The cytokinesis-blocked micronucleus assay as a novel biomarker for selection of lung cancer screening participants
Introduction
In 2002, the a Lung Screening Trial (NLST) was established to determine whether screening participants at high risk for lung cancer with low dose CT (LDCT) versus chest X-ray (CXR) would reduce lung cancer mortality rates among high risk smokers. In 2010, the study was terminated due to the compelling observation of 20.3% reduction in lung cancer mortality within the LDCT group compared to the CXR group (1,2). Considering that approximately 222,500 new cases of lung cancer and 155,870 deaths were expected in the US in 2017 (3), representing 25% of all cancer mortality potential benefits from screening trials such as the NLST in terms of total lives saved are immense. However, despite the dramatic results of the NLST, there are major concerns regarding screening with LDCT that include: possible harmful effects of cumulative radiation exposure from multiple CT scans; surgical complications in patients who prove not to have lung cancer and; the high false positive and negative rates.
Data from the NLST identified ~25% of the participants as having a positive finding for lung cancer of which 96% of the participants, after further diagnostic evaluation, were not lung cancer (1). Results from other screening trials reported similar findings to the NLST which underscores the need for better methods to accurately identify true positive lesions (4,5).
Despite this high false positive rate, the results from the NLST demonstrated that LDCT screening is effective in high risk individuals. NLST definition of high risk was limited to age (55–75 years) and pack years (smoked ≥30 pack-years and quit <15 years). With such criteria, over 9 million smokers meet NLST eligibility criteria, while an estimated 20.3 million smokers aged 55–74 years are considered NLST-ineligible. Kovalchik et al. (6) reported that in order to prevent one lung cancer death, 302 subjects have to be screened, however, by focusing on the individuals with highest pre-screening risk this number is reduced to 208 and the false positives per CT-prevented lung-cancer death were decreased from 108 to 78 in the three highest risk groups (6). This data reinforces the role for risk-based screening, and using individualized risk assessment instead of NLST entry criteria to increase efficiency of LDCT screening in early detection of lung cancer. With an estimated 94 million current and former smokers in the United States, there is an urgent need to improve screening detection outcome by identifying and validating markers of risk and early detection.
Although over 80% of lung cancers are attributed to tobacco exposure, only 15% of smokers develop lung cancer in their lifetimes which emphasize the role of genetic susceptibility on modulating the risk of developing the disease (7,8). This observation is a classic example of genetic host susceptibility as a risk modifier for development of cancer (9) and can explain the variation of individual susceptibility among current and former smokers (10). Therefore, the term “high-risk” as defined by the NLST (based on age and pack-years) needs refinement through the addition of robust and sensitive biomarkers that would allow the accurate identification of the true high-risk smokers that should be targeted in the lung screening programs.
Chromosome aberrations in peripheral blood lymphocytes have been shown to be a viable biomarker to measure both DNA damage and cancer risks (11-14). An increased frequency of aberrations has generally been considered indicative of subsequent cancer risk in humans, reflecting early biological effects of genotoxic carcinogens and individual cancer susceptibility (12,15).
The cytokinesis blocked micronucleus assay (CBMN) in human lymphocytes is one of the most commonly used methods for measuring DNA damage in the form of binucleated micronuclei (BN-MN) and nucleoplasmic bridges (BN-NPBs). The BN-MN originate from chromosome fragments or whole chromosomes that fail to engage with the mitotic spindle and therefore lag behind when the cell divides, and BN-NPBs originate from asymmetrical chromosome rearrangements and/or telomere end fusions (16,17). We have previously reported that the CBMN biomarker endpoints are strong predictors of lung cancer susceptibility (18-20). Furthermore, we have externally validated our findings in an independent lung cancer population and have extended the existing Spitz lung cancer risk prediction model (21) by adding the CBMN biomarker endpoints. Our results suggest a substantive increase in the overall discriminatory power of the Spitz model, with the greatest performance improvement observed among never-smokers followed by former and current smokers (22). These findings prompted the current investigation testing the utility of the CBMN endpoints in lung cancer screening participants. The goal of this study is to identify high-risk subgroups based on their genetic susceptibility to lung cancer rather than on the NLST study eligibility criteria of age and smoking status. Such an approach, using a robust and cost-effective assay, is highly appropriate for use in large population screening programs and would have immense public health impact.
Methods
Study participants
All study participants were recruited at The University of Texas M. D. Anderson Cancer Center (MDACC) under an approved institutional review board protocol entitled “Repository and research supplementation on the national lung screening trial in current and former smokers”. The goal of the study was to compile a comprehensive database of epidemiologic risk factors and clinical data on all study participants enrolled in the ACRIN arm of the NLST at the MDACC site. Risk factors that are not part of the NLST protocol were collected on all the study participants to serve as a resource for future studies. In addition, after signing an informed consent, all study participants donated a 10-mL blood sample at time of entry into the study (baseline, T0) for testing blood-based biomarkers of lung cancer. Since the NLST was a randomized trial comparing LDCT with CXR, eligible participants from both arms were included in the current study and their randomization status remained blinded until the termination of the NLST study.
PBL cultures for CBMN assay
The CBMN assay was performed using the cytochalasin B technique described by Fenech and Morley (23) and following recommendations from the International Collaborative Project on Micronucleus Frequency in Human Populations (HUMN Project) (24). Duplicate lymphocyte cultures were prepared for each study subject. Each culture contained 2.0×106 cells in 5 mL RPMI 1640 medium supplemented with 100 U/mL penicillin, 100 µg/mL streptomycin, 10% fetal bovine serum, and 2 mM L-glutamine (Gibco-Invitrogen, Carlsbad, CA, USA) and 1% phytohemagglutinin (Remel, Lenexa, KS, USA). At 44 hours after initiation, cells were blocked in cytokinesis by adding cytochalasin B (Sigma, St. Louis, MO, USA; final concentration 4 µg/mL). The total incubation time for all cultures was 72 hours. After incubation, the cells were fixed in 3:1 methanol: glacial acetic acid, dropped onto clean microscopic slides, air-dried and stained with Giemsa stain. For each sample, 1,000 binucleated (BN) cells were scored blindly using a Nikon E-400 light optical microscope following the scoring criteria outlined by HUMN Project (18-20). The endpoints scored were number of: (I) binucleated micronuclei (BN-MN) (reflective of chromosome fragments or whole chromosomes that lag behind when the cell divides); (II) binucleated nucleoplasmic bridges (BN-NPBs) (reflective of asymmetrical chromosome rearrangements and/or telomere end fusions (23-25). The CBMN assay was performed only one time which was at the time of entry into the study (T0).
NLST-imaging information
The imaging findings among the study participants were reported according to whether the participant was randomized to the LDCT arm or the CXR arm. At entry to the study, the baseline imaging findings (T0) for the LDCT arm were described as: negative, negative with minor abnormalities, negative with significant abnormalities not suspicious for lung cancer, positive 4–10 mm nodule suspicious for lung cancer, positive >10 mm nodule suspicious for lung cancer and inadequate study due to technical limitations. For the CXR arm, the baseline findings were described as: negative, negative with minor abnormalities, negative with significant abnormalities not suspicious for lung cancer, positive with nodule or mass suspicious for lung cancer, inadequate study due to technical limitations. The same classifications were used to define imaging results at year 1 follow-up (T1) and year 2 follow-up (T2) with the additional classification of positive, stable abnormalities potentially related to lung cancer. For this evaluation, participants with negative minor or negative significant abnormality not suspicious for lung cancer imaging results were combined with participants with negative imaging results. Participants were further defined as having a false positive at T0 or T1 if a participant was defined as positive, but further follow-up resulted in a negative or negative minor or negative significant abnormality conclusion.
Among those participants who had a negative imaging result at T0, we further characterized their end-of-screening study status (based on results of T1/T2 study) as: “negative-negative” if study result was negative/negative significant abnormality; or “negative-positive” if study result was positive (CXR arm) or positive 4–10 mm, positive >10 mm (LDCT arm).
After completion of the low dose CT screening component of the study (T0–T2); surveillance of participants continued for a median duration of 6.5 years (maximum of 7.4 years) and lung cancer diagnosis was captured during the long-term follow-up phase. Participants who were negative at T0 and negative at end of long-term follow-up phase were defined as “long-term follow-up negative”; participants who were negative at T0 and positive at end of long-term follow-up phase were defined as “long-term follow-up positive”.
Statistical analysis
All analyses were performed using the Intercooled Stata 15.0 statistical software package (Stata Corp., College Station, TX, USA). We used descriptive statistics to characterize the study population at baseline (T0), overall and by study arm by calculating mean and standard deviations (SD) of continuous variables and frequency and percentage for categorical variables. One-way analysis of variance (ANOVA) was used to compare mean baseline time of entry BN-NPB and BN-MN outcomes at baseline, separately, among T0 and T1 defined negative, positive and false positive participant groups. At T2, two-sample t-tests were used to compare mean baseline time of entry BN-NPB and BN-MN outcomes between T2 defined negative and positive groups. Two-sample t-tests were also used to compare mean BN-NPB and BN-MN outcomes at each time-point between participants between negative-negative and negative-positive groups. In addition, growth curve models were used to compare trajectories of change in BN-NPB and BN-MNs among the negative-negative and negative-positive participant groups. Two-sample t-tests were also used to compare mean baseline time of entry BN-NPB and BN-MN outcomes between participants with positive imaging finding or diagnosed with lung cancer during the long-term follow-up period and those who remained negative and no lung cancer diagnosis in the long-term follow-up period.
Results
Demographics and the study population
Seven-hundred and eighty-seven current and former smokers participated in the NLST MDACC study. A total of 642 study participants agreed to be included in the current ancillary study and consented to completing the risk factor questionnaire and donated blood. The number of participants with complete demographic, imaging and biomarker data at entry into the study (T0) was 641, including 311 in the CXR arm and 330 in the LDCT arm. Table 1 summarizes the patient population at time of entry into the study. The study included 319 current smokers, 300 former smokers and 22 recent quitters. The cohort consisted of 63.3% males and 36.7% females, with 89.1% being Caucasians, 6.7% African Americans, 3.4% Hispanic and 0.8% other. The average age of the cohort was 62.7 years old with 30.4% in the 55–59; 35.6% in the 60–64; 22.5% in 65–69 and 11.5% in 70–74 years old age groups.
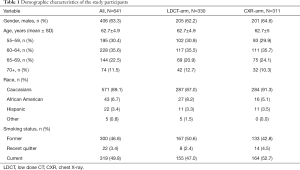
Full table
Frequencies of the baseline CBMN endpoints with imaging findings
Baseline BN-NPB endpoint and imaging findings
At T0, overall mean number of BN-NPBs among all the study participants was 1.4. The mean number of BN-NPBs was higher among the study participants (n=105) with T0 positive findings (regardless of imaging modality), mean ± standard error of the mean (SEM) =1.49±0.13 compared to those with T0 negative findings (n=535), mean ± SEM =1.38±0.05. However, when the positive findings group were further classified into positive and false positive, the level of BN-NPBs was higher among the positive findings group (n=11), mean ± SEM =2.09±0.53 compared to the false positive group (n=94), mean + SE =1.41±0.13. Although a positive trend was observed when comparing the extent of the BN-NPBs in the negative, false positive and positive groups, the results were not significant (P=0.1612; Table 2). Similar findings were observed when comparing each imaging modality (LDCT or CXR arms) separately.
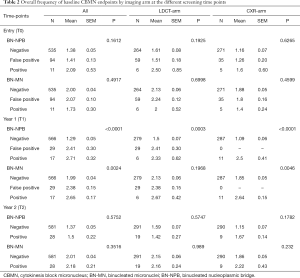
Full table
There was a significantly (P<0.0001) higher mean number of BN-NPBs among the study participants with T1 positive findings (n=17) (regardless of imaging modality), mean ± SEM =2.71±0.32 as compared to those with T1 negative findings (n=566), mean + SEM =1.29±0.05, but not those in the T1 false positive findings (n=29), mean + SE =2.41±0.30 (Table 2). Similar findings were observed when comparing each imaging modality separately. The overall mean number of BN-NPBs among the study participants with T2 positive findings (n=28) were not significantly different from the T2 negative findings (n=581) (Table 2). Similar results were observed when comparing each imaging modality separately.
Baseline BN-MN endpoint and imaging findings
At T0, overall mean number of BN-MNs was 2. The mean number of BN-MNs among the study participants with T0 positive findings (n=105) (regardless of imaging modality), mean ± SEM =2.03±0.10 as compared to those with T0 negative findings (n=535), mean ± SEM =2.00±0.04 (P>0.05). When the positive findings group was further classified into positive (n=11) and false positive (n=94), the mean ± SEM in the positive group =1.73±0.30, compared to the false positive 2.07±0.10 (P=0.4917; Table 2). Similar findings were observed when comparing each imaging modality separately.
There was a significantly (P=0.002) higher mean number of BN-MNs among the study participants with T1 positive findings (regardless of imaging modality), mean ± SEM =2.65±0.17 as compared to those with T1 negative findings, mean ± SEM =1.99±0.04, but not those in the T1 false positive findings, mean ± SEM =2.38±0.15 (Table 2). Similar findings were observed when comparing each imaging modality separately. The overall mean number of BN-MNs among the study participants with T2 positive findings mean ± SEM =2.18±0.21 were higher compared to those with T2 negative findings, mean ± SEM =2.01±0.04, but not significantly so (P=0.3516) (Table 2). Similar results were observed when comparing each imaging modality separately.
Change in CBMN endpoints across time among the study participants
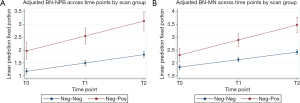
Growth curve models were used to compare expected trajectories of change in CBMN endpoints (BN-NPB and BN-MNs) among the study participants who were defined as negative-negative or negative-positive at the T0 compared to the end of screening study period. The models showed that the negative-negative participants (negative at T0 and remained negative at T2) had significantly lower mean number of BN-NPBs across all time points (all P<0.0001; Table 3) compared to negative-positive participants who were negative at T0 but positive at T2. Mean number of BN-NPBs showed an increase across time for both groups; however the magnitude of increase was significantly higher (P<0.0001) among the negative-positive participants compared to the negative-negative participants (Figure 1A). Similarly, the mean number of BN-MNs was significantly lower across all time points (P=0.0004 at T0 and P<0.0001 at T1 and T2; Table 3) among negative-negative participants compared to the negative-positive participants. The mean number of BN-MNs increased across time for both groups; however the magnitude of increase was significantly higher (P<0.0001) among the negative-positive compared to the negative-negative participants (Figure 1B).
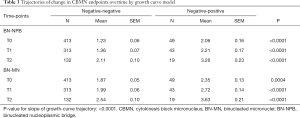
Full table
Frequency of CBMN endpoints and end of long-term follow-up
We investigated the association between the frequency of the CBMN endpoints at T0 and end of long-term follow-up phase results. Participants were classified into negative and positive groups. The negative group included participants who remained negative throughout the 3-year screening study and had no reported lung cancer at the end of the follow-up phase. The positive group, on the other hand, included those who had a positive finding during the 3-year screening study or had reported lung cancer at the end of the follow-up phase. Table 4 shows a significantly (P=0.0009) higher frequency in both CBMN endpoints at T0 with the mean ± SEM =1.68±0.11 vs. 1.31±0.05 for the BN-NPBs among the positive group (n=154) as compared to the negative group (n=486), and a significantly (P=0.0443) higher frequency for the BN-MNs with a mean ± SEM =2.14±0.08 vs. 1.97±0.04 in the positive and negative groups respectively.

Full table
Discussion
In 2013, the US Preventive Services Task Force recommended annual lung cancer screening with LDCT for high risk smokers and former smokers based on age (55–80 years) and smoking history (>30 pack-years in current smokers and <15 years of quitting in former smokers) (26). However, it has been suggested that lung cancer risk prediction models based on an individual’s risk should be used as a better targeted approach for selection of screening candidates (27,28). A crucial early event in carcinogenesis is the induction of DNA damage and genomic instability phenotype, which enables an initiated cell to evolve into a cancer cell by achieving a greater proliferative capacity (25). Such instability is mediated through chromosomal changes, at a gross level, and is therefore cytogenetically detected (29). We have previously extended an existing lung cancer risk prediction model with the addition of genomic instability endpoints and reported a substantive increase in the overall discriminatory power of the Spitz model (22). In the current study, we used the CBMN biomarker assay to measure the extent of DNA damage among pre-diagnostic smokers participating in an ancillary study that included NLST participants at the MD Anderson site. Several reasons prompted the selection of the CBMN endpoint for this study. First, cytogenetic assays are well-validated cancer risk biomarkers that reflect the exposure of an individual to clastogenic agents and individual’s repair capacity (11-15,30). Second, the CBMN assay in human lymphocytes is one of the most commonly used methods for measuring DNA damage and repair. Third, the CBMN assay is cost effective and is highly appropriate for use in large population screening studies. In this study, we tested the use of the CBMN endpoints as robust biomarkers to identify high-risk smokers that should be targeted in the lung screening programs based on their genetic susceptibility rather than on their age and smoking status. We showed that overall, the frequency of BN-NPBs and BN-MN endpoints (performed at time of entry into the study, T0) are significantly lower among study participants with negative imaging findings throughout the study period as compared to participants with positive imaging findings during the annual screenings and/or reported a lung cancer diagnosis at the end-of study follow-up period.
Eleven cases of lung cancer were diagnosed at time of entry into the study (T0). Although, we did not observe significant differences in CBMN endpoints between participants who were identified as having positive versus negative findings (on either LDCT or CXR arms), the frequency of the BN-NPBs was higher among the participants with positive findings as compared to those with false positive findings (Table 2). A similar frequency of BN-NPBs among the false positive and the negative groups was observed (Table 2) which highlights the sensitivity of BN-NPBs in discriminating between the cancer and non-cancer groups. The most striking results were observed between the CBMN endpoints and imaging findings in year one (T1), where seventeen incident lung cancers were diagnosed through screening in both imaging arms. Both the BN-NPB and the BN-MN endpoints were significantly higher among the participants who were identified with positive findings as compared to the false positive and negative finding groups (Table 2). These findings are supported by our previous lung cancer case control studies showing the high predictive values of the CBMN endpoints and lung cancer risk (20,22) as well as by other studies reporting the similar findings in other cancer sites (31-33). The results of the CBMN endpoints and imaging findings at T2, were higher among the positive findings group, however, the differences were not significant. A plausible explanation for the variation in level of significance over the screening time points may be due to the small sample size in each group as well as the relatively short period between evaluations. This is supported by the observed end of long term follow-up study findings that the extent of genomic instability among the participants who remained negative throughout the 3-year screening study and had no reported lung cancer at the end of the follow-up period was significantly lower that the those who had a positive finding during the 3-year screening study or had reported lung cancer at the end of the follow-up period.
It is well known that DNA repair capacity decreases as a function of age thus increasing the risk of cancer (34-36). Approximately 70% of participants included in our study were over the age of 60 at the time of entry into the study and therefore we expected an increase in the level of genomic instability through the course of the study period. We used Growth curve models to estimate the rate of increase in genomic instability over the course of the study period. Our results showed an increase in genomic instability among all study participants, however, the projected increase (as measured by the slope of growth curve trajectory) was significantly higher among those who were negative at T0 and positive by the end of screening study period as compared to those who remained negative. These findings provide additional support to the use of CBMN endpoints as predictors of genetically susceptible smokers that should be targeted for lung screening programs.
Although smoking is the main risk factor for developing lung cancer, up to 25% of all lung cancers occur in never-smokers (37,38). In the US it is estimated there are 17,000–26,000 annual deaths from lung cancer in never-smokers many of whom have either been exposed to second hand smoke or are genetically susceptible to development of lung cancer (39). Thus lung cancer among never-smokers is a significant public health need that needs to be addressed. There has been ongoing debate regarding recommending lung cancer screening among never-smokers at high risk of lung cancer (27,40-43). We have previously reported that the CBMN endpoints are predictive of lung cancer risk among never-smokers and that the extension of the Spitz model allowed a substantial improvement in the discriminatory power of the risk model in never-smokers (22). The simplicity, rapidity, and sensitivity of the CBMN assay make it a valuable tool for assessing genetic instability among populations that are not eligible for LDCT screening such as smokers younger than 50 years old and never-smokers and therefore prioritizing potential cases for surveillance programs.
Prevention of even 10% of annual deaths from lung cancer would save an estimated 17,000 lives, equivalent to all the annual deaths in the United States from ovarian cancer and almost all the annual deaths from brain cancers. The association between genetic instability and lung cancer CT screening outcomes could prove complementary for early detection of lung cancer and dramatically improves the performance of screening. Using the current lung cancer screening guidelines of 55 years of age with a 30-pack year smoking history, results in a very high false positive rate. While successful despite this high false positive rate, there are economical and psychological costs from high false positive rates that must be reduced. A blood biomarker that identifies individuals by their genetic susceptibility as true high risk of lung cancer would be a major asset that would complement CT lung cancer screening and would include those genetically susceptible to development of the disease and those outside of the current guidelines (susceptible young age smokers and never-smokers). Identifying the true high-risk individuals is expected to increase the number of lung cancer detected early while reducing the numbers of persons screened. Guidelines for appropriate follow-up of indeterminate lung nodules are based on radiologist experience and there are no scientific studies of which follow-up procedure results in the best outcome. Therefore, a sensitive biomarker-such as the CBMN-that improves the radiologist’s ability to predict the risk of a nodule being a true lung cancer would greatly improve the interpretation of screening studies. Subsequently, this would reduce unnecessary treatment, overall costs and emotional stress of false positive screens.
Our study has limitations, mainly the relatively small sample size at the different study time points which may have contributed to variation in our results; however, with the overall larger sample size, the data are consistently higher among the negative-positive versus the negative-negative participants. Compared to the overall NLST study participants, the number of subjects enrolled at the MD Anderson site was relatively small which prevented us from further stratifications. However, the data presented in this report warrant further investigation and validation in larger screening studies.
In order to identify the true high-risk individuals, it is imperative to consider validated biomarkers of risk. To our knowledge, our study is the first to assess genetic susceptibility in a high-risk lung cancer screening population using genetic instability markers. Such an approach could have immense public health significance by complementing CT screening in large populations as well as identifying high-risk subgroups that might benefit from increased screening surveillance that is not appropriate for low-risk individuals.
Acknowledgements
Funding: This study was supported in parts by NIH/NCI CA129050, RA El-Zein.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The study was approved by the Institutional Review Boards prior to subject enrollment. All subjects signed a study-specific, written informed consent form all participants.
References
- National lung screening trial research team, Aberle DR, Adams AM, et al. Reduced lung-cancer mortality with low dose computed tomographic screening. N Engl J Med 2011;365:395-409.
- Kramer BS, Berg CD, Aberle DR, et al. Lung cancer screening with low-dose helical CT: results from the national lung screening trial (NLST). J Med Screen 2011;18:109-11. [Crossref] [PubMed]
- American Cancer Society. Cancer Facts and Figures. Atlanta, GA: American Cancer Society, Inc, 2017.
- Swensen SJ, Jett JR, Hartman TE, et al. Lung cancer screening with CT: Mayo Clinic experience. Radiology 2003;226:756-61. [Crossref] [PubMed]
- Henschke CI, Yip R, Yankelevitz DF, et al. Computed tomography screening for lung cancer. Ann Intern Med 2013;159:156-7. [Crossref] [PubMed]
- Kovalchik SA, Tammemagi M, Berg CD, et al. Targeting of low-dose CT screening according to the risk of lung-cancer death. N Engl J Med 2013;369:245-54. [Crossref] [PubMed]
- Amos CI, Wu X, Broderick P, et al. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nature Genet 2008;40:616-22. [Crossref] [PubMed]
- Doll R, Peto R. Cigarette-smoking and bronchial carcinoma: dose and time relationships among regular smokers and lifelong non-smokers. J Epidemiol Community Health 1978;32:303-13. [Crossref] [PubMed]
- Hoffmann D, Hoffmann I, El Bayoumy K. Less harmful cigarette: a controversial issue. A tribute to Ernst L. Wynder. Chem Res Toxicol 2001;14:767-90. [Crossref] [PubMed]
- Sellers TA, Potter JD, Bailey-Wilson JE, et al. Lung cancer detection and prevention: evidence for an interaction between smoking and genetic predisposition. Cancer Res 1992;52:2694s-7s. [PubMed]
- Liou SH, Lung JC, Chen YH, et al. Increased chromosome-type chromosome aberration frequencies as biomarkers of cancer risk in a blackfoot endemic area. Cancer Res 1999;59:1481-4. [PubMed]
- Bonassi S, Hagmar L, Stromberg U, et al. Chromosomal aberrations in lymphocytes predict human cancer independently of exposure to carcinogens. European Study Group on Cytogenetic Biomarkers and Health. Cancer Res 2000;60:1619-25. [PubMed]
- Bonassi S, Znaor A, Norppa H, et al. Chromosomal aberrations and risk of cancer in humans: an epidemiologic perspective. Cytogenet Genome Res 2004;104:376-82. [Crossref] [PubMed]
- Smerhovsky Z, Landa K, Rossner P, et al. Risk of cancer in an occupationally exposed cohort with increased level of chromosomal aberrations. Environ Health Perspect 2001;109:41-5. [Crossref] [PubMed]
- Hagmar L, Bonassi S, Strömberg U, et al. Cancer predictive value of cytogenetic markers used in occupational health surveillance programs. Recent Results Cancer Res 1998;154:177-84. [Crossref] [PubMed]
- Umegaki K, Fenech M. Cytokinesis-block micronucleus assay in WIL2-NS cells: a sensitive system to detect chromosomal damage induced by reactive oxygen species and activated human neutrophils. Mutagenesis 2000;15:261-9. [Crossref] [PubMed]
- Stewénius Y, Gorunova L, Jonson T, et al. Structural and numerical chromosome changes in colon cancer develop through telomere mediated anaphase bridges, not through mitotic multipolarity. Proc Natl Acad Sci U S A 2005;102:5541-6. [Crossref] [PubMed]
- El-Zein RA, Schabath MB, Etzel CJ, et al. Cytokinesis-blocked micronucleus assay as a novel biomarker for lung cancer risk. Cancer Res 2006;66:6449-56. [Crossref] [PubMed]
- El-Zein RA, Fenech M, Lopez MS, et al. Cytokinesis-blocked micronucleas cytome assay biomarkers identify lung cancer cases amongst smokers. Cancer Epidemiol Biomarkers Prev 2008;17:1111-9. [Crossref] [PubMed]
- McHugh MK, Lopez MS, Ho CH, et al. Use of the cytokinesis-blocked micronucles assay to detect gender differences and genetic instability in a lung cancer case control study. Cancer Epidemiol Biomarkers Prev 2013;22:135-45. [Crossref] [PubMed]
- Spitz MR, Hong WK, Amos CI, et al. A risk model for prediction of lung cancer. J Natl Cancer Inst 2007;99:715-26. [Crossref] [PubMed]
- El-Zein RA, Lopez MS, D'Amelio AM Jr, et al. The cytokinesis-blocked micronucleus assay as a strong predictor of lung cancer: extension of a lung cancer risk prediction model. Cancer Epidemiol Biomarkers Prev 2014;23:2462-70. [Crossref] [PubMed]
- Fenech M, Morley AA. Measurement of micronuclei in lymphocytes. Mutat Res 1985;147:29-36. [Crossref] [PubMed]
- Fenech M, Chang WP, Kirsch-Volders M, et al. HUMN project: detailed description of the scoring criteria for the cytokinesis-block micronucleus assay using isolated human lymphocyte cultures. Mutat Res 2003;534:65-75. [Crossref] [PubMed]
- Fenech M. Chromosomal biomarkers of genomic instability relevant to cancer. Drug Discov Today 2002;7:1128-37. [Crossref] [PubMed]
- Moyer VA. U.S. Preventive Services Task Force. Screening for lung cancer: U.S. Preventive Services Task Force recommendation statement. Ann Intern Med 2014;160:330-8. [PubMed]
- Tammemägi MC, Church TR, Hocking WG, et al. Evaluation of the lung cancer risks at which to screen ever- and never-smokers: screening rules applied to the PLCO and NLST cohorts. PLoS Med 2014;11:e1001764. [Crossref] [PubMed]
- Ten Haaf K, Jeon J, Tammemägi MC, et al. Risk prediction models for selection of lung cancer screening candidates: A retrospective validation study. PLoS Med 2017;14:e1002277. [Crossref] [PubMed]
- Solomon E, Borrow J, Goddard AD. Chromosome aberrations and cancer. Science 1991;254:1153-60. [Crossref] [PubMed]
- Hsu TC, Johnston DA, Cherry LM, et al. Sensitivity to genotoxic effects of bleomycin in humans: possible relationship to environmental carcinogenesis. Int J Cancer 1989;43:403-9. [Crossref] [PubMed]
- Ionescu EM, Nicolaie T, Ionescu MA, et al. Predictive cytogenetic biomarkers for colorectal neoplasia in medium risk patients. J Med Life 2015;8:398-403. [PubMed]
- Bitgen N, Donmez-Altuntas H, Bayram F, et al. Increased micronucleus, nucleoplasmic bridge, nuclear bud frequency and oxidative DNA damage associated with prolactin levels and pituitary adenoma diameters in patients with prolactinoma. Biotech Histochem 2016;91:128-36. [Crossref] [PubMed]
- Pardini B, Viberti C, Naccarati A, et al. Increased micronucleus frequency in peripheral blood lymphocytes predicts the risk of bladder cancer. Br J Cancer 2017;116:202-10. [Crossref] [PubMed]
- Dollé ME, Giese H, Hopkins CL, et al. Rapid accumulation of genome rearrangements in liver but not in brain of old mice. Nat Genet 1997;17:431-4. [Crossref] [PubMed]
- Gorbunova V, Seluanov A, Mao Z, et al. Changes in DNA repair during aging. Nucleic Acids Res 2007;35:7466-74. [Crossref] [PubMed]
- Rübe CE, Fricke A, Widmann TA, et al. Accumulation of DNA damage in hematopoietic stem and progenitor cells during human aging. PLoS One 2011;6:e17487. [Crossref] [PubMed]
- Koo LC, Ho JH. Worldwide epidemiological patterns of lung cancer in nonsmokers. Int J Epidemiol 1990;19:S14-23. [Crossref] [PubMed]
- Parkin DM, Bray F, Ferlay J, et al. Global cancer statistics, 2002. CA Cancer J Clin 2005;55:74-108. [Crossref] [PubMed]
- Rivera GA, Wakelee H. Lung cancer in never-smokers. Adv Exp Med Biol 2016;893:43-57. [Crossref] [PubMed]
- McCarthy WJ, Meza R, Jeon J, et al. Chapter 6: Lung cancer in never-smokers: epidemiology and risk prediction models. Risk Anal 2012;32:S69-84. [Crossref] [PubMed]
- Silvestri GA, Nietert PJ, Zoller J, et al. Attitudes towards screening for lung cancer among smokers and their non-smoking counterparts. Thorax 2007;62:126-30. [Crossref] [PubMed]
- Tammemägi MC, Katki HA, Hocking WG, et al. Selection criteria for lung-cancer screening. N Engl J Med 2013;368:728-36. [Crossref] [PubMed]
- Subramanian J, Velcheti V, Gao F, et al. Presentation and stage specific outcomes of lifelong never-smokers with non-small cell lung cancer (NSCLC). J Thorac Oncol 2007;2:827-30. [Crossref] [PubMed]


