Lung cancer prediction using machine learning and advanced imaging techniques
Introduction
The demonstration of a 20% reduction in lung cancer mortality in the USA National Lung Screening Trial (NLST) (1) and the subsequent decision by the U.S. Centers for Medicare and Medicaid Services to provide Medicare coverage for lung cancer screening has paved the way for nationwide lung cancer screening in the USA.
This decision also underscored the pivotal role of low-dose computed tomography (LDCT) in the detection of lung cancer. However, one of the acknowledged downsides of LDCT based screening is its relatively high false positive rate. For example, the rate of positive screening tests in the NLST was approximately 27% in the first two rounds of the LDCT arm and 17% in the third year of screening. A screening CT was considered positive if it contained a non-calcified nodule of at least 4 mm in its long axis or other suspicious abnormalities were present. Over the three rounds, over 96% of such positive screens were false positives and 72% had some form of diagnostic follow-up.
To address this issue, the American College of Radiologists (ACR) Lung Imaging Reporting and Data System (Lung Rads™) tool (2) for standardized reporting of CT based lung cancer screening adopted a threshold for solid nodules of <6 mm for its category 2 where no additional diagnostic work-up is recommend and the subject is imaged again at annual screening. However, new nodules of 4 mm and greater are considered category 3 and a 6-month follow-up LDCT is recommended in recognition of their increased probability of malignancy.
The impact of Lung-RADS was analysed in a retrospective analysis of the NLST (3). Lung-RADS was shown to reduce the overall screening false positive rate to 12.8% and 5.3% at baseline and interval imaging respectively at the cost of a reduction of sensitivity from 93.5% in the NLST to 84.9% using Lung-RADS at baseline and 93.8% in the NLST and 84.9% using Lung-RADS after baseline. However, while Lung-RADS reduces the overall false positive rate, the false positive rate of positive screens, i.e., Lung-RADS 3 and above, remains very high at 93% at baseline and 89% after baseline; of 3,591 Lung-RADS 3 and above screens, 3,343 were false positives at baseline and of 2,858 Lung-RADS 3 and above screens after baseline 2,543 were false positives. Therefore, while the adoption of Lung-RADS can reduce the total number of benign nodules being worked-up within a screening programme, at a cost of just under 10% loss in sensitivity, there remain a very large number of benign nodules being investigated, and the nodule classification task remains a challenging one.
One approach to address this problem is to adopt computer aided diagnosis (CADx) technology as an aid to radiologists and pulmonary medicine physicians. Given an input CT and possible additional relevant patient meta-data, such techniques aim to provide a quantitative output related to the lung cancer risk.
One may consider the goal of such systems to be two-fold. First, to reduce the variability in assessing and reporting the lung cancer risk between interpreting physicians. Indeed, computer assisted approaches have been shown to improve consistency between physicians in a variety of clinical contexts, including nodule detection (4) and mammography screening (5) and one might expect such decision support tools could provide the same benefit in nodule classification. Second, CADx could improve classification performance by supporting the less experienced or non-specialised clinicians in assessing the risk of a particular nodule being malignant.
In this article, we review progress made towards the development and validation of lung cancer prediction models and nodule classification CADx software. While we do not intend this to be a comprehensive review, we do aim to provide an overview of the main approaches taken to date and outline some of the challenges that remain to bring this technology to routine clinical use.
Risk models
There have been a number of lung cancer risk models developed and validated that one may consider to be a form of CADx tool (6-9). Typically based on logistic regression, such tools aim to provide an overall risk of the patient having cancer based on patient meta-data such as age, sex and smoking history and nodule characteristics such as nodule size, morphology and growth, if a previous CT was available.
Although such tools currently require manual entry by the user, they do produce an objective lung cancer risk score which may be used in the decision-making process. However, despite their attraction and good performance, their adoption and performance as part of decision making has not been studied. The British Thoracic Society (BTS) guidelines on the management of incidentally detected pulmonary nodules (10), recommends the use of the Brock model (6). Anecdotally, many physicians report using them for patient communication only and feel that such models do not add a great deal to their clinical expertise. More specifically, questions remain as to the utility of such models when the patient population is different to that of the training data. It is clear, that for such models to be clinically useful, knowledge of the training data used is critical, and this also will determine the clinical scenarios in which they may be used. There are clearly significant differences in the pre-test probabilities of a nodule being malignant in different patient groups. For instance, patients with a current or prior history of malignancy are at significantly different risk of nodule malignancy than non-smokers with no significant prior history.
From a technical perspective, such models have a number of limitations. Foremost is the reliance on human interpretation of input variables such as nodule size, morphology and even the reliance on the patient’s own estimate of factors such as smoking history. For example, under the Brock model, a 1mm increase in the reported size of a 5 mm spiculated solid nodule in a 50-year-old female almost doubles its risk, from 0.98% to 1.89%. However, inter-radiologist variability in reporting nodule size is typically greater than this (11). Moreover, inter-reader variability in reporting morphology and nodule type is common even amongst experienced thoracic radiologists (12,13).
Some recent work to address this has been proposed by Ciompi et al. (14) where an automated system for the classification on nodules into solid, non-solid, part-solid, calcified, perifissural and spiculated types was proposed. Overall classification accuracy is reported to be within the inter-radiologist variability at 79.5% but this varies between 86% for solid and calcified nodules down to 43% for spiculated nodule classification. Of course, since the ground-truth classifications were provided by radiologist opinion, the performance at validation cannot be expected to improve on that. As the authors point out, the nodule types are radiologist developed concepts that, while useful for clinical purposes, lack a precise definition. The impact of the system’s output as an input to the Brock model was not reported and ultimately this approach should be judged on its ability to improve malignancy prediction.
Radiomics
The term Radiomics refers to the automatic extraction of quantitative features from medical images (15,16) and has been the subject of a great deal of investigation with applications including automated lesion classification, response assessment and therapy planning. Fundamentally, the Radiomic approach aims to turn image voxels into a set of numbers that characterize the biological property of interest such as lesion malignancy, tumour grade or therapy response.
Although research into, what are termed, Radiomics methods has seen an explosion in the last decade, the technical methods that it builds on have a very long history in the fields of computer vision and medical image understanding in the area of texture analysis. Indeed, many of the so-called Radiomic features are based on techniques that were first proposed in the 1970s (17) for the classification of textured images and have been largely superseded in the computer vision literature. Nevertheless, their application to medical image processing research has in some areas yielded some significant insights, in particular in how such quantitative features relate to tumour pheno- and genotypes. The idea that such advanced quantitative techniques may add to the qualitative clinical interpretation of radiologists is gaining momentum and is likely to move into mainstream clinical practice in the coming 5 to 10 years.
For a given application, the Radiomic approach proceeds in two phases—first a training or feature selection phase and then a second testing or application phase. The training phase typically proceeds as follows. First, a large set, typically some hundreds or thousands, of features are defined a-priori. Next, the features are extracted from a large corpus of training data where the object of interest, say a tumour, has been delineated such that a computer algorithm can extract the quantitative features automatically. Finally, a step known as feature selection is applied that aims to select a smaller subset, e.g., some tens of such features that efficiently captures the imaging characteristics of the biological phenomena of interest. For example, in the case of nodule classification into benign and malignant we may pick the features, either individually or in combination that perform the best at this task on the training data.
In the testing phase, the Radiomics are applied to a particular patient’s image, with the process being similar to the training phase but now the selected features are identified by the algorithm, extracted and then used to classify the patient.
Of course, both at training and testing steps, a classification algorithm will need to be defined to convert the Radiomics values into classifications. For small sets of individual features, we may simply use thresholds on the Radiomic features; however, for larger sets of features more sophisticated techniques from the field of machine learning, such as Support Vector Machines (SVMs) and Random Forests are typically used to yield better results. A very good review of Radiomics approaches applied to the classification of pulmonary nodules is provided in Wilson et al. (18).
One criticism of some of the earlier Radiomics work is the lack of independent training and validation data (19). Indeed, it is not unusual to find very high classification rates being reported based on the training data whereas it is well established within the machine learning literature that such results may be subject to “overfitting”—the apparent excellent performance that cannot be replicated on unseen and independent datasets. In fact, one measure of the goodness of a well-trained classifier is the difference in performance between training sets and test sets. This phenomenon has led to a generally over-optimistic view of the performance with area under the curve (AUC) numbers reported in the high 80s and 90s range that cannot be replicated on independent data.
The 2015 SPIE-AAPM-NCI LungX Lung Nodule Classification Challenge (20) was a first attempt at a Grand Challenge style competition and provided a sobering view of the actual real-life performance one might expect to see in clinical practice. Ten groups, including our own, submitted computer methods to classify nodules as benign or malignant. No additional training data was provided but a limited “calibration” dataset of ten cases was provided. Therefore, all groups were required to utilize either publicly available or their own proprietary datasets. Many of the methods used the Radiomic/texture feature extraction technique followed by a classification step.
AUCs ranged from 0.5 to 0.68 with only three of the methods outperforming random chance with statistical significance. Despite our classifier achieving the highest AUC and winning the competition, the performance was significantly below what we had seen on other independent datasets. In the next section, we provide some details of the system that have not been published previously along with some insights gained during the competition and subsequent analysis.
LungX winning entry
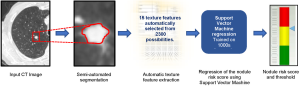
Figure 1 provides an overview of the main steps in the algorithm used in the winning entry. The software has four main steps at test time, i.e., when used to classify a nodule: (I) nodule segmentation, (II) texture feature extraction, (III) risk score regression and (IV) risk score thresholding.
The nodule segmentation is required because the subsequent step of feature extraction is applied to a region of interest (ROI) around the nodule. Each nodule was segmented in a semi-automated thresholding approach using a commercial software package (Mirada RTx, Mirada Medical Ltd.). The user first defined a spherical ROI around each nodule and then applied a fixed threshold to the ROI. Next, the user could adjust the threshold to improve the segmentation and finally, manual editing tools could be used to edit the segmentation to remove any voxels that did not correspond to the nodule of interest that the segmentation had included. Typically, adjacent vessels would need to be excluded in this manner. In later work, we replaced the semi-automated method with a more automated technique that did not require any user interaction other than to identify the centre and diameter of the nodule (21).
We extracted 15 texture features from two regions, the first inside the nodule segmentation and the second in a surrounding region defined automatically. Based on earlier work using our internal databases, we found that better performance could be achieved if the region inside the nodule was treated separately to the immediate surrounding parenchyma. The insight here is that the texture of the nodule carries separate information to the region in the nearby parenchyma and the very different ranges of Hounsfield units in each region would make it difficult for one set of texture features to capture the patterns. We believe this was a significant contribution to the performance of the system.
The 15 features were selected from a palette of over 1,300 classical texture features including Haralick (17), Gabor (22), along with simple measures such as mean, standard deviation and volume. We utilized a fully automated feature selection strategy that aimed to select a small subset of features that optimised classification performance over an in-house training dataset. Since it is computationally infeasible to test all combinations of the full palette of features, we utilized a sequential “greedy” algorithm that, starting with the optimal pair of features found by exhaustive search over all pairs of features, selected features one-by-one so as to maximise the performance over the training dataset at each step.
Finally, an SVM regression algorithm with a cubic kernel was trained using the libSVM library. The output of this step is a number between 0 and 1 that reflects the likelihood that a particular nodule is malignant.
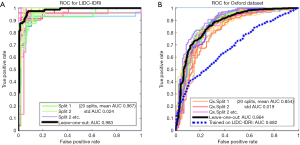
The training dataset we utilized for the competition was mostly derived from the Lung Image Database Consortium and Image Database Resource Initiative (LIDC-IDRI dataset) (23). This publicly available dataset comprises a wide variety of nodules and comes with multiple segmentations and likelihood of malignancy score estimated by expert clinicians. Nodules were included in our training set if at least three sets of clinician-drawn contours and corresponding likelihood-of-malignancy scores were included in the XML metadata. The malignancy scores are integers from one to five inclusive and are recorded per clinician. Only nodules whose malignancy scores were all below 3 (the benign set) or all above 3 (the malignant set) were included, yielding a labeled subset of 222 nodules overall for the LIDC-IDRI training set.
Figure 2 shows the Receiver Operating Characteristic (ROC) curves for the system as trained and tested on the LIDC-IDRI dataset using 20-way cross-validation. With such high AUCs, we were suspicious that the dataset was too easy to classify and so we trained and validated on a second dataset, PLAN, to examine the system’s performance further. The PLAN nodule database was built up from nodules collected from the Oxford University Hospitals NHS trust. This set consists of 709 nodules, 377 malignant and 328 benign, diagnosed either using histology or by 2-year stable follow-up. Using 20-way cross-validation, the average AUC was 0.854; the ROC curves are shown on the right of Figure 2. In the end, the system we submitted used both LIDC-IDRI and PLAN for feature selection but the SVM was trained only on the LIDC-IDRI dataset.
Convolutional neural networks and deep learning
Convolutional Neural Networks (CNN) trained using deep learning techniques have come to dominate pattern detection, recognition, segmentation and classification applications in both medical and non-medical fields. Indeed, where sufficient training data is available, CNNs have largely superseded the previous generation of Radiomic/texture analysis methods described above. In our own work, once we had collected and curated sufficiently large training sets by the end of 2016, our CNN based techniques started to outperform the previous state-of-the-art texture and SVM based method. While a detailed exposition of such techniques is beyond the scope of this article, it is worth understanding the main differences to previous methods and their advantages.
Feature learning vs. feature selection
Unlike Radiomic/texture analysis approaches, CNN techniques build features from scratch rather than selecting from a palette of engineered or pre-selected set that rely on the contextual knowledge of the algorithm developer.
Hierarchical features
The first few layers of a CNN typically comprise several layers of features allowing the network to learn the relationships between features in a much more sophisticated way than can be achieved with a single feature extraction stage. Consider this illustrative example: a texture feature, such as local entropy of the joint histogram, can be used to detect spiculations extending into the parenchyma. But a CNN can learn this and also learn that spiculations encompass the whole perimeter of the nodule and that this is a sign of a malignant nodule.
End-to-end learning
CNNs are typically trained “end-to-end” meaning that the entire network is trained to optimize the problem of interest, i.e., all the parameters of the network are adjusted until the peak classification rate is achieved. In contrast, each stage of the LungX texture-based approach that we developed had to be built and optimised individually and there was no guarantee that the entire pipeline would be optimal.
Segmentation-free
The CNN approach can operate without the nodule segmentation step because segmentation is handled in an implicit way within the algorithm. In subsequent analysis of our LungX algorithm, we found significant sensitivity of the prediction score to the segmentation step.
The Kaggle data science bowel 2017—lung cancer detection
The 2017 lung cancer detection data science bowel (DSB) competition hosted by Kaggle was a much larger two-stage competition than the earlier LungX competition with a total of 1,972 teams taking part. In stage 1, a large training dataset of 1,397 patients was provided comprising 362 with lung cancer and 1,035 without, along with an initial validation set of 198 patients. This validation set was used to produce the public stage 1 leader-board, using which the competitors could judge their performance. In stage 2, a further unseen dataset of 506 patients, on which the final competition results were judged, was made available for 7 days. This two-stage approach was used to avoid competitors inferring the test set labels using many entries. In contrast to the LungX competition, here the competitors needed to produce a completely automated pipeline, taking in a CT image and outputting a likelihood of cancer.
The results were judged using the log-loss function, popular on Kaggle competitions. Unlike AUC, the log-loss function penalizes more confident, but incorrect outputs, greater than less confident ones. All top three entries utilized CNNs trained using Deep Learning and scored within a few decimal places of each other, scoring 0.39755, 0.40117 and 0.40127, where a log-loss of zero corresponds to a perfect score. AUC-ROC results of 0.85 and 0.87 were subsequently reported for the top-two teams (24,25) respectively.
The winning entry (24), utilized a 3D Convolutional Neural Network training on a combination of DSB training data and the publicly available the dataset used in the LUNA16 nodule detection competition (26) which itself was derived from the LIDC-IDRI dataset (23). Since no nodules are identified in the validation and test datasets, a reliable automated nodule detection step is critical for correct classification. In fact, based on the subsequent write-ups from the winning teams (24,25), much of the effort was put into this step rather than the subsequent classification step.
Is size everything?
One interesting observation regarding the distribution of nodule sizes was made by the winning team. The LUNA16 dataset contained many more small nodules, (mean =8 mm), whereas the DSB datasets comprised many larger lesions (mean =14 mm), therefore the team had to adjust the training algorithm to compensate for this. Moreover, the distribution of nodule sizes between cancer and benign patients was reported to be very different; the malignant nodules were large and the benign were small. Hence predicting the diagnosis based on size alone would be expected to produce good results. The issue of size bias in training and test sets is a critical issue and one which we have studied in some depth.
It is well known from the risk model literature that the strongest predictor of a nodule’s malignancy, imaged at one point in time, is its size, whether expressed as its long axis, an average of the long and short axes or as a volume. The reason is quite simple: benign nodules are typically caused by processes that are self-limiting in size, e.g., inflamed lymph nodes, whereas malignant tumours have no such limits, and are constrained by other factors such as the duration of growth, the cell replication time, the ability of the tumour to invade adjacent structures, and its vascular and oxygen supply. Therefore, one might expect that nodule size, either implicitly or explicitly, will be included as part of any nodule CADx system.
However, additional differences in the size distribution of benign and malignant nodules may also occur due to selection bias in data collection. For example, a naive approach to collecting examples of malignant lesions might be to select all retrospective CTs for patients diagnosed with lung cancer and all retrospective CTs for patients with benign nodules. However, outside of a screening programme, most patients diagnosed with lung cancer present with symptoms prior to diagnostic imaging and hence are typically at a late stage and their nodules are consequently larger than benign nodules. A machine learning algorithm trained on such data would perform very poorly when applied to, for example, a screening application where the distribution of malignant nodule sizes is more similar to benign ones.
We explored this issue further by comparing the performance of a CADx system trained on size-matched and size unmatched data (27). Figure 3 illustrates the experiment. Two datasets were created from the US NLST. The first (A), comprising 640 solid nodules, was built to remove size as a discriminatory factor between benign and malignant; all malignant solid nodules between 4 and 20 mm diameter were selected, and for each, a benign solid nodule was selected that most closely matched it in diameter. Any malignant nodule for which an equivalently sized benign could not be found within 0.8 mm was rejected. Sizes were measured using automated volumetric segmentation. The second dataset (B), also comprising 640 subjects, included all malignant nodules in A but benign nodules were randomly selected following the empirical size distribution of the whole NLST dataset. Therefore, nodule size cannot be a discriminative factor in A but would be in B. Two nodule classifiers were built using texture features combined with an SVM classifier; this was utilized here because the small datasets prevented the use of a CNN model.
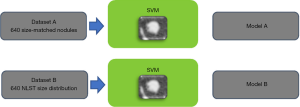
The average AUC for the classifier trained on dataset A was 0.70 whereas using size alone on the same dataset gave an AUC of 0.50 as would be expected. The AUC was 0.91 for the classifier trained on dataset B. This indicates that the classifier can learn morphological features that can discriminate between benign and malignant nodules and, moreover, that such features add approximately 0.2 AUC points to using size-alone.
Coincidentally, the performance on size matched data was very close to that we achieved on the LungX competition data (AUC: 0.70 and 0.68) which was subsequently revealed to have also used size-matched data in the test set (20).
Conclusions
We have provided an overview of the main approaches used for nodule classification and lung cancer prediction from CT imaging data. In our experience, given sufficient training data, the current state-of-the-art is achieved using CNNs trained with Deep Learning achieving a classification performance in the region of low 90s AUC points. When evaluating system performance, it is important to be aware of the limitations or otherwise of the training and validation data sets used, i.e., were the patients’ smokers or non-smokers, or were patients with a current or prior history of malignancy included.
Given an apparent acceptable level of performance, the next stage is to test such CADx systems in a clinical setting but before this can be done, we must first define the way in which the output of the CADx should be utilized in clinical decision making. Who should use such a system and how should it be integrated into their decisions? Should the algorithm produce an absolute risk of malignancy and how should this be expressed; should it be incorporated into clinical opinion and how much weight should clinicians or patients lend to it. Should the algorithms be incorporated into or designed to fit current guidelines such as Lung-RADS or the BTS guidelines? If nodules are followed over time, should the algorithm incorporate changes in nodule volume or should this be assessed separately? Is success defined by a reduction in the numbers of false positive scans defined as those needing further follow up or intervention, or by detecting all lung cancers and earlier than determined by following current guidelines? Who should be compared to the algorithm when determining its value? Should the comparison be experts or general radiologists, as it may be difficult to be significantly better than an expert but may be of substantial help to a generalist, and most scans are not interpreted by experts? Relatively little work has been done to address such questions.
Acknowledgments
The authors would like to thank the numerous research scientists and clinical staff involved in the project for their contributions: Sarim Ather, Djamal Boukerrouri, Amalia Cifor, Monica Enescu, Mark Gooding, William Hickes, Samia Hussain, Aymeric Larrue, Jean Lee, Heiko Peschl, Lyndsey Pickup, Shameema Stalin, Ambika Talwar, Eugene Teoh, Julien Willaime and Phil Whybra.
Funding: Part of this work was funded by Innovate UK project TSB 101676.
Footnote
Conflicts of Interest: T Kadir is CTO, Director and shareholder of Optellum Ltd. F Gleeson is a shareholder and advisor to Optellum Ltd.
References
- National Lung Screening Trial Research Team, Aberle DR, Adams AM, et al. Reduced Lung-Cancer Mortality with Low-Dose Computed Tomographic Screening. N Engl J Med 2011;365:395-409. [Crossref] [PubMed]
- Lung CT Screening Reporting & Data System. Available online: https://www.acr.org/Clinical-Resources/Reporting-and-Data-Systems/Lung-Rads
- Pinsky PF, Gierada DS, Black W, et al. Performance of Lung-RADS in the National Lung Screening Trial: A Retrospective Assessment. Ann Intern Med 2015;162:485-91. [Crossref] [PubMed]
- Awai K, Murao K, Ozawa A, et al. Pulmonary Nodules at Chest CT: Effect of Computer-aided Diagnosis on Radiologists Detection Performance. Radiology 2004;230:347-52. [Crossref] [PubMed]
- Freer TW, Ulissey MJ. Screening Mammography with Computer-aided Detection: Prospective Study of 12,860 Patients in a Community Breast Center. Radiology 2001;220:781-6. [Crossref] [PubMed]
- McWilliams A, Tammemagi MC, Mayo JR, et al. Probability of Cancer in Pulmonary Nodules Detected on First Screening CT. N Engl J Med 2013;369:910-9. [Crossref] [PubMed]
- Gould MK, Ananth L, Barnett PG, et al. A Clinical Model To Estimate the Pretest Probability of Lung Cancer in Patients With Solitary Pulmonary Nodules. Chest 2007;131:383-8. [Crossref] [PubMed]
- Swensen SJ, Silverstein MD, Ilstrup DM, et al. The probability of malignancy in solitary pulmonary nodules. Application to small radiologically indeterminate nodules. Arch Intern Med 1997;157:849-55. [Crossref] [PubMed]
- Deppen SA, Blume JD, Aldrich MC, et al. Predicting lung cancer prior to surgical resection in patients with lung nodules. J Thorac Oncol 2014;9:1477-84. [Crossref] [PubMed]
- Callister ME, Baldwin DR, Akram AR, et al. British Thoracic Society guidelines for the investigation and management of pulmonary nodules. Thorax 2015;70 Suppl 2:ii1-54. [Crossref] [PubMed]
- Revel MP, Bissery A, Bienvenu M, et al. Are two-dimensional CT measurements of small noncalcified pulmonary nodules reliable? Radiology 2004;231:453-8. [Crossref] [PubMed]
- Bartlett EC, Walsh SL, Hardavella G, et al. Interobserver Variation in Characterisation of Incidentally-Detected Pulmonary Nodules: An International, Multicenter Study. Available online: http://4wcti.org/2017/SS5-3.cgi
- Zinovev D, Feigenbaum J, Furst J, et al. Probabilistic lung nodule classification with belief decision trees. Conf Proc IEEE Eng Med Biol Soc 2011;2011:4493-8. [PubMed]
- Ciompi F, Chung K, van Riel SJ, et al. Towards automatic pulmonary nodule management in lung cancer screening with deep learning. Sci Rep 2017;7:46479. [Crossref] [PubMed]
- Aerts HJ, Velazquez ER, Leijenaar RT, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun 2014;5:4006. [Crossref] [PubMed]
- Lambin P, Rios-Velazquez E, Leijenaar R, et al. Extracting more information from medical images using advanced feature analysis. Eur J Cancer 2012;48:441-6. [Crossref] [PubMed]
- Haralick RM, Shanmugam K, Dinstein I. Textural Features for Image Classification. IEEE Trans Syst Man Cybern Syst 1973;3:610-21. [Crossref]
- Wilson R, Devaraj A. Radiomics of pulmonary nodules and lung cancer. Transl Lung Cancer Res 2017;6:86-91. [Crossref] [PubMed]
- Chalkidou A, O’Doherty MJ, Marsden PK. False Discovery Rates in PET and CT Studies with Texture Features: A Systematic Review. PLoS One 2015;10:e0124165. [Crossref] [PubMed]
- Armato SG 3rd, Drukker K, Li F, et al. LUNGx Challenge for computerized lung nodule classification. J Med Imaging (Bellingham) 2016;3:044506. [Crossref] [PubMed]
- Willaime JM, Pickup L, Boukerroui D, et al. Impact of segmentation techniques on the performance of a CT texture-based lung nodule classification system. Available online: https://posterng.netkey.at/esr/viewing/index.php?module=viewing_poster&task=&pi=135229
- Lee TS. Image Representation Using 2D Gabor Wavelets. IEEE Trans Pattern Anal Mach Intell 1996;18:1-13.
- Armato SG 3rd, McLennan G, Bidaut L, et al. The Lung Image Database Consortium (LIDC) and Image Database Resource Initiative (IDRI): a completed reference database of lung nodules on CT scans. Med Phys 2011;38:915-31. [Crossref] [PubMed]
- Li Y, Chen KZ, Sui XZ, et al. Establishment of a mathematical prediction model to evaluate the probability of malignancy or benign in patients with solitary pulmonary nodules. Beijing Da Xue Xue Bao Yi Xue Ban 2011;43:450-4. [PubMed]
- Hammack D. Forecasting Lung Cancer Diagnoses with Deep Learning. Available online: https://raw.githubusercontent.com/dhammack/DSB2017/master/dsb_2017_daniel_hammack.pdf
- Setio AA, Traverso A, de Bel T, et al. Validation, comparison, and combination of algorithms for automatic detection of pulmonary nodules in computed tomography images: The LUNA16 challenge. Med Image Anal 2017;42:1-13. [Crossref] [PubMed]
- Pickup L, Declerck J, Munden R, et al. MA 14.13 Nodule Size Isn't Everything: Imaging Features Other Than Size Contribute to AI Based Risk Stratification of Solid Nodules. J Thorac Oncol 2017;12:S1860-1. [Crossref]