLC expressing the wild type EGFR protein by IHC (119). The US FDA also approved necitumumab for use in combination with cisplatin/gemcitabine chemotherapy for the first line treatment of metastatic squamous NSCLC, but EGFR protein expression by IHC is not a requirement for the treatment (120).
A small fraction (2–4%) of NSCLC harbors BRAF mutations. A BRAF V600E-mutation specific antibody has been developed and proven useful in detecting BRAF V600E mutations in colon cancer (121), but hardly detects any of the proteins encoded by non-V600E mutations (122,123). In NSCLC 40–50% of the BRAF mutations are non-V600E (124), thus BRAF V600E-mutation specific IHC does not appear to be useful.
NSCLC with MET amplification or exon 14 skipping mutation could be treated with a MET inhibitor, crizotinib (or others) in clinical trials. High MET protein expression by IHC appears to be associated with MET FISH positivity and amplification, but there is a significant overlap between FISH/amplification positive and negative cases. In addition, MET exon 14 skipping mutations are proportionally much rare (125), thus, MET IHC does not seem to be an efficient method for the detection of MET alterations amenable for MET inhibition.
Pre-analytical variables for IHC
The stability of proteins can be affected by multiple pre-analytic variables, including ischemia time, fixative type, fixation time, archive conditions, and age of archived material (126). Thus, the successful implementation of IHC-based assays in general depends on pre-analytic tissue handling (127) as well as the antigen retrieval and detection systems. An ischemia time from excision to the initiation of fixation should be short (as short as possible), and biopsies should immediately be immersed in fixative for 6–48 h. Formalin (neutral buffered formalin) is historically the preferred and most common fixative used in the practice of histopathology (128). Consequently, the majority of pathology laboratories typically perform the initial validation of IHC protocols on FFPE tissue. Decalcifying solutions used for bony specimens vary in their effects on retention and integrity of nucleic acids and proteins. Thus, results of IHC on decalcified specimens are unpredictable because of wide variations in specimen types and sizes, fixation time, and the particular solution(s) used (129). Similarly, alcohol fixation used for cytology specimens including alcohol-fixed cell blocks decreases IHC accuracy by causing loss or decrease of immunogenicity when IHC protocols optimized with FFPE tissue samples are used (126).
In order to improve the accuracy of IHC performed on samples fixed in these fixatives, in the US, the guidelines from the College of American Pathologists (CAP) Pathology and Laboratory Quality Center recommend that pathology laboratories test a sufficient number of such cases to ensure that assays consistently achieve expected results (129). It is of particular importance since at least 30–40% of advanced NSCLCs are diagnosed by cytology alone. Rigorous validation and protocol optimization should be performed in each laboratory that performs IHC on cytology specimens (e.g., alcohol-fixed cell blocks, air-dried smears, formalin-post fixed specimens) (126). Irrespective of specimen types, the CAP guideline recommends examination of at least 20 positive and 20 negative samples for initial analytic validation of predictive marker assays, while a minimum of 10 positive and 10 negative tissues are sufficient for non-predictive marker assays (129).
Special note on pre-analytical variables for PD-L1 IHC assays
The PD-L1 IHC assays have not been validated for decalcified tissue (130,131), thus PD-L1 IHC on decalcified tissues should be avoided when another tissue sample is available. Specimen age for PD-L1 testing should be less than 3 years, since antigenicity may drop significantly in those older than 3 years (132). In addition, in our experience, the antigen retrieval conditions (citrate buffer pH6 vs. citrate buffer pH8 or EDTA pH9) have been shown to significantly affect intensity and rate of positivity for the clone E1L3N (unpublished observation). These differences may alter the outcome of the test, leading to alternative PD-L1 scoring around a given cut-off threshold.
Importantly, the use of cytology samples for PD-L1 IHC is currently not recommended, due to the lack of rigorous validation for this purpose (131). The study by Rebelatto and colleagues has also shown that 95% alcohol, AFA and PRFER are unacceptable fixatives for IHC with the SP263 clone (133). Furthermore, an evaluation of PD-L1 positive immune cells with the Ventana SP142 assay will likely be more challenging in cytology specimens, since the lack of tissue architecture precludes distinction of the relevant immune cells within the tumor area from immune cells outside of the tumor boundaries that are considered irrelevant for PD-L1 scoring. Pre-existing lymphocytes in a fine-needle aspirate of a lymph node also precludes an optimal immune cell scoring (96).
When a cytology specimen is the only available sample, however, a cell block may be used for PD-L1 testing as long as it is FFPE and contains a sufficient number of tumor cells (minimum 50–100) (Figure 3C,D) (134-136). Several studies have reported high concordance on PD-L1 expression between cell blocks and matched histological specimens and/or comparable PD-L1 expression among cell blocks, small biopsies and resections in a prospective cohort (134-136). The results of these studies suggest that cytological materials are as good as histological materials for the tumor cell analysis, but large-scale validation studies are warranted to establish PD-L1 IHC testing on cytology specimens.
Conclusions
Given the increasing number of targeted therapies available for advanced NSCLC patients in clinic, predictive biomarker testing has never been more important. IHC is a widely available and technically less challenging method to perform and interpret, can provide clinically meaningful results very quickly and is more cost efficient than molecular assays. Thus, the use of IHC to detect predictive biomarkers, in particular gene rearrangements, in NSCLC has been well established. It is important, however, to understand the performance of multiple antibody clones, pros and cons of IHC platforms and various scoring systems to design an optimal algorithm for predictive biomarker testing. In addition, given the recent FDA approval of an anti PD-1 agent as a first line therapy for advanced NSCLC patients, PD-L1 IHC testing has become routine in pathology laboratories in the US and some other countries. The one drug-one predictive biomarker assay concept, however, has brought unique challenges to pathology and oncology communities, since operating five different PD-L1 IHC assays to support the use of five drugs is extremely difficult in any laboratory from both the practical/regulatory and financial perspective. With this background, several recent studies evaluated and compared the analytical/technical performance of clinical trial assays and/or LDTs leading to the optimism that harmonization of PD-L1 IHC assays may be feasible, if rigorous optimization and validation are performed for LDTs that are used to identify patients who will likely respond to anti PD-1/PD-L1 agents. Finally, appropriate pre-analytical tissue handling and selection of optimal tissue samples are the keys to successful IHC, not only for predictive biomarkers, but also for protein expression in general. Of particular note, the use of cytology smear and cell blocks for PD-L1 IHC is not currently recommended, but cell blocks that are made following appropriate protocols may be used with caution.
Acknowledgements
The author thanks Mr. Cris Kenudson for editorial support.
Funding: This work was supported by a Stand Up To Cancer-American Cancer Society Dream Team Translation Research Grant.
Footnote
Conflicts of Interest: M Mino-Kenudson has served as a consultant for Merrimack Pharmaceuticals, H3 Biomedicine, and Advanced Cell Diagnostics and as an advisory board member for Roche.
References
- Schiller JH, Harrington D, Belani CP, et al. Comparison of four chemotherapy regimens for advanced non-small-cell lung cancer. N Engl J Med 2002;346:92-8. [Crossref] [PubMed]
- Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med 2009;361:947-57. [Crossref] [PubMed]
- Pao W, Miller V, Zakowski M, et al. EGF receptor gene mutations are common in lung cancers from "never smokers" and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci U S A 2004;101:13306-11. [Crossref] [PubMed]
- Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med 2004;350:2129-39. [Crossref] [PubMed]
- Paez JG, Janne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 2004;304:1497-500. [Crossref] [PubMed]
- Available online: https://www.accessdata.fda.gov/drugsatfda_docs/label/2011/202570s000lbl.pdf
- Available online: https://www.fda.gov/MedicalDevices/ProductsandMedicalProcedures/InVitroDiagnostics/ucm301431.htm
- Available online: https://www.fda.gov/drugs/informationondrugs/approveddrugs/ucm490391.htm
- Bergethon K, Shaw AT, Ou SH, et al. ROS1 rearrangements define a unique molecular class of lung cancers. J Clin Oncol 2012;30:863-70. [Crossref] [PubMed]
- Kwak EL, Bang YJ, Camidge DR, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med 2010;363:1693-703. [Crossref] [PubMed]
- Shaw AT, Hsu PP, Awad MM, et al. Tyrosine kinase gene rearrangements in epithelial malignancies. Nat Rev Cancer 2013;13:772-87. [Crossref] [PubMed]
- Mino-Kenudson M, Chirieac LR, Law K, et al. A novel, highly sensitive antibody allows for the routine detection of ALK-rearranged lung adenocarcinomas by standard immunohistochemistry. Clin Cancer Res 2010;16:1561-71. [Crossref] [PubMed]
- Rimkunas VM, Crosby KE, Li D, et al. Analysis of receptor tyrosine kinase ROS1-positive tumors in non-small cell lung cancer: identification of a FIG-ROS1 fusion. Clin Cancer Res 2012;18:4449-57. [Crossref] [PubMed]
- Gulley JL, Rajan A, Spigel DR, et al. Avelumab for patients with previously treated metastatic or recurrent non-small-cell lung cancer (JAVELIN Solid Tumor): dose-expansion cohort of a multicentre, open-label, phase 1b trial. Lancet Oncol 2017;18:599-610. [Crossref] [PubMed]
- Garassino M, Vansteenkiste J, Kim JH, et al. Durvalumab in ≥3rd-Line Locally Advanced or Metastatic, EGFR/ALK Wild-Type NSCLC: Results from the Phase 2 ATLANTIC Study J Thorac Oncol 2017;12:S10-1. [Crossref]
- Fehrenbacher L, Spira A, Ballinger M, et al. Atezolizumab versus docetaxel for patients with previously treated non-small-cell lung cancer (POPLAR): a multicentre, open-label, phase 2 randomised controlled trial. Lancet 2016;387:1837-46. [Crossref] [PubMed]
- Rittmeyer A, Barlesi F, Waterkamp D, et al. Atezolizumab versus docetaxel in patients with previously treated non-small-cell lung cancer (OAK): a phase 3, open-label, multicentre randomised controlled trial. Lancet 2017;389:255-65. [Crossref] [PubMed]
- Borghaei H, Paz-Ares L, Horn L, et al. Nivolumab versus Docetaxel in Advanced Nonsquamous Non-Small-Cell Lung Cancer. N Engl J Med 2015;373:1627-39. [Crossref] [PubMed]
- Garon EB, Rizvi NA, Hui R, et al. Pembrolizumab for the treatment of non-small-cell lung cancer. N Engl J Med 2015;372:2018-28. [Crossref] [PubMed]
- Herbst RS, Baas P, Kim DW, et al. Pembrolizumab versus docetaxel for previously treated, PD-L1-positive, advanced non-small-cell lung cancer (KEYNOTE-010): a randomised controlled trial. Lancet 2016;387:1540-50. [Crossref] [PubMed]
- Travis WD, Brambilla E, Nicholson AG, et al. The 2015 World Health Organization Classification of Lung Tumors: Impact of Genetic, Clinical and Radiologic Advances Since the 2004 Classification. J Thorac Oncol 2015;10:1243-60. [Crossref] [PubMed]
- Tsao MS, Hirsch FR, Yatabe Y. IASLC atlas of ALK and ROS1 testing in lung cancer. 2nd ed. Colorado: Editorial Rx Press, 2016.
- Soda M, Choi YL, Enomoto M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature 2007;448:561-6. [Crossref] [PubMed]
- Fan L, Feng Y, Wan H, et al. Clinicopathological and demographical characteristics of non-small cell lung cancer patients with ALK rearrangements: a systematic review and meta-analysis. PLoS One 2014;9:e100866. [Crossref] [PubMed]
- Ying J, Guo L, Qiu T, et al. Diagnostic value of a novel fully automated immunochemistry assay for detection of ALK rearrangement in primary lung adenocarcinoma. Ann Oncol 2013;24:2589-93. [Crossref] [PubMed]
- Zheng Z, Liebers M, Zhelyazkova B, et al. Anchored multiplex PCR for targeted next-generation sequencing. Nat Med 2014;20:1479-84. [Crossref] [PubMed]
- Murakami Y, Mitsudomi T, Yatabe Y. A Screening Method for the ALK Fusion Gene in NSCLC. Front Oncol 2012;2:24. [Crossref] [PubMed]
- Yi ES, Boland JM, Maleszewski JJ, et al. Correlation of IHC and FISH for ALK gene rearrangement in non-small cell lung carcinoma: IHC score algorithm for FISH. J Thorac Oncol 2011;6:459-65. [Crossref] [PubMed]
- Wallander ML, Geiersbach KB, Tripp SR, et al. Comparison of reverse transcription-polymerase chain reaction, immunohistochemistry, and fluorescence in situ hybridization methodologies for detection of echinoderm microtubule-associated proteinlike 4-anaplastic lymphoma kinase fusion-positive non-small cell lung carcinoma: implications for optimal clinical testing. Arch Pathol Lab Med 2012;136:796-803. [Crossref] [PubMed]
- Hofman P, Ilie M, Hofman V, et al. Immunohistochemistry to identify EGFR mutations or ALK rearrangements in patients with lung adenocarcinoma. Ann Oncol 2012;23:1738-43. [Crossref] [PubMed]
- McLeer-Florin A, Moro-Sibilot D, Melis A, et al. Dual IHC and FISH testing for ALK gene rearrangement in lung adenocarcinomas in a routine practice: a French study. J Thorac Oncol 2012;7:348-54. [Crossref] [PubMed]
- Lopes LF, Bacchi CE. Anaplastic lymphoma kinase gene rearrangement in non-small-cell lung cancer in a Brazilian population. Clinics (Sao Paulo) 2012;67:845-7. [Crossref] [PubMed]
- To KF, Tong JH, Yeung KS, et al. Detection of ALK rearrangement by immunohistochemistry in lung adenocarcinoma and the identification of a novel EML4-ALK variant. J Thorac Oncol 2013;8:883-91. [Crossref] [PubMed]
- Cabillic F, Gros A, Dugay F, et al. Parallel FISH and immunohistochemical studies of ALK status in 3244 non-small-cell lung cancers reveal major discordances. J Thorac Oncol 2014;9:295-306. [Crossref] [PubMed]
- Paik JH, Choe G, Kim H, et al. Screening of anaplastic lymphoma kinase rearrangement by immunohistochemistry in non-small cell lung cancer: correlation with fluorescence in situ hybridization. J Thorac Oncol 2011;6:466-72. [Crossref] [PubMed]
- Kim H, Yoo SB, Choe JY, et al. Detection of ALK gene rearrangement in non-small cell lung cancer: a comparison of fluorescence in situ hybridization and chromogenic in situ hybridization with correlation of ALK protein expression. J Thorac Oncol 2011;6:1359-66. [Crossref] [PubMed]
- Park HS, Lee JK, Kim DW, et al. Immunohistochemical screening for anaplastic lymphoma kinase (ALK) rearrangement in advanced non-small cell lung cancer patients. Lung Cancer 2012;77:288-92. [Crossref] [PubMed]
- Sholl LM, Weremowicz S, Gray SW, et al. Combined use of ALK immunohistochemistry and FISH for optimal detection of ALK-rearranged lung adenocarcinomas. J Thorac Oncol 2013;8:322-8. [Crossref] [PubMed]
- Martinez P, Hernandez-Losa J, Montero MA, et al. Fluorescence in situ hybridization and immunohistochemistry as diagnostic methods for ALK positive non-small cell lung cancer patients. PLoS One 2013;8:e52261. [Crossref] [PubMed]
- Minca EC, Portier BP, Wang Z, et al. ALK status testing in non-small cell lung carcinoma: correlation between ultrasensitive IHC and FISH. J Mol Diagn 2013;15:341-6. [Crossref] [PubMed]
- Zhou J, Zhao J, Sun K, et al. Accurate and economical detection of ALK positive lung adenocarcinoma with semiquantitative immunohistochemical screening. PLoS One 2014;9:e92828. [Crossref] [PubMed]
- Shan L, Lian F, Guo L, et al. Combination of conventional immunohistochemistry and qRT-PCR to detect ALK rearrangement. Diagn Pathol 2014;9:3. [Crossref] [PubMed]
- Wang Q, Zhao L, Yang X, et al. Antibody 1A4 with routine immunohistochemistry demonstrates high sensitivity for ALK rearrangement screening of Chinese lung adenocarcinoma patients: A single-center large-scale study. Lung Cancer 2016;95:39-43. [Crossref] [PubMed]
- Gruber K, Kohlhaufl M, Friedel G, et al. A novel, highly sensitive ALK antibody 1A4 facilitates effective screening for ALK rearrangements in lung adenocarcinomas by standard immunohistochemistry. J Thorac Oncol 2015;10:713-6. [Crossref] [PubMed]
- Conklin CM, Craddock KJ, Have C, et al. Immunohistochemistry is a reliable screening tool for identification of ALK rearrangement in non-small-cell lung carcinoma and is antibody dependent. J Thorac Oncol 2013;8:45-51. [Crossref] [PubMed]
- Jiang L, Yang H, He P, et al. Improving Selection Criteria for ALK Inhibitor Therapy in Non-Small Cell Lung Cancer: A Pooled-Data Analysis on Diagnostic Operating Characteristics of Immunohistochemistry. Am J Surg Pathol 2016;40:697-703. [Crossref] [PubMed]
- Blackhall FH, Peters S, Bubendorf L, et al. Prevalence and clinical outcomes for patients with ALK-positive resected stage I to III adenocarcinoma: results from the European Thoracic Oncology Platform Lungscape Project. J Clin Oncol 2014;32:2780-7. [Crossref] [PubMed]
- Cutz JC, Craddock KJ, Torlakovic E, et al. Canadian anaplastic lymphoma kinase study: a model for multicenter standardization and optimization of ALK testing in lung cancer. J Thorac Oncol 2014;9:1255-63. [Crossref] [PubMed]
- Ibrahim M, Parry S, Wilkinson D, et al. ALK Immunohistochemistry in NSCLC: Discordant Staining Can Impact Patient Treatment Regimen. J Thorac Oncol 2016;11:2241-7. [Crossref] [PubMed]
- Wynes MW, Sholl LM, Dietel M, et al. An international interpretation study using the ALK IHC antibody D5F3 and a sensitive detection kit demonstrates high concordance between ALK IHC and ALK FISH and between evaluators. J Thorac Oncol 2014;9:631-8. [Crossref] [PubMed]
- Available online: https://www.accessdata.fda.gov/cdrh_docs/pdf14/p140025c.pdf
- Savic S, Diebold J, Zimmermann AK, et al. Screening for ALK in non-small cell lung carcinomas: 5A4 and D5F3 antibodies perform equally well, but combined use with FISH is recommended. Lung Cancer 2015;89:104-9. [Crossref] [PubMed]
- Nakamura H, Tsuta K, Yoshida A, et al. Aberrant anaplastic lymphoma kinase expression in high-grade pulmonary neuroendocrine carcinoma. J Clin Pathol 2013;66:705-7. [Crossref] [PubMed]
- Popat S, Gonzalez D, Min T, et al. ALK translocation is associated with ALK immunoreactivity and extensive signet-ring morphology in primary lung adenocarcinoma. Lung Cancer 2012;75:300-5. [Crossref] [PubMed]
- Rodig SJ, Mino-Kenudson M, Dacic S, et al. Unique clinicopathologic features characterize ALK-rearranged lung adenocarcinoma in the western population. Clin Cancer Res 2009;15:5216-23. [Crossref] [PubMed]
- Yoshida A, Tsuta K, Watanabe S, et al. Frequent ALK rearrangement and TTF-1/p63 co-expression in lung adenocarcinoma with signet-ring cell component. Lung Cancer 2011;72:309-15. [Crossref] [PubMed]
- Takeuchi K, Choi YL, Togashi Y, et al. KIF5B-ALK, a novel fusion oncokinase identified by an immunohistochemistry-based diagnostic system for ALK-positive lung cancer. Clin Cancer Res 2009;15:3143-9. [Crossref] [PubMed]
- Alì G, Proietti A, Pelliccioni S, et al. ALK rearrangement in a large series of consecutive non-small cell lung cancers: comparison between a new immunohistochemical approach and fluorescence in situ hybridization for the screening of patients eligible for crizotinib treatment. Arch Pathol Lab Med 2014;138:1449-58. [Crossref] [PubMed]
- Conde E, Suarez-Gauthier A, Benito A, et al. Accurate identification of ALK positive lung carcinoma patients: novel FDA-cleared automated fluorescence in situ hybridization scanning system and ultrasensitive immunohistochemistry. PLoS One 2014;9:e107200. [Crossref] [PubMed]
- Gao X, Sholl LM, Nishino M, et al. Clinical Implications of Variant ALK FISH Rearrangement Patterns. J Thorac Oncol 2015;10:1648-52. [Crossref] [PubMed]
- Jurmeister P, Lenze D, Berg E, et al. Parallel screening for ALK, MET and ROS1 alterations in non-small cell lung cancer with implications for daily routine testing. Lung Cancer 2015;87:122-9. [Crossref] [PubMed]
- Lantuejoul S, Rouquette I, Blons H, et al. French multicentric validation of ALK rearrangement diagnostic in 547 lung adenocarcinomas. Eur Respir J 2015;46:207-18. [Crossref] [PubMed]
- Marchetti A, Di Lorito A, Pace MV, et al. ALK Protein Analysis by IHC Staining after Recent Regulatory Changes: A Comparison of Two Widely Used Approaches, Revision of the Literature, and a New Testing Algorithm. J Thorac Oncol 2016;11:487-95. [Crossref] [PubMed]
- Pekar-Zlotin M, Hirsch FR, Soussan-Gutman L, et al. Fluorescence in situ hybridization, immunohistochemistry, and next-generation sequencing for detection of EML4-ALK rearrangement in lung cancer. Oncologist 2015;20:316-22. [Crossref] [PubMed]
- Tantraworasin A, Lertprasertsuke N, Kongkarnka S, et al. Retrospective study of ALK rearrangement and clinicopathological implications in completely resected non- small cell lung cancer patients in Northern Thailand: role of screening with D5F3 antibodies. Asian Pac J Cancer Prev 2014;15:3057-63. [Crossref] [PubMed]
- von Laffert M, Stenzinger A, Hummel M, et al. ALK-FISH borderline cases in non-small cell lung cancer: Implications for diagnostics and clinical decision making. Lung Cancer 2015;90:465-71. [Crossref] [PubMed]
- Zwaenepoel K, Van Dongen A, Lambin S, et al. Detection of ALK expression in non-small-cell lung cancer with ALK gene rearrangements--comparison of multiple immunohistochemical methods. Histopathology 2014;65:539-48. [Crossref] [PubMed]
- Wang J, Cai Y, Dong Y, et al. Clinical characteristics and outcomes of patients with primary lung adenocarcinoma harboring ALK rearrangements detected by FISH, IHC, and RT-PCR. PLoS One 2014;9:e101551. [Crossref] [PubMed]
- Ilie MI, Bence C, Hofman V, et al. Discrepancies between FISH and immunohistochemistry for assessment of the ALK status are associated with ALK 'borderline'-positive rearrangements or a high copy number: a potential major issue for anti-ALK therapeutic strategies. Ann Oncol 2015;26:238-44. [Crossref] [PubMed]
- Kim H, Xu X, Yoo SB, et al. Discordance between anaplastic lymphoma kinase status in primary non-small-cell lung cancers and their corresponding metastases. Histopathology 2013;62:305-14. [Crossref] [PubMed]
- Salido M, Pijuan L, Martínez-Avilés L, et al. Increased ALK gene copy number and amplification are frequent in non-small cell lung cancer. J Thorac Oncol 2011;6:21-7. [Crossref] [PubMed]
- Manning G, Whyte DB, Martinez R, et al. The protein kinase complement of the human genome. Science 2002;298:1912-34. [Crossref] [PubMed]
- Rikova K, Guo A, Zeng Q, et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell 2007;131:1190-203. [Crossref] [PubMed]
- Roskoski R Jr. ROS1 protein-tyrosine kinase inhibitors in the treatment of ROS1 fusion protein-driven non-small cell lung cancers. Pharmacol Res 2017;121:202-12. [Crossref] [PubMed]
- Gainor JF, Shaw AT. Novel targets in non-small cell lung cancer: ROS1 and RET fusions. Oncologist 2013;18:865-75. [Crossref] [PubMed]
- Bubendorf L, Büttner R, Al-Dayel F, et al. Testing for ROS1 in non-small cell lung cancer: a review with recommendations. Virchows Arch 2016;469:489-503. [Crossref] [PubMed]
- Yoshida A, Tsuta K, Wakai S, et al. Immunohistochemical detection of ROS1 is useful for identifying ROS1 rearrangements in lung cancers. Mod Pathol 2014;27:711-20. [Crossref] [PubMed]
- Boyle TA, Masago K, Ellison KE, et al. ROS1 immunohistochemistry among major genotypes of non-small-cell lung cancer. Clin Lung Cancer 2015;16:106-11. [Crossref] [PubMed]
- Acquaviva J, Wong R, Charest A. The multifaceted roles of the receptor tyrosine kinase ROS in development and cancer. Biochim Biophys Acta 2009;1795:37-52. [PubMed]
- Li C, Fang R, Sun Y, et al. Spectrum of oncogenic driver mutations in lung adenocarcinomas from East Asian never smokers. PLoS One 2011;6:e28204. [Crossref] [PubMed]
- Mescam-Mancini L, Lantuejoul S, Moro-Sibilot D, et al. On the relevance of a testing algorithm for the detection of ROS1-rearranged lung adenocarcinomas. Lung Cancer 2014;83:168-73. [Crossref] [PubMed]
- Sholl LM, Sun H, Butaney M, et al. ROS1 immunohistochemistry for detection of ROS1-rearranged lung adenocarcinomas. Am J Surg Pathol 2013;37:1441-9. [Crossref] [PubMed]
- Cha YJ, Lee JS, Kim HR, et al. Screening of ROS1 rearrangements in lung adenocarcinoma by immunohistochemistry and comparison with ALK rearrangements. PLoS One 2014;9:e103333. [Crossref] [PubMed]
- Rogers TM, Russell PA, Wright G, et al. Comparison of methods in the detection of ALK and ROS1 rearrangements in lung cancer. J Thorac Oncol 2015;10:611-8. [Crossref] [PubMed]
- Shan L, Lian F, Guo L, et al. Detection of ROS1 gene rearrangement in lung adenocarcinoma: comparison of IHC, FISH and real-time RT-PCR. PLoS One 2015;10:e0120422. [Crossref] [PubMed]
- Cao B, Wei P, Liu Z, et al. Detection of lung adenocarcinoma with ROS1 rearrangement by IHC, FISH, and RT-PCR and analysis of its clinicopathologic features. Onco Targets Ther 2015;9:131-8. [Crossref] [PubMed]
- Wu J, Lin Y, He X, et al. Comparison of detection methods and follow-up study on the tyrosine kinase inhibitors therapy in non-small cell lung cancer patients with ROS1 fusion rearrangement. BMC Cancer 2016;16:599. [Crossref] [PubMed]
- Kao HL, Yeh YC, Lin CH, et al. Diagnostic algorithm for detection of targetable driver mutations in lung adenocarcinomas: Comprehensive analyses of 205 cases with immunohistochemistry, real-time PCR and fluorescence in situ hybridization methods. Lung Cancer 2016;101:40-7. [Crossref] [PubMed]
- Selinger CI, Li BT, Pavlakis N, et al. Screening for ROS1 gene rearrangements in non-small-cell lung cancers using immunohistochemistry with FISH confirmation is an effective method to identify this rare target. Histopathology 2017;70:402-11. [Crossref] [PubMed]
- Yoshida A, Kohno T, Tsuta K, et al. ROS1-rearranged lung cancer: a clinicopathologic and molecular study of 15 surgical cases. Am J Surg Pathol 2013;37:554-62. [Crossref] [PubMed]
- Herbst RS, Soria JC, Kowanetz M, et al. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature 2014;515:563-7. [Crossref] [PubMed]
- Topalian SL, Hodi FS, Brahmer JR, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med 2012;366:2443-54. [Crossref] [PubMed]
- Brahmer J, Reckamp KL, Baas P, et al. Nivolumab versus Docetaxel in Advanced Squamous-Cell Non-Small-Cell Lung Cancer. N Engl J Med 2015;373:123-35. [Crossref] [PubMed]
- Available online: https://www.fda.gov/Drugs/InformationOnDrugs/ApprovedDrugs/ucm279174.htm
- Reck M, Rodriguez-Abreu D, Robinson AG, et al. Pembrolizumab versus Chemotherapy for PD-L1-Positive Non-Small-Cell Lung Cancer. N Engl J Med 2016;375:1823-33. [Crossref] [PubMed]
- Mino-Kenudson M. Programmed death-ligand 1 immunohistochemistry testing for non-small cell lung cancer in practice. Cancer 2017;125:521-8. [PubMed]
- Tsao MS, Kerr KM, Dacic S, et al. IASLC Atlas of PD-L1 Immunohistochemistry Testing in Lung Cancer. COlorado: Editorial Rx Press, 2017.
- Available online: https://www.nccn.org/professionals/physician_gls/PDF/nscl.pdf
- Ratcliffe MJ, Sharpe A, Midha A, et al. Agreement between Programmed Cell Death Ligand-1 Diagnostic Assays across Multiple Protein Expression Cutoffs in Non-Small Cell Lung Cancer. Clin Cancer Res 2017;23:3585-91. [Crossref] [PubMed]
- Scheel AH, Dietel M, Heukamp LC, et al. Harmonized PD-L1 immunohistochemistry for pulmonary squamous-cell and adenocarcinomas. Mod Pathol 2016;29:1165-72. [Crossref] [PubMed]
- Hirsch FR, McElhinny A, Stanforth D, et al. PD-L1 Immunohistochemistry Assays for Lung Cancer: Results from Phase 1 of the Blueprint PD-L1 IHC Assay Comparison Project. J Thorac Oncol 2017;12:208-22. [Crossref] [PubMed]
- Neuman T, London M, Kania-Almog J, et al. A Harmonization Study for the Use of 22C3 PD-L1 Immunohistochemical Staining on Ventana's Platform. J Thorac Oncol 2016;11:1863-8. [Crossref] [PubMed]
- Ameratunga M, Asadi K, Lin X, et al. PD-L1 and Tumor Infiltrating Lymphocytes as Prognostic Markers in Resected NSCLC. PLoS One 2016;11:e0153954. [Crossref] [PubMed]
- Huynh TG, Morales-Oyarvide V, Campo MJ, et al. Programmed Cell Death Ligand 1 Expression in Resected Lung Adenocarcinomas: Association with Immune Microenvironment. J Thorac Oncol 2016;11:1869-78. [Crossref] [PubMed]
- Igarashi T, Teramoto K, Ishida M, et al. Scoring of PD-L1 expression intensity on pulmonary adenocarcinomas and the correlations with clinicopathological factors. ESMO Open 2016;1:e000083. [Crossref] [PubMed]
- Inaguma S, Wang Z, Lasota J, et al. Comprehensive Immunohistochemical Study of Programmed Cell Death Ligand 1 (PD-L1): Analysis in 5536 Cases Revealed Consistent Expression in Trophoblastic Tumors. Am J Surg Pathol 2016;40:1133-42. [Crossref] [PubMed]
- Koh J, Go H, Keam B, et al. Clinicopathologic analysis of programmed cell death-1 and programmed cell death-ligand 1 and 2 expressions in pulmonary adenocarcinoma: comparison with histology and driver oncogenic alteration status. Mod Pathol 2015;28:1154-66. [Crossref] [PubMed]
- Parra ER, Behrens C, Rodriguez-Canales J, et al. Image Analysis-based Assessment of PD-L1 and Tumor-Associated Immune Cells Density Supports Distinct Intratumoral Microenvironment Groups in Non-small Cell Lung Carcinoma Patients. Clin Cancer Res 2016;22:6278-89. [Crossref] [PubMed]
- Paulsen EE, Kilvaer TK, Khanehkenari MR, et al. Assessing PDL-1 and PD-1 in Non-Small Cell Lung Cancer: A Novel Immunoscore Approach. Clin Lung Cancer 2017;18:220-33. [Crossref] [PubMed]
- Sheffield BS, Fulton R, Kalloger SE, et al. Investigation of PD-L1 Biomarker Testing Methods for PD-1 Axis Inhibition in Non-squamous Non-small Cell Lung Cancer. J Histochem Cytochem 2016;64:587-600. [Crossref] [PubMed]
- Smith J, Robida MD, Acosta K, et al. Quantitative and qualitative characterization of Two PD-L1 clones: SP263 and E1L3N. Diagn Pathol 2016;11:44. [Crossref] [PubMed]
- Tang Y, Fang W, Zhang Y, et al. The association between PD-L1 and EGFR status and the prognostic value of PD-L1 in advanced non-small cell lung cancer patients treated with EGFR-TKIs. Oncotarget 2015;6:14209-19. [Crossref] [PubMed]
- Uruga H, Bozkurtlar E, Huynh TG, et al. Programmed Cell Death Ligand (PD-L1) Expression in Stage II and III Lung Adenocarcinomas and Nodal Metastases. J Thorac Oncol 2017;12:458-66. [Crossref] [PubMed]
- Rimm DL, Han G, Taube JM, et al. A Prospective, Multi-institutional, Pathologist-Based Assessment of 4 Immunohistochemistry Assays for PD-L1 Expression in Non-Small Cell Lung Cancer. JAMA Oncol 2017;3:1051-8. [Crossref] [PubMed]
- Adam J, Rouquette I, Damotte D, et al. Multicentric French harmonization study for PD-L1 IHC testing in NSCLC. J Thorac Oncol 2017;12:S11-2. [Crossref]
- Cooper WA, Russell PA, Cherian M, et al. Intra- and Interobserver Reproducibility Assessment of PD-L1 Biomarker in Non-Small Cell Lung Cancer. Clin Cancer Res 2017;23:4569-77. [Crossref] [PubMed]
- Rehman JA, Han G, Carvajal-Hausdorf DE, et al. Quantitative and pathologist-read comparison of the heterogeneity of programmed death-ligand 1 (PD-L1) expression in non-small cell lung cancer. Mod Pathol 2017;30:340-9. [Crossref] [PubMed]
- Chen Z, Liu HB, Yu CH, et al. Diagnostic value of mutation-specific antibodies for immunohistochemical detection of epidermal growth factor receptor mutations in non-small cell lung cancer: a meta-analysis. PLoS One 2014;9:e105940. [Crossref] [PubMed]
- Available online: http://www.ema.europa.eu/docs/en_GB/document_library/EPAR_-_Public_assessment_report/human/003886/WC500202696.pdf
- Heist RS. First-Line Systemic Therapy for Non-Small Cell Lung Cancer. Hematol Oncol Clin North Am 2017;31:59-70. [Crossref] [PubMed]
- Bledsoe JR, Kamionek M, Mino-Kenudson M. BRAF V600E immunohistochemistry is reliable in primary and metastatic colorectal carcinoma regardless of treatment status and shows high intratumoral homogeneity. Am J Surg Pathol 2014;38:1418-28. [Crossref] [PubMed]
- Ilie M, Long E, Hofman V, et al. Diagnostic value of immunohistochemistry for the detection of the BRAFV600E mutation in primary lung adenocarcinoma Caucasian patients. Ann Oncol 2013;24:742-8. [Crossref] [PubMed]
- Sasaki H, Shimizu S, Tani Y, et al. Usefulness of immunohistochemistry for the detection of the BRAF V600E mutation in Japanese lung adenocarcinoma. Lung Cancer 2013;82:51-4. [Crossref] [PubMed]
- Nguyen-Ngoc T, Bouchaab H, Adjei AA, et al. BRAF Alterations as Therapeutic Targets in Non-Small-Cell Lung Cancer. J Thorac Oncol 2015;10:1396-403. [Crossref] [PubMed]
- Tong JH, Yeung SF, Chan AW, et al. MET Amplification and Exon 14 Splice Site Mutation Define Unique Molecular Subgroups of Non-Small Cell Lung Carcinoma with Poor Prognosis. Clin Cancer Res 2016;22:3048-56. [Crossref] [PubMed]
- Zhou F, Moreira AL. Lung Carcinoma Predictive Biomarker Testing by Immunoperoxidase Stains in Cytology and Small Biopsy Specimens: Advantages and Limitations. Arch Pathol Lab Med 2016;140:1331-7. [Crossref] [PubMed]
- Gown AM. Diagnostic Immunohistochemistry: What Can Go Wrong and How to Prevent It. Arch Pathol Lab Med 2016;140:893-8. [Crossref] [PubMed]
- Thavarajah R, Mudimbaimannar VK, Elizabeth J, et al. Chemical and physical basics of routine formaldehyde fixation. J Oral Maxillofac Pathol 2012;16:400-5. [Crossref] [PubMed]
- Fitzgibbons PL, Bradley LA, Fatheree LA, et al. Principles of analytic validation of immunohistochemical assays: Guideline from the College of American Pathologists Pathology and Laboratory Quality Center. Arch Pathol Lab Med 2014;138:1432-43. [Crossref] [PubMed]
- Available online: http://www.accessdata.fda.gov/cdrh_docs/pdf15/P150013c.pdf
- Available online: http://www.accessdata.fda.gov/cdrh_docs/pdf16/P160002c.pdf
- Midha A, Sharpe A, Scott M, et al. PD-L1 expression in advanced NSCLC: Primary lesions versus metastatic sites and impact of sample age. J Clin Oncol 2016;34:3025.
- Rebelatto MC, Midha A, Mistry A, et al. Development of a programmed cell death ligand-1 immunohistochemical assay validated for analysis of non-small cell lung cancer and head and neck squamous cell carcinoma. Diagn Pathol 2016;11:95. [Crossref] [PubMed]
- Heymann JJ, Pagan C, Crapanzano J, et al. PD-L1 expression in non-small cell lung carcinoma (NSCLC): feasibility of cytology and its comparison with resection and small biopsies. Mod Pathol 2017;30:479A.
- Russell-Goldman E, Sholl LM, Vivero M. Cytology-histologic correlation of PD-L1 immunohistochemistry in lung carcinomas. Mod Pathol 2017;30:114A.
- Skov BG, Skov T. Paired Comparison of PD-L1 Expression on Cytologic and Histologic Specimens From Malignancies in the Lung Assessed With PD-L1 IHC 28-8pharmDx and PD-L1 IHC 22C3pharmDx. Appl Immunohistochem Mol Morphol 2017;25:453-9. [Crossref] [PubMed]


