Genome-wide copy number analyses of samples from LACE-Bio project identify novel prognostic and predictive markers in early stage non-small cell lung cancer
Introduction
Lung cancer is the leading cause of cancer death worldwide. Non-small cell lung cancer (NSCLC), accounting for 85% of all lung cancers, has a 5-year survival of 59% for early resectable disease, but only 15% for cancers in advances stages (1). However, great differences within individual stages suggest the existence of unknown tumor factors. In the era of personalized medicine, the assessment of prognostic factors is crucial for individual treatment decision making. The activation of oncogenes (i.e., EGFR and KRAS) and the inhibition of tumor-suppressors (TP53) drive tumor progression. While targeting some of these genes is a promising therapeutic strategy in adenocarcinoma (ADC), most lung cancers lack proven (targetable) driver genes and identification of additional ones is critical. Recent developments of genome-wide profiling have identified new genes, but the studies reported to date are underpowered or lack a control arm. Bass et al. (2) profiled 40 esophageal squamous cell carcinomas (SCC) (29 primary and 11 cell lines) and 47 primary lung SCC for DNA copy number (CN) change. They reported that SOX2 (chr.3q26.33) was significantly amplified and that it was a lineage-survival oncogene by knockdown experiments in cell lines. However, the small sample size hindered assessment of the prognostic value of CN aberrations (CNA). The Cancer Genome Atlas (TCGA) recruited 10,000 samples from 33 cancer types and profiled alterations from genomic DNA, RNA, and protein. However, due to the inclusion criteria (≥70% tumor cellularity), advanced stages were underrepresented. Furthermore, the samples used in these studies were snap-frozen tissues whereas most of the samples in clinical settings are formalin-fixed and paraffin-embedded (FFPE). Thus, identifying prognostic markers from FFPE samples may be clinically relevant.
The Lung Adjuvant Cisplatin Evaluation (LACE-Bio) project comprises FFPE samples from four LACE adjuvant chemotherapy (ACT) trials and evaluated the prognostic and predictive role of biomarkers including ERCC1 (3), tumor infiltrating lymphocytes (TILs) (4), mucin (5), beta-tubulin (6), KRAS (7), EGFR (8) and TP53 (9). Importantly, 1,013 samples from three trials were profiled for their DNA CNAs. Since the trials were randomized and controlled, the data were fit for evaluating markers associated with the magnitude of ACT benefit.
Methods
Patients and samples
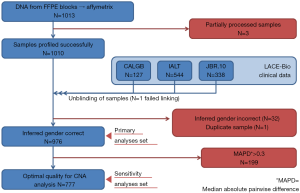
The LACE-Bio2 consortium includes patients from four pivotal trials comparing platinum-based ACT to observation after complete resection of stage I–III NSCLC (10-15). Of these, 1,013 patients from three trials had FFPE samples available, whereas samples in one trial (15) were exhausted. All individual trials including tissue collection for future research were approved by institutional review boards at each participating site.
DNA isolation and profiling
DNA was successfully extracted from 976 FFPE samples using the AllPrep DNA/RNA FFPE Kit (Quagen, Germantown, MD, USA), and profiled using the OncoScan CNV Plus Assay (ThermoFisher, Carlsbad, California, USA), a molecular inversion probe SNP assay (16). The platform algorithm delivered the median of the absolute values of all pairwise differences (MAPD) (17,18) as quality metrics; 777 samples with MAPD ≤0.3 were classified as optimal quality.
Statistical analyses
The data were normalized relative to a pool of reference normal samples and segmented using circular binary segmentation (19,20). Minimal recurrent regions were identified via the CGHregions algorithm (21). Tumor clonal composition number was estimated by using the OncoClone composition program (22). The primary endpoint was disease-free survival (DFS). Secondary endpoints were overall survival (OS) and lung-cancer specific survival (LCSS). CNAs were correlated to endpoints using Cox models stratified by trial and adjusted for treatment and clinicopathological factors. The regression models estimated the hazard ratio HRgain for a 2-fold higher CN, with HRloss = 1/HRgain the relative hazard for a 2-fold lower CN. The predictive role of CNAs was estimated by further adding a treatment-by-CN interaction to the models. We performed univariate (by region) and two multivariate analyses (stepwise selection and penalized regression) (23,24). Q values were used to correct P values for multiple comparisons (25).
Preplanned sensitivity analyses included: histologic subgroups (ADC vs. SCC), optimal quality subgroup. CN differences between histologies were assessed by t-tests, with P values corrected via step-down multiple testing procedures (26,27). We compared results to those from our reanalysis of the TCGA (28,29) using exactly the same method. Known tumor suppressors and oncogenes were obtained from literature (30).
The association of the number of breakpoints (BPs), quantifying chromosomal instability, with clinicopathological factors was tested in univariate analyses, then in multivariate log-linear models. The association of chromosomal instability and of clonality with outcomes and treatment effect was studied in Cox models.
Full details of statistical methods are provided in the supplementary material.
Results
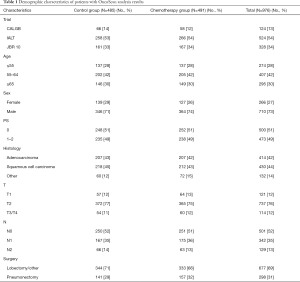
Three samples (Figure S1) were partially processed; 1 failed linkage to the clinical database; the inferred gender of 32 patients was incorrect; 1 sample was duplicate. In total, 976 samples were analyzed: 414 (42%) ADC, 430 (44%) SCC, 132 (14%) other NSCLC; 485 were in the control and 491 in the ACT groups (Table 1).
Full table
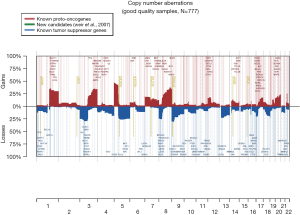
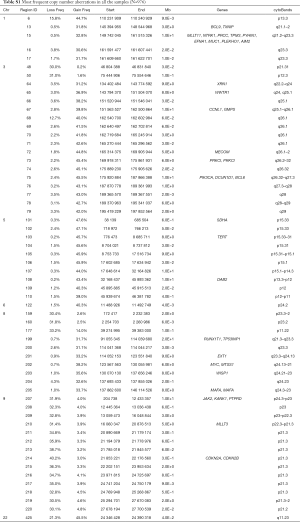
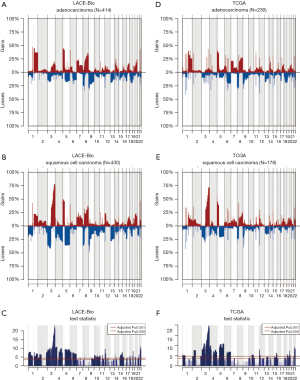
The 217,611 array probes were grouped into 431 common-CN regions; 253 regions (59%) were ≤3 Mb, 340 (79%) were ≤10 Mb (Figure 1); 166 regions had a loss (177 a gain) in ≥10% patients. On average, patients had 94 CNAs (standard deviation 69), 51 gains and 43 losses.
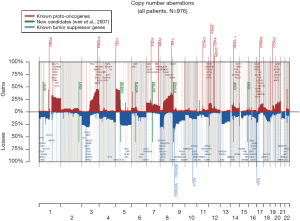
The most frequent CN gains (Table S1) were in 1q21–23, 3q22–26, 5p13–15, 6p24, 8q21–24, 22q11, containing genes TERT, PIK3CA, MECOM, CCNL1 among others. The most frequent CN losses were in chromosomes 3p21.31, 8p23, and 9p21.3, containing CDKN2A/B. These results remained consistent in the optimal quality samples subset (N=777; Figure S2, Table S2).
Full table

Full table
The CN profile was heterogeneous across histology and results were confirmed in our reanalysis of the TCGA data (Figure S3). The frequency of 195 regions (49% were ≤3 Mb and 71% ≤10 Mb; Table S3) was significantly different between ADC and SCC (Q≤0.05). The most significant differences were: more gains in 3q (including genes PIK3CA, MECOM, CCNL1), 22q (NF2, PDGFB) and 12p (KRAS) in SCC; more losses in 3p (RASSF1), 4 (PTTG2, NKX2-1), and 5q in SCC.

Full table
Copy-number aberrations associated with prognosis
The median follow-up for DFS (510 events) was 5.3 years. In univariate analyses (Table 2), 14 focal regions (11 ≤3 Mb, 14 ≤10 Mb) in loci 7p11–12, 9p21, 18q12, 19p11–13 were prognostic (P≤0.005) with Q≤0.142. Losses associated with shorter DFS were in: 8 regions in 9p21.3 (loss frequency: 31–40%, including CDKN2A/B), with HRloss =1.5 (95% CI: 1.2–1.9) (P<0.001, Q=0.02); one region in 19p13 [STK11, 11%, HRloss =2.4 (1.3–4.3), P=0.005, Q=0.15]; one in 18q12.1 [12%, HRloss =1.6 (1.2–2.3), P=0.004, Q=0.12]. Other seemingly deleterious losses were found in 19p11–13 (MLLT1, SH3GL1, TCF3, VAV1). Gains in 7p11–12 (frequency: 17%) were associated with shorter DFS [HRgain =2.0 (1.2–3.2), P=0.005, Q=0.14]. Two of these regions (7p12.3 and 9p21.3) remained significant in multivariate analyses (Table S4), which also suggested a benefit [HRloss =0.32 (0.16–0.61), P<0.001] for losses in a region in 1p31–36 (9.8%), including EPS15, FGR, JUN, LCK, PAX7, STIL, TAL1, NBL1, EPHB2, MUTYH, ARNT. Penalized regression confirmed the prognostic role of the region in 9p21.3, plus another one containing CDKN2A/B (Table S5).
Full table

Full table
Full table
The median follow-up for OS (451 events) was 5.3 years. The above-mentioned CN losses in 9p21, 18q12, 19p13 were also prognostic of shorter OS (P≤0.005, Q≤0.092; Table 2), together with 5 additional regions in 9p21.1, 18q12.1, and 19p12–13 (ELL). One further focal region on 14q23.1 (8.5% of losses, 89% of gains) was prognostic for OS (P=0.002, Q=0.079), with HRloss =2.2 (1.3–3.6), corresponding to HRgain =0.46 (0.28–0.76). The prognostic role of a region in 9p12.3 was confirmed in multivariate analyses (Table S4), together with the possible benefit for gains in 3q26 [MECOM, 45%, HRgain =0.55 (0.38–0.79), P=0.001]. Penalized regression (Table S5) did not select any region for OS.
The median follow-up for LCSS (427 events) was 5.0 years. Results were similar to DFS, with the addition of one region in chr8, for which gains (17%) were associated with longer LCSS [HRgain =0.51 (0.32–0.82), P=0.005, Q=0.13]. In multivariate analyses (Table S4), two of the three regions associated with DFS (chr3 and 9) were also associated with LCSS, in addition to regions in 6p24.2 [HRgain =1.3 (1.1–1.5), P=0.002], 8p23 [HRloss =0.53 (0.34–0.83), P=0.005], 19p13 [MLLT1, SH3GL1, TCF3, VAV1; HRloss =3.7 (1.7–7.7), P<0.001], and 20q11.21 [HRgain =0.44 (0.24–0.81), P=0.009]. Penalized regression (Table S5) selected 17 prognostic regions for LCSS on chr1 (EPS15, FGR, JUN, LCK, PAX7, STIL, TAL1, NBL1, EPHB2, MUTYH, NBL1, ARNT), chr9 (CDKN2A/B), chr12 (FGF6, ING4), chr19 (MLLT1, SH3GL1, TCF3, VAV1), and chr20 (HCK).
Copy-number aberrations associated with the effect of ACT
The average ACT effect on DFS estimated within the 976 patients with CN data was HRACT =0.85 (0.71–1.0) (P=0.06). Univariate analyses (Table 3) identified five regions in 14q32.33 as potentially predictive of better response to ACT (P<0.05), but with very high Q values. The effect of CNAs in these regions was similar. CN loss in one region in 14q32.33 had HRloss for interaction of 0.42 (0.22–0.83) (P=0.012, Q=0.010), corresponding to HRgain for interaction of 2.4 (1.2–4.6). This means that, given a treatment effect (ACT vs. control) of HR[ACT|CN=2] =0.85 for a patient with CN=2, such an effect is stronger for a patient with CN=1 (HR[ACT|CN=1] =0.42×0.85=0.36) and reversed with CN=4 (HR[ACT|CN=4] =2.4×0.85=2.0). The predictive role of this region was the only confirmed in multivariate analyses (Table S6), with HRloss for interaction of 0.39 (0.20–0.79) (P=0.009).

Full table

Full table
The average effect of ACT on OS was HRACT=0.95 (0.79–1.1) (P=0.58). At a raw P<0.05, 5 regions were possibly associated to the ACT effect for OS, but with very high Q values (Table 3). One region in 8p23.2 showed a treatment effect enhanced for the 31.8% of patients with a CN loss [HRloss for interaction 0.73 (0.57–0.95), P=0.019, Q=0.76), meaning that the HR for a patient with CN=1 was HR[ACT|CN=1] =0.73×0.95=0.69. An adjacent region in 8p23 was selected in multivariate analyses (Table S6), with a similar effect [HRloss for interaction 0.42 (0.19–0.93), P=0.032). In univariate analyses, 3 regions in chr10 (BMI1, NET1, MAP3K8, BMI1, MLLT10, ZMYND11, RET, RASSF4) with 5–7% losses and 4–5% gains showed predictive effects with HRloss for interaction 0.26–0.28 and CNgain for interaction 3.6–3.9. One region in 15q26 (losses: 4.7%, gains: 4.5%) had HRloss for interaction 0.21 (0.06–0.71) (P=0.012, Q=0.76) corresponding to HRgain for interaction 4.8 (1.4–16). In multivariate analyses (Table S6) one region in 14q32.33 was predictive (P=0.006), with HRloss for interaction 0.35 (0.17–0.74) and HRgain for interaction 2.8 (1.4–6.0).
The average effect of ACT on LCSS was HRACT =0.83 (0.68–1.0) (P=0.05). Three of the above-mentioned regions in 14q32 predictive of ACT effect for DFS were also predictive for LCSS (Table 3). Two additional regions in 20q11.21 (gain frequency: 20%) had possibly significant interaction with ACT, with HRgain for interaction 5.6 (1.9–16) (P=0.002, Q=0.57) and 5.9 (1.9–19) (P=0.003, Q=0.57), respectively. Two of them (14q32 and 20q11) were confirmed in multivariate analyses (Table S6).
Penalized regression did not select any predictive region for either endpoint.
Sensitivity analyses
The results within the optimal quality sample subgroup (Tables S7-S10) were consistent with those of the whole population. Table S11 shows the genomic regions for which the prognostic effect was significantly different between ADC and SCC (interaction P<0.005). CN gains in two regions in 1q23–31 (FCGR2B, PBX1, TPR, LHX4, CDC73) were associated to shorter DFS in ADC [HR =2.8 (1.3–5.8) and 2.3 (1.1–4.7)] and longer DFS in SCC [HR =0.44 (0.18–1.1) and 0.53 (0.27–1.0)]. One of these regions showed similar results for LCSS. Similar results were observed for 3 regions in 7p11 (also for LCSS and including EGFR), one in 7q11, one in 11p14 (also for OS), and one in 20q11, with increased risk in ADC and reduced risk in SCC for CN gains. Of note, only one region (chr11p14) had quite low interaction Q-value and only for OS (Q=0.056). Conversely, CN gains in 3 further regions [1p13, 4p12–15 (PTTG2), 4q27] were associated to longer OS in ADC [HR =0.50 (0.19–1.3), 0.20 (0.07–0.57), and 0.51 (0.28–0.92), respectively] than in SCC [HR =2.4 (1.0–5.6), 2.1 (0.9–4.8), and 1.6 (0.97–2.6), respectively].

Full table

Full table
Full table

Full table

Full table
Chromosomal instability
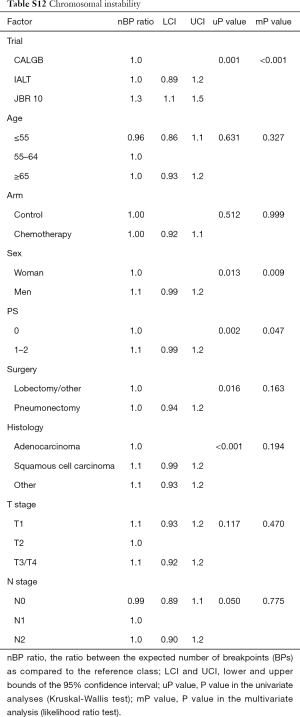
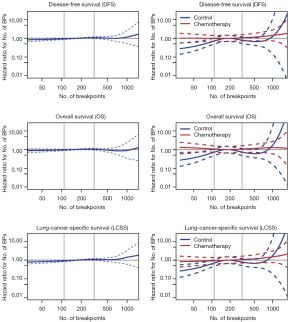
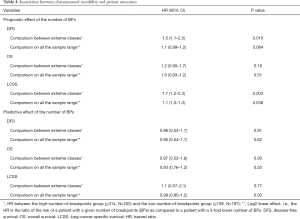
The number of BPs was heterogeneous across trials, higher for men and possibly for high performance status (Table S12). Patients with a very high number of BPs (≥314) had shorter DFS than patients with very few (≤109) [HR =1.5 (1.1–2.0), P=0.015). This result was weaker for OS [HR =1.2 (0.90–1.7), P=0.19), but stronger for LCSS [HR =1.7 (1.2–2.3), P=0.003). Flexible models (Figure S4) in all patients showed that the BP effect can be considered log-linear. Such an effect was HR =1.1 (0.99–1.2, P=0.084, Table 4) on DFS for a patient as compared to another having a two times fewer BPs; this log-linear effect was similar LCSS [HR =1.1 (1.0–1.3), P=0.036) and statistically not significant on OS [HR =1.0 (0.93–1.2), P=0.51). The treatment effect was independent of the number of BPs both when comparing extreme groups (HR range: 0.96 to 1.1, P range: 0.78 to 0.93) and in terms of log-linear effects (HR: 0.93 to 0.99, P: 0.53 to 0.93).
Full table
Full table
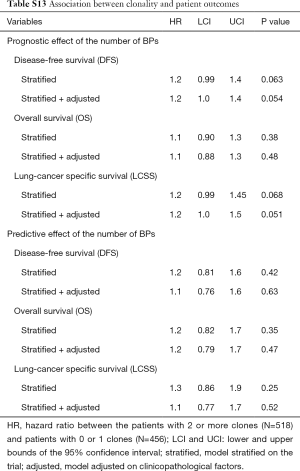
Clonality
Patients with 2+ clones (N=518) had shorter DFS and LCSS [HR =1.2 (1.0–1.4 and 1.0–1.3), Table S13] than patients with 0–1 clones (N=456). This result was statistically non-significant (P=0.054 and 0.051, respectively) notably for OS [HR =1.1 (0.88–1.3), P=0.48]. The treatment effect was not associated to clonality [P=0.63 (DFS), 0.47 (OS), 0.52 (LCSS)].
Full table
Discussion
Increased understanding of the genomic changes of NSCLC facilitates the identification of prognostic and predictive biomarkers and provides vital information for personalized therapy, potentially allowing tailored treatments for individual patients. We utilized NSCLC FFPE samples from the LACE-Bio project to profile DNA CNAs. The most frequent CN gains were found on 1p13, 1q21, 3q22–26, 5p13–15, 6p24, and 22q11, the most frequent losses on 3p21.31, 8p23, and 9p21.3. The more focal and less frequent losses might be due to harder identification of losses in tumors with stromal cell contamination. Telomerase reverse transcriptase (TERT), among the most frequently amplified genes, is the catalytic subunit of the enzyme telomerase; its overexpression has been associated with poor prognosis (31). Among the loss genes, cyclin-dependent kinase Inhibitor 2A (CDKN2A), reported to be deleted in many tumors including lung cancer (32), codes for two proteins, p16 (or p16INK4a) and p14arf, which act as tumor suppressors by regulating the cell cycle.
The different spectrum of CNAs between ADC and SCC has been reported previously (33,34). Genes such as PIK3CA (33) and PDGFB (35) were amplified in lung SCC. Cyclin L (CCNL1) has been identified as oncogene in head and neck cancer (36). Mutations in CHEK2 (37) and NF2 (38) have been reported to be associated with SCC. CN loss and promoter hypermethylation of RASSF1 was reported in SCCHN (39) and in early stage NSCLC (40). NKX2-1 amplification was significantly less frequent than in ADC (33).
Our analyses confirmed some of the prognostic genes reported in the literature, such as shorter survival with CN loss of CDKN2A/B (32). In the present study, CDKN2A/B CN loss occurred in 40% of the cases and was significantly associated with shorter DFS. CDKN2A/B CN loss was also prognostic in ADC. Copy number loss of the tumor suppressor STK11 (or LKB1) has been associated with increased risk of brain metastasis (41). We were not able to confirm this due to incomplete reporting of metastatic sites. NSCLC patients with STK11 exon 1 or 2 mutations have shorter survival (42). A recent meta-analysis (14 studies, 1915 patients with solid tumors) revealed that decreased expression of STK11 was a prognostic factor [HR =2.2 (1.5–3.2), P<0.001] (43). In the present study, STK11 CN loss was found in 11% of samples and was significantly associated with shorter DFS [HR =2.4 (1.3–4.3), P=0.005]. We also identified novel prognostic genes, such as FSTL3, which encodes a secreted glycoprotein, and transcriptional factors MLLT1, SH3GL1, and TCF3, and the guanine nucleotide exchange factor (GEF) gene VAV1. Its overexpression significantly increased the risk of death [HR =1.81 (1.39–2.36), P<0.001) (44). However, in the present study, the CN loss frequency of the region containing these genes was 9%. Additional studies on their prognostic value are warranted.
The LACE-bio study has the unique possibility to identify biomarkers that predict efficacy of ACT in NSCLC by comparison to observation arms. Three regions had significant differences in multivariate analyses between the two study arms, but they came with high false discovery rate (Table S6). Particularly, 8p23.3–2 losses were significantly associated with increased ACT efficacy for OS. The frequent gains of 20q11.21 strongly were associated with no benefit from ACT for LCSS. This deleterious effect from ACT was in strong contrast with the small group (0.8%) of patients with 20q11.21 loss where ACT lead to a notably high survival benefit. The 20q11.21 region is rich in genes that might have a potential role in cancer such as HCK (tyrosine kinase), BCL2L1 (apoptotic regulator), MAPRE1 and TPX2 (microtubule associated factors), DNMT3B (epigenetic modifier) and transcriptional regulators. It is even more striking to find the p53 and DNA damage-regulated gene named PDRG1 in 20q11.21. PDRG1 is an oncogene in lung cancer cell lines, is selectively regulated by DNA damaging agents such as UV, and promotes radioresistance (45,46). Whatsoever, its exact role in mediating resistance to ACT in NSCLC remains to be confirmed. Finally, the 14q32.33 region also had differential HR (loss predictive of ACT efficacy, gain predictive of inefficacy), but the proportion of patients with losses and gains were equally high (10.8% and 11.4% respectively), making interpretation more difficult in the context of prediction of ACT efficacy.
In exploratory analyses in the LACE-Bio2 samples, the prognosis of patients with very high chromosomal instability was significantly worse than for patients with very low, independently of the clinical factors. Chromosomal instability could likely be associated with the risk of relapses rather than to death. We found no association with the magnitude of the ACT effect.
The LACE-Bio data and tissue bank provided a valuable source for studying the prognostic and predictive role of the CN of genomic regions in stage I–III NSCLC. These large-scale genome-wide analyses were consistent with previous results and provide new candidate prognostic markers. Furthermore, as the data come from randomized controlled trials, we propose new markers which could predict the effect of ACT.
Detailed bioinformatics and statistical methods
Bioinformatics pre-processing
The CGH data were normalized relative to an internal pool of 390 reference normal tissues, segmented using circular binary segmentation (CBS) (17,18), and minimal recurrent regions were identified via the CGHregions algorithm (19). We planned to discard regions with <20 CNAs. The proportion of probes on the X-chromosome with called allelic imbalance allowed inferring patient gender. The inferred gender was compared to the actual gender and inconsistent samples were discarded.
Endpoints
The primary endpoint was:
- Disease-free survival (DFS), defined as the time from randomization to first recurrence (loco-regional or distant) or death from any cause.
Secondary endpoints were:
- Overall survival (OS), defined as the time from randomization to death from any cause, and;
- Lung-cancer specific survival (LCSS), defined as the time from randomization to death from lung cancer. Death without evidence of cancer relapse was treated as censoring for LCSS.
Statistical analyses
The called copy number (CN) of each region was correlated to survival endpoints via Cox models stratified by trial and adjusted for treatment arm, patient age, sex, performance status, histology, type of surgery, T, and N stage. The CN entered in the regression models as log2(CN). Thus, the estimated hazard ratio (HRgain) expresses the relative hazard for a 2-fold higher CN of a given region. Its reciprocal (HRloss = 1/HRgain) is the relative hazard for a 2-fold lower CN. To evaluate the predictive role of CNAs, a treatment-by-log2(CN) interaction was further added. In both the prognostic and the predictive models, the Cox model was stratified by trial and adjusted for treatment arm and clinical variables.
We performed both univariate (each region separately) and multivariate analyses (several regions jointly). The P values were corrected to control the false discovery rate [Q values (20)]. Multivariate models were built by stepwise selection (αin =0.10 and αout =0.01) and using a penalized regression approach (21,22) with lasso penalty for prognostic analyses and adaptive lasso for predictive analyses.
Preplanned sensitivity analyses were
The analyses were repeated, in addition to the entire study population, within the following subgroups:
- Histological subtypes (ADC vs. SCC);
- Optimal quality subgroup (MAPD ≤0.3).
The significance of the CN differences between histologic subtypes was assessed by t-tests; the P values were corrected via step- down multiple testing procedures (23,24). We compared the obtained results to those from TCGA (25,26). Known tumor suppressor genes and oncogenes were obtained from previously published results (27).
Chromosomal instability
The number of breakpoints (BPs) in the CN was used as measure of chromosomal instability. Its association with clinicopathological factors was first tested in univariate analyses (Kruskal-Wallis tests), then in a multivariate analysis using a log-linear quasi-Poisson model. Its association with outcomes and treatment effect was studied in Cox models comparing the 20% of patients with the highest number of BPs (≥314) to the 20% of patients with the lowest (≤109).
Software
The bioinformatics pre-processing and the statistical analyses were performed using R software v3.3, with the following packages: biospear, CGHbase, CGHcall, CGHregions, DNAcopy, glmnet, gplots, parallel, qvalue, scales, survival, TxDb.Hsapiens.UCSC.hg19.knownGene, XLConnect.
Acknowledgements
The authors would like to acknowledge Ni Liu (Princess Margaret Cancer Centre), Nicolas Lemaitre (Institut Albert Bonniot) and Shakeel Virk (Canadian Cancer Trials Group) for technical assistance. Grants from the US NCI R01 grant, Ligue Nationale Contre le Cancer (France), le Programme National d’Excellence Spécialisé cancer du poumon de l’Institut National du Cancer (INCa) (France), Canadian Cancer Society, the Gustave Roussy Foundation, the Princess Margaret Cancer Foundation and the European contract EU-FP7 Curelung.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The study was approved by institutional review boards (No. UHN 04-0333-T).
References
- Howlader N, Noone A, Krapcho M, et al. SEER Cancer Statistics Review, 1975-2014. Bethesda, MD; 2017. Available online: https://seer.cancer.gov/csr/1975_2014/. Based on November 2016 SEER data submission, posted to the SEER web site, April 2017.
- Bass AJ, Watanabe H, Mermel CH, et al. SOX2 is an amplified lineage-survival oncogene in lung and esophageal squamous cell carcinomas. Nat Genet 2009;41:1238-42. [Crossref] [PubMed]
- Friboulet L, Olaussen KA, Pignon JP, et al. ERCC1 Isoform Expression and DNA Repair in Non–Small-Cell Lung Cancer. N Engl J Med 2013;368:1101-10. [Crossref] [PubMed]
- Brambilla E, Le Teuff G, Marguet S, et al. Prognostic Effect of Tumor Lymphocytic Infiltration in Resectable Non-Small-Cell Lung Cancer. J Clin Oncol 2016;34:1223-30. [Crossref] [PubMed]
- Graziano SL, Lacas B, Vollmer R, et al. Cross-validation analysis of the prognostic significance of mucin expression in patients with resected non-small cell lung cancer treated with adjuvant chemotherapy: Results from IALT, JBR.10 and ANITA. Lung Cancer 2013;82:149-55. [Crossref] [PubMed]
- Reiman T, Lai R, Veillard AS, et al. Cross-validation study of class III beta-tubulin as a predictive marker for benefit from adjuvant chemotherapy in resected non-small-cell lung cancer: analysis of four randomized trials. Ann Oncol 2012;23:86-93. [Crossref] [PubMed]
- Shepherd FA, Domerg C, Hainaut P, et al. Pooled Analysis of the Prognostic and Predictive Effects of KRAS Mutation Status and KRAS Mutation Subtype in Early-Stage Resected Non-Small-Cell Lung Cancer in Four Trials of Adjuvant Chemotherapy. J Clin Oncol 2013;31:2173-81. [Crossref] [PubMed]
- Shepherd FA, Lacas B, Le Teuff G, et al. Pooled Analysis of the Prognostic and Predictive Effects of TP53 Comutation Status Combined With KRAS or EGFR Mutation in Early-Stage Resected Non–Small-Cell Lung Cancer in Four Trials of Adjuvant Chemotherapy. J Clin Oncol 2017;35:2018-27. [Crossref] [PubMed]
- Tsao M-S, Marguet S, Le Teuff G, et al. Subtype Classification of Lung Adenocarcinoma Predicts Benefit From Adjuvant Chemotherapy in Patients Undergoing Complete Resection. J Clin Oncol 2015;33:3439-46. [Crossref] [PubMed]
- Arriagada R, Bergman B, Dunant A, et al. Cisplatin-Based Adjuvant Chemotherapy in Patients with Completely Resected Non–Small-Cell Lung Cancer. N Engl J Med 2004;350:351-60. [Crossref] [PubMed]
- Arriagada R, Dunant A, Pignon JP, et al. Long-Term Results of the International Adjuvant Lung Cancer Trial Evaluating Adjuvant Cisplatin-Based Chemotherapy in Resected Lung Cancer. J Clin Oncol 2010;28:35-42. [Crossref] [PubMed]
- Winton T, Livingston R, Johnson D, et al. Vinorelbine plus cisplatin vs. observation in resected non-small-cell lung cancer. N Engl J Med 2005;352:2589-97. [Crossref] [PubMed]
- Butts CA, Ding K, Seymour L, et al. Randomized phase III trial of vinorelbine plus cisplatin compared with observation in completely resected stage IB and II non-small-cell lung cancer: updated survival analysis of JBR-10. J Clin Oncol 2010;28:29-34. [Crossref] [PubMed]
- Strauss GM, Herndon JE, Maddaus MA, et al. Adjuvant Paclitaxel Plus Carboplatin Compared With Observation in Stage IB Non-Small-Cell Lung Cancer: CALGB 9633 With the Cancer and Leukemia Group B, Radiation Therapy Oncology Group, and North Central Cancer Treatment Group Study Groups. J Clin Oncol 2008;26:5043-51. [Crossref] [PubMed]
- Douillard JY, Rosell R, De Lena M, et al. Adjuvant vinorelbine plus cisplatin versus observation in patients with completely resected stage IB–IIIA non-small-cell lung cancer (Adjuvant Navelbine International Trialist Association [ANITA]): a randomised controlled trial. Lancet Oncol 2006;7:719-27. [Crossref] [PubMed]
- Foster JM, Oumie A, Togneri FS, et al. Cross-laboratory validation of the OncoScan® FFPE Assay, a multiplex tool for whole genome tumour profiling. BMC Med Genomics 2015;8:5. [Crossref] [PubMed]
- Wang Y, Carlton VE, Karlin-Neumann G, et al. High quality copy number and genotype data from FFPE samples using Molecular Inversion Probe (MIP) microarrays. BMC Med Genomics 2009;2:8. [Crossref] [PubMed]
- Xie T, d’Ario G, Lamb JR, et al. A Comprehensive Characterization of Genome-Wide Copy Number Aberrations in Colorectal Cancer Reveals Novel Oncogenes and Patterns of Alterations. Corvalan AH, ed. PLoS One 2012;7:e42001.
- Olshen AB, Venkatraman ES, Lucito R, et al. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics 2004;5:557-72. [Crossref] [PubMed]
- Venkatraman ES, Olshen AB. A faster circular binary segmentation algorithm for the analysis of array CGH data. Bioinformatics 2007;23:657-63. [Crossref] [PubMed]
- van de Wiel MA, Van Wieringen WN. CGHregions: Dimension reduction for array CGH data with minimal information loss. Cancer Inform 2007;3:55-63. [Crossref] [PubMed]
- Sakai K, Ukita M, Schmidt J, et al. Clonal composition of human ovarian cancer based on copy number analysis reveals a reciprocal relation with oncogenic mutation status. Cancer Lett 2017;405:22-8. [Crossref] [PubMed]
- Ternès N, Rotolo F, Michiels S. Empirical extensions of the lasso penalty to reduce the false discovery rate in high-dimensional Cox regression models. Stat Med 2016;35:2561-73. [Crossref] [PubMed]
- Ternès N, Rotolo F, Heinze G, et al. Identification of biomarker-by-treatment interactions in randomized clinical trials with survival outcomes and high-dimensional spaces. Biom J 2017;59:685-701. [Crossref] [PubMed]
- Storey JD. A direct approach to false discovery rates. J R Stat Soc Ser B 2002;64:479-98. (Statistical Methodol). [Crossref]
- Westfall PH, Young SS. Resampling-Based Multiple Testing: Examples and Methods for P-Value Adjustment. New York, NY: John Wiley & Sons, 1993.
- Dudoit S, Shaffer JP, Boldrick JC. Multiple Hypothesis Testing in Microarray Experiments. Stat Sci 2003;18:71-103. [Crossref]
- Cancer Genome Atlas Research Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature 2012;489:519-25. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014;511:543-50. [Crossref] [PubMed]
- Weir BA, Woo MS, Getz G, et al. Characterizing the cancer genome in lung adenocarcinoma. Nature 2007;450:893-98. [Crossref] [PubMed]
- Aviel-Ronen S, Coe BP, Lau SK, et al. Genomic markers for malignant progression in pulmonary adenocarcinoma with bronchioloalveolar features. Proc Natl Acad Sci 2008;105:10155-60. [Crossref] [PubMed]
- Nobori T, Miura K, Wu DJ, et al. Deletions of the cyclin-dependent kinase-4 inhibitor gene in multiple human cancers. Nature 1994;368:753-6. [Crossref] [PubMed]
- Inoue Y, Matsuura S, Kurabe N, et al. Clinicopathological and Survival Analysis of Japanese Patients with Resected Non-Small-Cell Lung Cancer Harboring NKX2-1, SETDB1, MET, HER2, SOX2, FGFR1, or PIK3CA Gene Amplification. J Thorac Oncol 2015;10:1590-600. [Crossref] [PubMed]
- Lee E, Moon JW, Wang X, et al. Genomic Copy Number Signatures Uncovered a Genetically Distinct Group from Adenocarcinoma and Squamous Cell Carcinoma in Non-Small Cell Lung Cancer. Hum Pathol 2015;46:1111-20. [Crossref] [PubMed]
- Donnem T, Al-Shibli K, Al-Saad S, et al. Prognostic Impact of Fibroblast Growth Factor 2 in Non-small Cell Lung Cancer: Coexpression with VEGFR-3 and PDGF-B Predicts Poor Survival. J Thorac Oncol 2009;4:578-85. [Crossref] [PubMed]
- Redon R, Hussenet T, Bour G, et al. Amplicon mapping and transcriptional analysis pinpoint cyclin L as a candidate oncogene in head and neck cancer. Cancer Res 2002;62:6211-7. [PubMed]
- Wang Y, McKay JD, Rafnar T, et al. Rare variants of large effect in BRCA2 and CHEK2 affect risk of lung cancer. Nat Genet 2014;46:736-41. [Crossref] [PubMed]
- Yoo NJ, Park SW, Lee SH. Mutational analysis of tumour suppressor gene NF2 in common solid cancers and acute leukaemias. Pathology 2012;44:29-32. [Crossref] [PubMed]
- Hogg RP, Honorio S, Martinez A, et al. Frequent 3p allele loss and epigenetic inactivation of the RASSF1A tumour suppressor gene from region 3p21.3 in head and neck squamous cell carcinoma. Eur J Cancer 2002;38:1585-92. [Crossref] [PubMed]
- Buckingham L, Penfield Faber L, Kim A, et al. PTEN, RASSF1 and DAPK site-specific hypermethylation and outcome in surgically treated stage I and II nonsmall cell lung cancer patients. Int J Cancer 2010;126:1630-9. [PubMed]
- Zhao N, Wilkerson MD, Shah U, et al. Alterations of LKB1 and KRAS and risk of brain metastasis: Comprehensive characterization by mutation analysis, copy number, and gene expression in non-small-cell lung carcinoma. Lung Cancer 2014;86:255-61. [Crossref] [PubMed]
- Pécuchet N, Laurent-Puig P, Mansuet-Lupo A, et al. Different prognostic impact of STK11 mutations in non-squamous non-small-cell lung cancer. Oncotarget 2017;8:23831-40. [Crossref] [PubMed]
- Xiao J, Zou Y, Chen X, et al. The Prognostic Value of Decreased LKB1 in Solid Tumors: A Meta-Analysis. PLoS One 2016;11:e0152674. [Crossref] [PubMed]
- Qi Y, Kong FM, Deng Q, et al. Clinical significance and prognostic value of Vav1 expression in Non-small cell lung cancer. Am J Cancer Res 2015;5:2491-7. [PubMed]
- Jiang L, Luo X, Shi J, et al. PDRG1, a novel tumor marker for multiple malignancies that is selectively regulated by genotoxic stress. Cancer Biol Ther 2011;11:567-73. [Crossref] [PubMed]
- Tao Z, Chen S, Mao G, et al. The PDRG1 is an oncogene in lung cancer cells, promoting radioresistance via the ATM-P53 signaling pathway. Biomed Pharmacother 2016;83:1471-77. [Crossref] [PubMed]