Prognostic markers in lung cancer: is it ready for prime time?
Cancer prognostic markers are patient or tumor characteristics that predict outcome (usually survival) independent of the treatment (1). Thus, they are usually identified and validated in patients who receive no or surgical therapy only. The goal of identifying prognostic markers is to define patient subpopulations with significantly different anticipated outcomes, who might benefit from different therapies. Good prognostic patients may not require additional treatment beyond the primary surgical resection, while poor prognostic patients may derive improved survival benefit from adjuvant therapy. Therefore, prognostic markers could potentially be “drivers” of cancer progression. In turn, these markers could themselves represent therapeutic targets.
Predictive markers, on the other hand, are patient or tumor characteristics that predict benefit from specific treatments (either in terms of tumor shrinkage or survival). In other words, the differences in tumor response or survival benefit between treated versus untreated patients will be significantly different in those with or without the predictive marker (e.g., a mutation). In contrast, the effect of treatment is not expected to be different in patient groups distinguished by a prognostic marker only. The validation of prognostic marker can be established by using data from retrospective series, while the validation of predictive marker should be done in a controlled clinical trial, in which the effect of the marker can be tested in both the treated and placebo groups.
Prognostic markers can be proteins, mRNAs or miRNAs or the gene itself. For the latter, mutations, gene copy number aberrations and single nucleotide variation could potentially also be prognostic. Most markers that have been extensively studied are proteins, which are typically assessed by immunohistochemistry (IHC). However, the high-throughput profiling techniques in cancer genome have led to the identification of mRNA and miRNA prognostic signatures. Proteomic signatures generated by mass spectrometry are also emerging (2).
In lung cancer, prognostic markers are most relevant to early-stage (I-IIIA) non-small cell lung cancer (NSCLC) patients, who are potentially curable by complete surgical resection. However, the prognostic significance of a marker should also be assessed during the validation of a predictive marker, as the apparent benefit from a specific therapy could merely reflect the inherently prognostic value of the marker. As an example, VeriStrat (2) is a mass spectrometry-derived proteomic signature, which was initially reported as capable of stratifying advanced NSCLC patients for their responses to epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors gefitinib and erlotinib. In two cohorts of patients treated by these TKIs, respectively, the VeriStrat “good” patients demonstrated a significantly longer time to progression and overall survival than the VeriStrat “poor” patients, even after adjustment for other clinical factors. A subsequent retrospective study appeared to validate the independent predictiveness of VeriStrat to erlotinib for progression-free survival (P=0.011) and overall survival (P=0.017) in a randomized phase II trial of first-line therapy with gemcitabine, erlotinib, or the combination in elderly patients (>70 years) (3). When tested in 3 “control” advanced NSCLC patient cohorts (total n=158) who did not receive any TKI treatment, VeriStrat signature was found not to be prognostic. However, all these studies were conducted in patients treated by a single therapy. When VeriStrat was tested in the samples from NCIC CTG BR.21 trial, a randomized placebo-controlled study of erlotinib in previously treated advanced NSCLC patients, erlotinib treatment prolonged survival in both VeriStrat “good” and “poor” patient groups, indicating the lack of predictive value of VeriStrat for erlotinib treatment (4). Importantly the VeriStrat “poor” group had poorer survival in the placebo group patients, consistent with VeriStrat being a prognostic marker (4).
Single gene/protein prognostic markers
Most lung cancer prognostic markers reported are proteins evaluated by IHC. Despite >500 reported studies, not a single protein marker has as yet been validated sufficiently for clinical use (5). For most markers, the results from various studies have been inconsistent. This could largely be accounted for by the lack of standardization in the IHC methods used, including the source and quality of the antibodies used, the staining protocol, scoring algorithm, and statistical approach to analyse the data. Inconsistent results could also be due to the small sample size in some studies, for which cases included are less representative. Institutional and publication biases could also play an important role. As an example, from 1987 to 2005 there were 15 reported studies on the prognostic value of cyclin D1 (CCND1) (6-20). Five studies identified CCND1 overexpression as a negative prognostic marker (6,8,9,14,16), while three other studies associated it with better prognosis (11,18,20); the remaining seven reported no association (Table 1). It is noted that the source of antibody varied from laboratory generated to commercial sources, and different antibody dilutions and scoring cut-offs for positive staining were used (Table 1). Overall, no conclusive result on the prognostic value of CCND1 could be made from these studies (5).
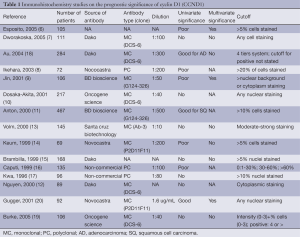
Full table
The most credible prognostic markers reported have been based on samples of patients who were involved in large multi-institutional studies, especially randomized placebo-controlled treatment trials. The advantages of these cohorts include more uniform and better-defined patient characteristics, as well as the ability to test the predictive value of the markers for benefit from adjuvant chemotherapy. The Lung Adjuvant Cisplatin Evaluation-Biology (LACE-Bio) studies are organized by investigators from the four seminal adjuvant chemotherapy trials: the International Adjuvant Lung Cancer (IALT), Adjuvant Navelbine International Trialist Association (ANITA), Cancer and Leukemia Group B (CALGB) 9633, and NCIC Clinical Trials Group (CTG) JBR.10. The goal of LACE-Bio studies include cross validation or pooled analyses of promising prognostic and predictive markers reported by one or more of the member groups. The NCIC CTG group initially reported that high β-tubulin (bTub III) expression by IHC was a poor prognostic marker for recurrence-free survival (RFS) and borderline prognostic for overall survival (OS) in surgery-alone patients, as well as being predictive for survival benefit from adjuvant chemotherapy (21). When the marker was tested in the pooled data set of the other 3 trials (total n=1149), the poor prognostic value of high bTubIII was validated [hazard ratio (HR): 1.27; 95% confidence interval (CI): 1.07-1.51; P=0.008 for OS and HR: 1.30; 95% CI: 1.11- 1.53; P<0.001 for RFS] (22). However, interaction between bTubIII expression and chemotherapy was not significant, which indicates that high bTubIII is not predictive of benefit from adjuvant chemotherapy (22).
One of the most celebrated prognostic and predictive markers for early-stage NSCLC is the Excision Repair Cross-Complementation group (ERCC1) protein, a critical component of nucleotide excision repair mechanism for DNA damage induced by cisplatin. The ERCC1 protein expression was evaluated by IHC in 761 of 1,867 patients involved in the IALT trial (23). High ERCC1 expression was found to be a good prognostic marker (adjusted HR: 0.66; 95% CI: 0.49-0.90; P=0.009) in surgery-alone patients, but adjuvant chemotherapy benefit was seen only in ERCC1-low (negative) patients (23). However, subsequent LACE-Bio cross validation study failed to establish ERCC1 as a predictive marker for adjuvant chemotherapy using the same yet a different batch of ERCC1 antibody (clone 8F1) (24). The group has tested 16 commercially available ERCC1 antibodies and found none of the 16 antibodies could distinguish among the four ERCC1 protein isoforms, whereas only one isoform produced a protein that had full capacities for nucleotide excision repair and cisplatin resistance (24). The result highlights the pitfall of IHC studies using antibodies that have not been characterized rigorously for their properties as well as quality.
Meta-analysis is a cost-effective practice for increasing the sample size and statistical power by combining results of comparable studies or trials. Quite a few meta-analyses have been performed and showed potential prognostic value of HER-2, p53, Ki-67, and Bcl-2, however, with potential institutional and publication biases, caution should be taken to interpret conclusions from meta-analyses. For example, KRAS mutation has been reported as a marker of poor prognosis by a meta-analysis (HR: 1.35; 95% CI: 1.16-1.56) (25). However, in a recent pooled analysis of 1536 LACE-Bio patients, KRAS mutation was not validated as a prognostic marker in NSCLC (HR: 1.18; 95% CI: 0.97-1.44; P= 0.09), nor in adenocarcinoma patient alone (HR: 1.0; 95% CI: 0.78-1.28, P=1.00) (26). Furthermore, contrary to the original finding in the JBR.10 patients, KRAS mutation was also not predictive of benefit from adjuvant chemotherapy (26).
Multigene prognostic markers
To date, the large numbers of studies have reported that the prognostic HRs of single marker have reached up to 1.5-1.7. Kwiatkowski et al. (27) and D’Amico et al. (28) previously demonstrated that multiple cumulative markers may better stratify prognosis compared to a single marker. The invention of microarray technologies has made it possible to explore the prognostic significance of thousands of markers using genome-wide high-throughput and computational approaches. Initial studies were conducted mainly on mRNA expression markers, as the technology was initially developed for this molecule. To date, more than 35 such studies have been reported (29), a large number showing that gene expression signature may stratify early stage NSCLC, or its subtypes (e.g., adenocarcinoma or squamous cell carcinoma), patients with different prognosis or survival outcome.
Since 2005, reports on expression prognostic markers have also included validation in independent cohorts, mostly using published microarray data sets. This was facilitated by the requirement by most high-impact journals that authors make their microarray data publicly available either through their own institute website, such as the Broad Institute (http://www.broadinstitute.org/) or by depositing to publicly repositories, such as the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) or ArrayExpress (https://www.ebi.ac.uk/arrayexpress/). This requirement has allowed greater level of transparency on gene expression signatures, as independent validation and verification could be conducted. Over the years, as most studies selected to use the platforms developed and commercialized by Affymetrix (Santa Clara, CA), Illumina (San Diego, CA) and Agilent (Santa Clara, CA) and as Bioconductor http://www.bioconductor.org/) was developed based on R, an open source statistical software, to analyze microarray data, significant standardization of microarray analyses has occurred. The Sweave function (http://stat.ethz.ch/R-manual/R-devel/library/utils/html/Sweave.html) and the new development of Knitr function (http://yihui.name/knitr/) in R integrates R code into LaTeX, HTML, Markdown, AsciiDoc, and reStructuredText documents which enables creating dynamic reports and making the data mining process even more transparent and reproducible. As many scientifically rational approaches have been developed and used by investigators to identify gene signatures associated with survival outcome, numerous signatures have been reported. Some are large gene set signatures made up of hundreds of genes, whereas many others are trimmed down to less than 20 genes through optimization process. Although most of these signatures have been validated in one or more independent patient cohort microarray data sets, overlaps between the genes sets have consistently been minimal. This has raised question on the robustness of gene expression signatures as a reliable biomarker. Nevertheless, a permutation study using a common data set has shown that it is statistically possible to identify numerous equally significant prognostic signatures (30). However, validation of prognostic signatures in multiple independent patient cohorts can be extremely challenging, as the signature discovery algorithms that are applied to small data sets (hundreds) containing disproportionately large number (thousands) of data elements may easily introduce data over-fitting, thus difficulty to reproduce in independent data sets (31). Furthermore, independent data sets may also carry institutional biases related to the sample selection, as well as other patient and population demographic features.
Clinically applicable prognostic gene signatures
Several features may facilitate the application of prognostic gene signature in the clinical setting to assist in management of NSCLC patients. Aside from the signatures being validated in multiple independent patient cohorts, the technique to assay the signatures should also be implementable in clinical laboratories, according to the regulatory body approved protocols, such as the Clinical Laboratory Improvement Amendments (CLIA). As the standard pathology practice process tissue into formalin-fixed and paraffin-embedded (FFPE) blocks, technologies that favor the use of FFPE samples would fast-track the adoption of the signature for clinical use. Last but not least, in order for a prognostic signature to assist oncologists in selecting patients for adjuvant chemotherapy, the signature should be predictive, such that the “high risk” patients (as identified by the signature) would likely benefit from the postsurgical chemotherapy, and “low risk” patients (who do not benefit and could potentially be harmed by chemotherapy) would be spared the toxicity and cost. In this context, a few signatures are worthy of highlighting.
A 15-gene prognostic signature was established from microarray expression analysis of snap-frozen tumor samples from 133 Canadian patients who participated in the JBR.10 trial (32). These included 62 patients who were treated by surgery alone, and 71 patients who received adjuvant chemotherapy. This stage-independent prognostic signature was developed from the data of surgery-only patients (adjusted HR: 18.00; 95% CI: 5.78-56.05; P<0.001) and was validated in 4 independent published microarray data (total 356 stage IB to II patients without adjuvant treatment), with HR ranging from 1.96 to 3.57 (32). This was more recently further validated in another independent cohort (HR: 1.92; 95% CI: 1.15-3.23; P= 0.012) (33). More importantly, when the signature was applied to JBR.10 patients who received adjuvant chemotherapy, the “high risk” patients demonstrated improved survival (HR: 0.33; 95% CI: 0.17-0.63; P<0.001), whereas low-risk patients did not (HR: 3.67; 95% CI: 1.22-11.06; P=0.013; interaction P<0.001). The predictiveness of the signature was validated by quantitative polymerase chain reaction (qPCR) in 30 JBR.10 patients (19 with surgery only, 11 with adjuvant chemotherapy) who did not have their tumor samples examined by microarray. However, the predictiveness of the signature has not been independently validated, as there are no microarray data sets available from other randomized adjuvant chemotherapy trials for testing. Furthermore, the validation and application of this signature in FFPE samples remain to be demonstrated.
A 14-gene expression was developed using qPCR directly on DNA isolated from FFPE tumor samples of 361 non-squamous NSCLC patients resected at the University of California, San Francisco (UCSF, Table 2) (34). The assay was then independently validated in a masked cohort of 433 patients with stage I non-squamous NSCLC resected at Kaiser Permanente Division of Research (KPDOR), and on a cohort of 1006 patients with stage I-III non-squamous NSCLC resected in several leading cancer centers that are part of the China Clinical Trials Consortium (CCTC). The signature reported a 5-year overall survival of 71.4% (95% CI: 60.5-80.0) in low-risk, 58.3% (95% CI: 48.9-66.6) in intermediate-risk, and 49.2% (95% CI: 42.2-55.8) in high-risk patients (Ptrend=0.0003) at KPDOR. Similar analysis of the CCTC cohort indicated 5-year overall survivals of 74.1% (95% CI: 66.0-80.6) in low-risk, 57.4% (95% CI: 48.3-65.5) in intermediate-risk, and 44.6% (95% CI: 40.2-48.9) in high risk patients (Ptrend<0.0001). Multivariate analysis in both cohorts indicated that no standard clinical risk factors could account for, or provide the prognostic information derived from tumor gene expression. As the signature was developed and tested using qPCR in FFPE samples, its transfer to clinical testing was facilitated and it is already commercially available as the Pervenio Lung RS Test (Life Technologies, Inc, Grand Island, NY). In addition, the assay recently showed prognostic value for small <2-cm node-negative stage IA patients. In this subset of patients, similar to those likely to be identified in emerging computed tomography screening programs for lung cancer, the assay identified in pathologically confirmed stage IA patients, ~25% of patients who had a survival of ~50% versus a >90% survival for low risk patients (39). Importantly, the signature was equally prognostic in patients who did (HR: 2.31; 95% CI: 1.29-4.24) or did not (HR: 2.42; 95% CI: 1.88-3.11) receive adjuvant chemotherapy, suggesting it is primarily a prognostic marker (34). However, to test the predictive value of this assay, a large 1500-patient prospective stage III global trial is now underway to randomize Pervenio Lung RS Test identified “high-risk” stage I patients to receive adjuvant cisplatin based adjuvant chemotherapy versus observation (current standard of care) (40).
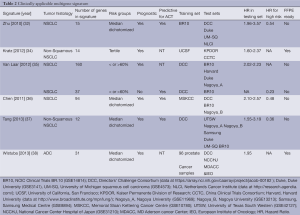
Full table
The ChipDx is claimed by the author as an “online gene expression based diagnostic system, the creation and delivery of clinically-useful diagnostic and prognostic oncology assays”. It published two signatures (35), one is a prognostic signature with 160 genes, identified from 332 stage I-III NSCLC from the Directors’ Challenge Consortium cohort (DCC, total n=442) and tested in 264 stage I-II NSCLC, compiling from subsets of 5 NSCLC cohorts [JBR10, total n=133; Duke, total n=89; a data set from the Harvard University (Harvard), total n=139, and a data set from Nagoya University (Nagoya_A), total n=163, Table 2] (35). The other is a predictive signature made up of 37 genes, identified from 88 stage I-III NSCLC patients treated with adjuvant chemo- or/and radio-therapy in the DCC cohort, and tested in 109 stage I-II NSCLC from JBR.10 (32,41). The 160-gene prognostic signature was able to stratify 90 high risk patients with significant poorer survival (HR: 2.80; 95% CI: 1.83-4.28, P<0.0001) after adjustment for other prognostic factors. The 37-gene predictive signature was able to separate 70 responders from the other 39 non-responders in the test set. Among the 70 responders, the adjuvant chemotherapy significantly increased survival (HR: 0.23; 95% CI: 0.08-0.61, P=0.0032) after the adjustment of age, gender, stage and histology whereas in the 39 non-responder, no significant difference in survival by adjuvant chemotherapy was observed (HR: 0.55; 95% CI: 0.15-2.04, P=0.38). However, there was no report on the interaction term.
The malignancy-risk gene signature was originally developed for breast cancer and contained a large number of proliferative genes (36,42). The investigators tested their signature in the DCC (31), another data set from Nagoya University (Nagoya_B, n=117) (43) and JBR.10 (32) datasets (Table 2). As the signature genes were identified by Affymetrix U133A platform and testing was performed on data obtained using the Agilent platform, cross-platform mapping was used to identify one hundred and sixteen probe sets to represent 87 genes for the validation. The malignancy risk score was the summed products of gene expressions and their weights in the first component, then was median dichotomized to define high and low risk groups, as they were used in the breast cancer. The signature was able to classify NSCLC patients without adjuvant chemotherapy with significant difference in survival (HR: 2.10; 95% CI: 1.26-3.51, Plog-rank=0.004 in DCC, HR: 2.17; 95% CI: 1.22-3.68, Plog-rank=0.007 in Nagoya_B, and HR: 2.57; 95% CI: 1.17-5.64, Plog-rank=0.01 in JBR.10). Furthermore, in the high risk group in JBR.10, the authors observed a significant improvement in survival by adjuvant chemotherapy (HR: 0.48; 95% CI: 0.24-0.96, Plog-rank=0.03). In contrast, adjuvant chemotherapy non-significantly decreased patients’ survival in the low risk group. Nevertheless, the interaction between risk group and adjuvant chemotherapy was significant (Pinteraction=0.02) indicated that adjuvant chemotherapy might benefit high risk group but not the low risk group.
The University of Texas South Western (UTSW) 12-gene signature (37) was derived from the DCC data set (31). The investigators first identified 797 genes that were univariately associated with patients’ 5-year overall survival and then through a partial correlation matrix to obtained 18-hub genes. The 18-hub genes was further trimmed down to a 12-gene signature by incorporating data from synthetical lethality study with paclitaxel and genetic aberrations in Tumorscape. The signature was validated in silico in 5 independent cohorts, UTSW (37), Duke (44), Samsung Medical Center (45), Nagoya_A (43), Nagoya_B (46) but not in squamous cell carcinoma. Additionally, the 12-gene signature was tested in 2 cohorts of NSCLC with adjuvant chemotherapy: UTSW (n=176 NSCLC) (37) and the JBR.10 (n=90, NSCLC) (32). Adjuvant chemotherapy appeared to prolong survival only in the high risk group (HR: 0.34; 95% CI: 0.13-0.86; P=0.017 for the UTSW and HR: 0.36; 95% CI: 0.13-0.97, P=0.038 for the JBR.10) but not in low risk groups (37).
The cell cycle proliferation (CCP) score (https://myriadpro.com/lung-cancer/myriad-myplan-lung-cancer/) was originally derived from FFPE samples of prostate cancer by RT-qPCR (47). The investigators utilized 96 commercially available prostate cancer samples to select signature from 126 cell cycle related genes. Thirty-one genes were selected as a CCP signature based on their correlation with the mean expression of the entire 126 genes (47). Wistuba et al. (38) validated the CCP (31-gene) in 3 lung ADC cohorts: DCC (HR: 2.02; 95% CI: 1.29-3.17, P=0.0022, n=442, profiled with Affymetrix U133A, Table 2) (31), data set from the National Cancer Center Hospital of Japan (NCCHJ, HR: 2.16; 95% CI: 1.32-3.53, P=0.0026, n=226 profiled with U133 plus2, Table 2) (48), and a jointed cohort of a total of 381 FFPE NSCLC patient samples from MD Anderson Cancer Center (MDACC, n=207) and European Institute of Oncology (IEO, n=174) (HR: 1.92; 95% CI: 1.18-3.10, Table 2) by qPCR, after adjustments for other prognostic factors (38).
Other molecular prognostic signatures
As mentioned previously, extensive analysis to date has not established the significant prognostic value of KRAS or p53 mutation. Interestingly, several studies have consistently demonstrated that epidermal growth factor receptor (EGFR) tyrosine kinase mutation is a good prognostic marker for both early and advanced-stage patients (49-52). This may potentially account for the generally better prognosis of Asian NSCLC patients. However, a recent large study in early-stage NSCLC patients did not show an independent prognostic value of EGFR mutation in Asian (Korean) patients (53). There are as yet no gene copy changes (e.g., amplification) that have been reported as showing prognostic value. In contrast, many investigators have recently reported the prognostic significance of microRNA (miRNA) or its signatures in NSCLC patients (54-58). These studies remain preliminary, as extensive independent validations to the scale of mRNA signatures have not been performed. The miRNA as a prognostic marker is highly attractive for two reasons: (I) there are less miRNA species and single miRNA may control the expression or function of multiple genes, thus, they are more likely to function as master regulatory elements in gene function, and (II) miRNA assay can easily be performed on FFPE samples, as they are of short sequences and are more stable.
Future outlook
During the past decade, we have witnessed the rapid translation of advances in the molecular understanding of lung cancer into clinics, as in the development of targeted therapies and the use of molecular markers to select patients for such treatment. Testing for EGFR mutations and anaplastic lymphoma kinase (ALK) gene rearrangement is now becoming standard for personalizing therapies in advanced NSCLC patients. With the current pace of advances being witnessed, it is almost certain that molecular prognostication would one day be integrated into standard pathologic diagnosis to improve the management, treatment, and survival of early-stage NSCLC patients, just as it has become standard in other solid organ cancers such as breast cancer and colon cancer. Successful practice in this field is the incorporation of molecular markers into the histological classification system of lung cancers (59).
Acknowledgements
Grand support: This study was supported by the Canadian Cancer Society Research Institute grant #020527, the Ontario Ministry of Health and Long Term Care, and the Princess Margaret Hospital Foundation. M.-S. Tsao holds the M. Qasim Choksi Chair in Lung Cancer Translational Research
Disclosure: The authors declare no conflict of interest. The authors are part holders of a patent on the 15-gene signature.
References
- Shepherd FA, Tsao MS. Unraveling the mystery of prognostic and predictive factors in epidermal growth factor receptor therapy. J Clin Oncol 2006;24:1219-20; author reply 1220-1. [PubMed]
- Taguchi F, Solomon B, Gregorc V, et al. Mass spectrometry to classify non-small-cell lung cancer patients for clinical outcome after treatment with epidermal growth factor receptor tyrosine kinase inhibitors: a multicohort cross-institutional study. J Natl Cancer Inst 2007;99:838-46. [PubMed]
- Stinchcombe TE, Roder J, Peterman AH, et al. A retrospective analysis of VeriStrat status on outcome of a randomized phase II trial of first-line therapy with gemcitabine, erlotinib, or the combination in elderly patients (age 70 years or older) with stage IIIB/IV non-small-cell lung cancer. J Thorac Oncol 2013;8:443-51. [PubMed]
- Carbone DP, Ding K, Roder H, et al. Prognostic and predictive role of the VeriStrat plasma test in patients with advanced non-small-cell lung cancer treated with erlotinib or placebo in the NCIC Clinical Trials Group BR.21 trial. J Thorac Oncol 2012;7:1653-60. [PubMed]
- Zhu CQ, Shih W, Ling CH, et al. Immunohistochemical markers of prognosis in non-small cell lung cancer: a review and proposal for a multiphase approach to marker evaluation. J Clin Pathol 2006;59:790-800. [PubMed]
- Esposito V, Baldi A, De Luca A, et al. Cell cycle related proteins as prognostic parameters in radically resected non-small cell lung cancer. J Clin Pathol 2005;58:734-9. [PubMed]
- Dworakowska D, Jassem E, Jassem J, et al. Prognostic value of cyclin D1 overexpression in correlation with pRb and p53 status in non-small cell lung cancer (NSCLC). J Cancer Res Clin Oncol 2005;131:479-85. [PubMed]
- Ikehara M, Oshita F, Ito H, et al. Expression of cyclin D1 but not of cyclin E is an indicator of poor prognosis in small adenocarcinomas of the lung. Oncol Rep 2003;10:137-9. [PubMed]
- Jin M, Inoue S, Umemura T, et al. Cyclin D1, p16 and retinoblastoma gene product expression as a predictor for prognosis in non-small cell lung cancer at stages I and II. Lung Cancer 2001;34:207-18. [PubMed]
- Dosaka-Akita H, Hommura F, Mishina T, et al. A risk-stratification model of non-small cell lung cancers using cyclin E, Ki-67, and ras p21: different roles of G1 cyclins in cell proliferation and prognosis. Cancer Res 2001;61:2500-4. [PubMed]
- Anton RC, Coffey DM, Gondo MM, et al. The expression of cyclins D1 and E in predicting short-term survival in squamous cell carcinoma of the lung. Mod Pathol 2000;13:1167-72. [PubMed]
- Nguyen VN, Mirejovský P, Mirejovský T, et al. Expression of cyclin D1, Ki-67 and PCNA in non-small cell lung cancer: prognostic significance and comparison with p53 and bcl-2. Acta Histochem 2000;102:323-38. [PubMed]
- Volm M, Koomägi R. Relevance of proliferative and pro-apoptotic factors in non-small-cell lung cancer for patient survival. Br J Cancer 2000;82:1747-54. [PubMed]
- Keum JS, Kong G, Yang SC, et al. Cyclin D1 overexpression is an indicator of poor prognosis in resectable non-small cell lung cancer. Br J Cancer 1999;81:127-32. [PubMed]
- Brambilla E, Moro D, Gazzeri S, et al. Alterations of expression of Rb, p16(INK4A) and cyclin D1 in non-small cell lung carcinoma and their clinical significance. J Pathol 1999;188:351-60. [PubMed]
- Caputi M, De Luca L, Papaccio G, et al. Prognostic role of cyclin D1 in non small cell lung cancer: an immunohistochemical analysis. Eur J Histochem 1997;41:133-8. [PubMed]
- Kwa HB, Michalides RJ, Dijkman JH, et al. The prognostic value of NCAM, p53 and cyclin D1 in resected non-small cell lung cancer. Lung Cancer 1996;14:207-17. [PubMed]
- Au NH, Cheang M, Huntsman DG, et al. Evaluation of immunohistochemical markers in non-small cell lung cancer by unsupervised hierarchical clustering analysis: a tissue microarray study of 284 cases and 18 markers. J Pathol 2004;204:101-9. [PubMed]
- Burke L, Flieder DB, Guinee DG, et al. Prognostic implications of molecular and immunohistochemical profiles of the Rb and p53 cell cycle regulatory pathways in primary non-small cell lung carcinoma. Clin Cancer Res 2005;11:232-41. [PubMed]
- Gugger M, Kappeler A, Vonlanthen S, et al. Alterations of cell cycle regulators are less frequent in advanced non-small cell lung cancer than in resectable tumours. Lung Cancer 2001;33:229-39. [PubMed]
- Sève P, Lai R, Ding K, et al. Class III beta-tubulin expression and benefit from adjuvant cisplatin/vinorelbine chemotherapy in operable non-small cell lung cancer: analysis of NCIC JBR.10. Clin Cancer Res 2007;13:994-9. [PubMed]
- Reiman T, Lai R, Veillard AS, Paris E, et al. Cross-validation study of class III beta-tubulin as a predictive marker for benefit from adjuvant chemotherapy in resected non-small-cell lung cancer: analysis of four randomized trials. Ann Oncol 2012;23:86-93. [PubMed]
- Olaussen KA, Dunant A, Fouret P, et al. DNA repair by ERCC1 in non-small-cell lung cancer and cisplatin-based adjuvant chemotherapy. N Engl J Med 2006;355:983-91. [PubMed]
- Friboulet L, Olaussen KA, Pignon JP, et al. ERCC1 isoform expression and DNA repair in non-small-cell lung cancer. N Engl J Med 2013;368:1101-10. [PubMed]
- Mascaux C, Iannino N, Martin B, et al. The role of RAS oncogene in survival of patients with lung cancer: a systematic review of the literature with meta-analysis. Br J Cancer 2005;92:131-9. [PubMed]
- Shepherd FA, Domerg C, Hainaut P, et al. Pooled analysis of the prognostic and predictive effects of KRAS mutation status and KRAS mutation subtype in early-stage resected non-small-cell lung cancer in four trials of adjuvant chemotherapy. J Clin Oncol 2013;31:2173-81. [PubMed]
- Kwiatkowski DJ, Harpole DH Jr, Godleski J, et al. Molecular pathologic substaging in 244 stage I non-small-cell lung cancer patients: clinical implications. J Clin Oncol 1998;16:2468-77. [PubMed]
- D’Amico TA, Massey M, Herndon JE 2nd, et al. A biologic risk model for stage I lung cancer: immunohistochemical analysis of 408 patients with the use of ten molecular markers. J Thorac Cardiovasc Surg 1999;117:736-43. [PubMed]
- Zhu CQ, Pintilie M, John T, et al. Understanding prognostic gene expression signatures in lung cancer. Clin Lung Cancer 2009;10:331-40. [PubMed]
- Boutros PC, Lau SK, Pintilie M, et al. Prognostic gene signatures for non-small-cell lung cancer. Proc Natl Acad Sci U S A 2009;106:2824-8. [PubMed]
- Director’s Challenge Consortium for the Molecular Classification of Lung Adenocarcinoma1, Shedden K, Taylor JM, et al. Gene expression-based survival prediction in lung adenocarcinoma: a multi-site, blinded validation study. Nat Med 2008;14:822-7.
- Zhu CQ, Ding K, Strumpf D, et al. Prognostic and predictive gene signature for adjuvant chemotherapy in resected non-small-cell lung cancer. J Clin Oncol 2010;28:4417-24. [PubMed]
- Der SD, Sykes J, Pintilie M, et al. Validation of a histology-independent prognostic gene signature for early-stage, non-small-cell lung cancer including stage IA patients. J Thorac Oncol 2014;9:59-64. [PubMed]
- Kratz JR, He J, Van Den Eeden SK, et al. A practical molecular assay to predict survival in resected non-squamous, non-small-cell lung cancer: development and international validation studies. Lancet 2012;379:823-32. [PubMed]
- Van Laar RK. Genomic signatures for predicting survival and adjuvant chemotherapy benefit in patients with non-small-cell lung cancer. BMC Med Genomics 2012;5:30. [PubMed]
- Chen DT, Hsu YL, Fulp WJ, et al. Prognostic and predictive value of a malignancy-risk gene signature in early-stage non-small cell lung cancer. J Natl Cancer Inst 2011;103:1859-70. [PubMed]
- Tang H, Xiao G, Behrens C, et al. A 12-gene set predicts survival benefits from adjuvant chemotherapy in non-small cell lung cancer patients. Clin Cancer Res 2013;19:1577-86. [PubMed]
- Wistuba II, Behrens C, Lombardi F, et al. Validation of a proliferation-based expression signature as prognostic marker in early stage lung adenocarcinoma. Clin Cancer Res 2013;19:6261-71. [PubMed]
- Kratz JR, Van den Eeden SK, He J, et al. A prognostic assay to identify patients at high risk of mortality despite small, node-negative lung tumors. JAMA 2012;308:1629-31. [PubMed]
- Tsao MS, Jablons DM. Molecular prognostication of non-small cell lung cancer. Semin Thorac Cardiovasc Surg 2013;25:4-7. [PubMed]
- Winton T, Livingston R, Johnson D, et al. Vinorelbine plus cisplatin vs. observation in resected non-small-cell lung cancer. N Engl J Med 2005;352:2589-97. [PubMed]
- Chen DT, Nasir A, Culhane A, et al. Proliferative genes dominate malignancy-risk gene signature in histologically-normal breast tissue. Breast Cancer Res Treat 2010;119:335-46. [PubMed]
- Tomida S, Takeuchi T, Shimada Y, et al. Relapse-related molecular signature in lung adenocarcinomas identifies patients with dismal prognosis. J Clin Oncol 2009;27:2793-9. [PubMed]
- Bild AH, Yao G, Chang JT, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature 2006;439:353-7. [PubMed]
- Lee ES, Son DS, Kim SH, et al. Prediction of recurrence-free survival in postoperative non-small cell lung cancer patients by using an integrated model of clinical information and gene expression. Clin Cancer Res 2008;14:7397-404. [PubMed]
- Takeuchi T, Tomida S, Yatabe Y, et al. Expression profile-defined classification of lung adenocarcinoma shows close relationship with underlying major genetic changes and clinicopathologic behaviors. J Clin Oncol 2006;24:1679-88. [PubMed]
- Cuzick J, Swanson GP, Fisher G, et al. Prognostic value of an RNA expression signature derived from cell cycle proliferation genes in patients with prostate cancer: a retrospective study. Lancet Oncol 2011;12:245-55. [PubMed]
- Okayama H, Kohno T, Ishii Y, et al. Identification of genes upregulated in ALK-positive and EGFR/KRAS/ALK-negative lung adenocarcinomas. Cancer Res 2012;72:100-11. [PubMed]
- Eberhard DA, Johnson BE, Amler LC, et al. Mutations in the epidermal growth factor receptor and in KRAS are predictive and prognostic indicators in patients with non-small-cell lung cancer treated with chemotherapy alone and in combination with erlotinib. J Clin Oncol 2005;23:5900-9. [PubMed]
- Riely GJ, Politi KA, Miller VA, et al. Update on epidermal growth factor receptor mutations in non-small cell lung cancer. Clin Cancer Res 2006;12:7232-41. [PubMed]
- Sasaki H, Shimizu S, Endo K, et al. EGFR and erbB2 mutation status in Japanese lung cancer patients. Int J Cancer 2006;118:180-4. [PubMed]
- Kosaka T, Yatabe Y, Onozato R, et al. Prognostic implication of EGFR, KRAS, and TP53 gene mutations in a large cohort of Japanese patients with surgically treated lung adenocarcinoma. J Thorac Oncol 2009;4:22-9. [PubMed]
- Kim HR, Kim DJ, Kang DR, et al. Fibroblast growth factor receptor 1 gene amplification is associated with poor survival and cigarette smoking dosage in patients with resected squamous cell lung cancer. J Clin Oncol 2013;31:731-7. [PubMed]
- Bogner PN, Patnaik SK, Pitoniak R, et al. Lung cancer xenografting alters microRNA profile but not immunophenotype. Biochem Biophys Res Commun 2009;386:305-10. [PubMed]
- Raponi M, Dossey L, Jatkoe T, et al. MicroRNA classifiers for predicting prognosis of squamous cell lung cancer. Cancer Res 2009;69:5776-83. [PubMed]
- Hu Z, Chen X, Zhao Y, et al. Serum microRNA signatures identified in a genome-wide serum microRNA expression profiling predict survival of non-small-cell lung cancer. J Clin Oncol 2010;28:1721-6. [PubMed]
- Tan X, Qin W, Zhang L, et al. A 5-microRNA signature for lung squamous cell carcinoma diagnosis and hsa-miR-31 for prognosis. Clin Cancer Res 2011;17:6802-11. [PubMed]
- Duncavage E, Goodgame B, Sezhiyan A, et al. Use of microRNA expression levels to predict outcomes in resected stage I non-small cell lung cancer. J Thorac Oncol 2010;5:1755-63. [PubMed]
- Travis WD, Brambilla E, Noguchi M, et al. International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society: international multidisciplinary classification of lung adenocarcinoma: executive summary. Proc Am Thorac Soc 2011;8:381-5. [PubMed]


