
Metabolic profiles of serum samples from ground glass opacity represent potential diagnostic biomarkers for lung cancer
Introduction
Lung cancer is the most prevalent cause of cancer death, being responsible for 1.3 million deaths and about one-fifth of cancer-related deaths annually all over the world. Since the initial diagnoses of the majority of lung cancer cases are at advanced stages and have a grim prognosis, the relative 5-year survival rate is a mere 10–15% (1,2). Early diagnosis is extremely important to the success of clinical interventions because the relative 5-year survival rate of lung cancer patients diagnosed in the early stages is remarkably better when compared with the advanced lung cancer patients (3). Recently, data from the National Lung Screening Trial (NLST), which assessed the diagnosis of lung cancer through low-dose computed tomography (LDCT), demonstrated that LDCT could reduce lung cancer mortality by 20% and total mortality by 7% (4-6). Therefore, LDCT of lung cancer is the most efficacious noninvasive medical test to reduce worldwide cancer mortality of lung cancer. However, a major concern was the high prevalence (96%) of false positives that might have led to “over-diagnosis” (4,7), especially of ground glass opacity (GGO). GGO, defined as “hazy increased opacity of lung, with preservation of bronchial and vascular margins”, can be observed in preinvasive lung cancer and in benign conditions such as hemorrhage, inflammation, and focal interstitial fibrosis (8,9). GGO shows a slight increase in density, a cloud-like shape and a limitation of less than 3 cm. Studies on radiological pathological correlation have shown that GGO represents pathological lepidic growth and consolidation on CT represents pathological invasive components. It is problematic to use GGO in discriminating between benign and cancerous conditions, and this issue has led to excessive use of invasive procedures, over-treatment, and an overall increase in anxiety. Additional tests, preferably of the non-invasive type, and exhibiting high sensitivity and specificity, are crucially needed to complement LDCT.
There are more available biomarkers found in the blood capable of enhancing the power of early lung cancer detection than in any other source. Blood analysis by the use of analytical methods has yielded several clinically valuable biomarkers of lung cancer (10) that can complement LDCT and represent a major advance in implementing lung cancer screening. Several discovery platforms that can identify markers to provide complementary assessment need to be explored, as single biomarkers are unlikely to perform sufficiently for implementation in screening.
The metabolomic analysis is able to profile small molecules in a variety of human biofluids and tissue activities in the body and thus has been established as a platform for gaining new insights into the pathology of cancer on the cellular, tissue, and organ level (11,12). Currently, no analytical method provides a comprehensive analysis of numerous endogenous metabolites required for metabonomics. The main advantages of liquid chromatography-mass spectrometry (LC-MS) which include its wide dynamic range, the possibility of small sample volumes, and ease of automation for a large sample series, make LC-MS an ideal analytical method or metabolite profiling (13,14). Metabonomics by liquid chromatography–hybrid quadrupole time-of-flight mass spectrometry (LC-Q-TOF-MS) has also been recently shown to be reliable (15,16). By coupling with several multivariate statistical methods, including principal component analysis (PCA) and partial least squares discriminant analysis (PLS-DA), the latest information from spectroscopic data can recognize potential biomarkers for cancers and ailments.
Using a metabolomics approach, the early detection of lung cancer may be implemented by revealing new biomarkers associated with diagnosis and prognosis (12,17,18). In the present study, LC-Q-TOF-MS profiling of serum from subjects’ GGOs was explored as a means to establish a lung cancer-related metabolic signature, which could provide novel insights about differentiating between benign and malignant GGO. We hypothesize that the candidate metabolomics biomarkers identified will be helpful for early detection of lung cancer.
Methods
Patient population and collection of patient samples
This prospective study was approved by the Ethics Committee of The 211th Hospital of Chinese People’s Liberation Army. The 211th Hospital of Chinese People’s Liberation Army performs low-dose CT-scan screening for high-risk smoking cases with a family history of lung cancer. Lung cancer (n=34) cases for this study were confirmed by histopathological examination (Table 1) from March 2014 to January 2017. Controls (n=39) were healthy persons with normal low-dose CT-scan screening who were included at The 211th Hospital of Chinese People’s Liberation Army between March 2014 and December 2017. No participants in this study were taking any medications, and none were suffering from metabolic diseases such as liver diseases, kidney diseases, or any other types of cancer. We have collected the blood samples of the patients from fasting venous at the next morning after admission of the patient. The blood samples were obtained by routine venipuncture, transported, and then centrifuged less than 4 hours after collection at room temperature. After that, the isolated serum samples were stored at −80 °C for further analysis.
Full table
Sample preparation
All the serum samples were thawed at 4 °C for 50 min and centrifuged at 4,000 ×g for 10 min at 4 °C after vortexing for 10 s. The upper aliquot solution (200 µL) of serum samples was transferred to a clean 2-mL centrifuge tube, and then acetonitrile (1,000 µL) was added. After that, the samples were vortexed for 2 min and centrifuged at 12,000 ×g for 15 min at 4 °C. The upper solution (1,000 µL) was transferred to a clean 2-mL centrifuge tube and then evaporated to dryness over a heat block at 35 °C under nitrogen gas. Two-hundred µL acetonitrile/water (1:3, v/v) was added into the residue of the upper solution (1,000 µL), vortexed for 1 min, and centrifuged at 12,000 ×g for 15 min at 4 °C. The supernatant (200 µL) was transferred to an autosampler vial and injected into the LC-Q-TOF-MS (6530 series; Agilent Technologies, Santa Clara, CA, USA) apparatus for analysis. Equal amounts of supernatant samples from all samples were mixed for quality control (QC).
Chromatography
Each 10-µL aliquot of sample was injected into a 2.1×100 mm (1.8 mm) ZORBAX SB-C18 column (Agilent Technologies, Santa Clara, CA, USA) and then was rapidly resolved by liquid chromatography (6530 series; Agilent Technologies, Santa Clara, CA, USA). Electron spray ionization in positive mode (ESI+) of the mobile phase was constituted by 0.1% formic acid (phase A) and water containing 0.1% formic acid (phase B), while electron spray ionization in negative mode (ESI−) was formed with acetonitrile (phase A) and water (phase B). The protocols for the linear mobile phase gradient were as follows: 95% A held for 1 min, decreased to 2% A by 10 min, held at 2% A until 13 min, increased to 95% A by 13.1 min, and held at 95% A until 20 min. The flow rate of the mobile phase was 0.3 mL/min at 40 °C.
MS
The Agilent 6530-Q-TOF MS apparatus (6530 series; Agilent Technologies, Santa Clara, CA, USA) operating in ESI+ or ESI− mode was used to perform MS. The capillary voltage was set at 4.0 kV for ESI+ and 3.5 kV for ESI−. Nitrogen was applied as the desolvation gas at a flow rate of 10 L/min. The desolvation temperature was 350 °C. The centroid data were obtained with the full scan mode [mass-to-charge ratio (m/z) =50–1,000].
Data preprocessing and annotation
By using MassHunter Qualitative Analysis Software (Agilent Technologies B.04.00), the raw data were converted into mzdata-format files and further imported to the XCMS package in R (3.0.2) for preprocessing. The analyses were performed by the default XCMS parameter settings, with the following exceptions: xcms Set (fwhm, 10), group (minfrac, 0.5; bw, 30), and rector (method, “obiwarp”). After that, a data matrix was generated that included results of retention time, m/z values, and peak intensity. By using the CAMERA in R (3.0.2), isotope peaks were annotated. Meanwhile, adducts and fragments in the peak lists were generated in the same way (19).
Statistical analysis
The grouping trends and outliers were at first detected by using PCA, and the significance (P<0.05) of each metabolite was then determined by applying the Wilcoxon rank-sum test (20). A PLS-DA was applied to identify the differences in metabolites between lung cancer cases and controls (20). The supervised model and avoid overfitting were validated by permutation tests with 100 iterations (21). Parameters describing the variable importance in the projection (VIP) for each metabolite were calculated based on the PLS-DA model. The metabolic biomarkers were detected with thresholds of P values and VIP values of 0.05 and 1, respectively. The Wilcoxon rank-sum test was applied to the R platform (3.0.2). The PCA and PLS-DA were performed via SIMCA-P (version 11.5; Umetrics, Malmö, Sweden).
Results
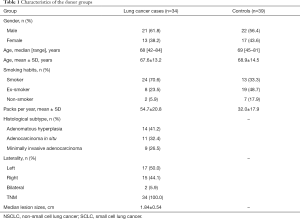
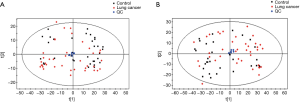
Detailed characteristics of the participants for the respective cohorts are provided in Table 1. There were no significant differences in the baseline characteristics. The metabolic analysis revealed numerous metabolic differences between lung cancer and healthy controls. We have found 1,409 kinds of metabolites in ESI+ mode and 891 in ESI− mode in this study. The PCA performed on all the samples revealed that the QC samples were tightly clustered in the PCA score plots (Figure 1), which showed stability and repeatability of the sample analysis sequence. All of the statistically significant ions were analyzed (P<0.05 and VIP >1; Figure 2) by the application of the ESI+ and ESI− modes. The differences between lung cancer and healthy controls were identified by using a supervised PLS-DA model. There was an obvious separation between lung cancer cases and healthy controls present in the ESI+ mode (Figure 2A) and ESI− mode (Figure 2B), which were present in the PLS-DA score plot. The PLS-DA models contained two predictive components in the ESI+ mode (R2X =0.426; R2Ycum =0.634; Q2cum =0.428) and two components in the ESI− mode (R2X =0.488; R2Ycum =0.572; Q2cum =0.468). Permutation tests consisting of 100 iterations and containing two predictive components were also used (22). As shown in Figure 2C,D, the validity of the supervised models, were further confirmed. The results showed that the permuted Q2cum values were lower than the original values in almost all cases.
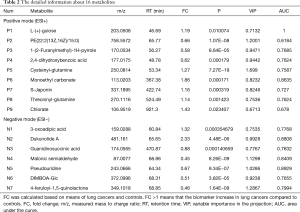
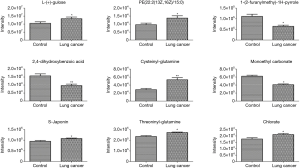
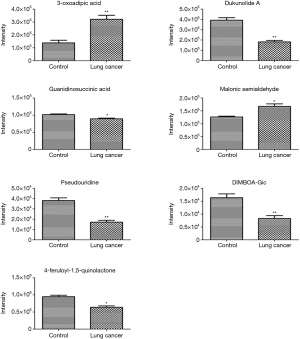
The discriminatory metabolites contributing to the differences between lung cancer cases and healthy controls were revealed by analyzing the VIP values. Then, on the basis of false discovery rate and VIP thresholds of 0.05 and 1, respectively, biomarker candidates were selected from differential ions for subsequent metabolite identification (23,24). In total, we identified 8 metabolites in the ESI+ mode and 7 metabolites in the ESI− mode (Table 2). L-(+)-gulose, phosphatidylethanolamine (PE)(22:2(13Z,16Z)/15:0), cysteinyl-glutamine, S-japonin, threoninyl-glutamine, chlorate, 3-oxoadipic acid, dukunolide A, and malonic semialdehyde levels were observed to be elevated in the serum samples of lung cancer cases compared with those of healthy controls (Figures 3,4). By contrast, 1-(2-furanylmethyl)-1H-pyrrole, 2,4-dihydroxybenzoic acid, monoethyl carbonate, guanidinosuccinic acid, pseudouridine, DIMBOA-Glc, and 4-feruloyl-1,5-quinolactone levels were lower in serum samples of lung cancer cases when compared with those of healthy controls (Figures 3,4).
Full table
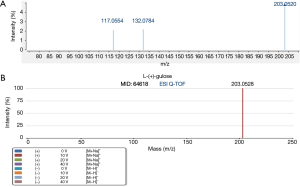
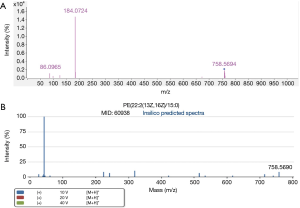
L-(+)-gulose and PE(22:2(13Z,16Z)/15:0) were identified based on accurate mass data, retention time, experimental MS/MS spectra, and library MS/MS spectra. Figures 5,6 are representative identification procedures based on MS/MS spectral matching.
The involved biochemical pathways mapped in the Human Metabolome Database (HMDB) (25) and the Kyoto Encyclopedia of Genes and Genomes (KEGG) (26) included the glycosylphosphatidylinositol (GPI)-anchor biosynthesis, beta-alanine metabolism, propanoate metabolism, inositol phosphate metabolism, and glycerophospholipid metabolism.
Discussion
Lung cancer has the highest incidence rate of cancer-related death worldwide each year. Histological examination plays a prominent part in cancer diagnosis through invasive procedures that include surgical resection, biopsy, and mediastinoscopy. However, these invasive procedures are often unsuitable for early diagnosis. LDCT screening is particularly effective in the early detection of lung cancer but is marred by high false positive rates. Therefore, determining whether a GGO is malignant or benign upon presentation and choosing the appropriate optimal course of management is still challenging. In this regard, less invasive and more cost-effective diagnostic tests with the same reliability as histology are needed.
In the current study, metabolic perturbations in serum that could distinguish healthy participants from those with malignant GGOs were investigated by using a metabolomics approach. Metabolomics features that could be mirrored in blood metabolome of lung cancer patients were in the blood (27,28). In this study, metabolite abundance adjusted for the mentioned above covariates to minimize potential biases were evaluated because physical characteristics, including gender and age, could affect metabonomics results (29). We detected significant metabolic differences between malignant GGO by LDCT and healthy participants and identified 16 metabolites as potential biomarkers for lung cancer. Of note, the metabolites accounting for the differences between the serum of malignant GGO patients and healthy participants were matched with known human metabolites in the HMDB or KEGG, and these results were further confirmed by a manual search for similarities between the annotated and the library spectra.
Compared to subjects with healthy participants, participants that presented malignant GGOs showed elevated levels of serum metabolites, particularly, L-(+)-gulose, PE(22:2(13Z,16Z)/15:0), cysteinyl-glutamine, and threoninyl-glutamine. Moreover, elevation in these specific PEs is consistent with past research (30). This shows that tumorigenesis is accompanied by lipid metabolism. PE binding proteins (PEBPs) have already been proven to increase secretion and work on modulating the development, invasion, and metastatic potential of tumors in lung cancer (31-33). Fahrmann et al. also found that levels of PEs decreased after surgical removal of malignant nodules (30). In addition, elevation in PEs of lung cancer patients exhibited drastic changes in lipid profiles (34,35).
Amino acids constitute one of the most significant metabolite changes, and there are a few studies that have reported amino acid differences in the serum of non-small cell lung cancer (NSCLC) patients (36,37). The metabolite changes in our study were consistent with reports and Cascino et al., who identified the increase of amino acids glutamine and arginine in the blood of lung cancers patients (38). L-(+)-gulose is an L-hexose sugar and an intermediate in the biosynthesis of L-ascorbate (vitamin C). It can be oxidized to L-guluno-1-4-lactone and produced by the hydrolysis of L-gulose-1-P. Many animals, including fish, amphibians, reptiles, and bird species can synthesize ascorbic acid by several biosynthesis pathways. We have grounds to believe that lung cancer cells acquire the capability to synthesize vitamin C. Given this, L-(+)-gulose might be seen as an alternative to tumor markers for malignant GGO.
In conclusion, our study was able to identify putative markers of malignant GGO in serum, although the interpretation of the metabolic information retrieved is somewhat speculative. Indeed, while we have assessed the important factors such as gender, age, and smoking habits, other unmeasured or uncontrolled factors, including lifestyle, body mass index, and diet have not been examined. It is difficult to control all multiple factors of metabonomic studies applied to humans. Here, the power of identifying biomarkers with the potential clinical application for LC-Q-TOF-MS was shown by using metabolomics. In future work, we will acquire the utility of metabolomics biomarkers by using a larger patient cohort and combine this with other clinical diagnostic methods such as magnetic resonance (MR) imaging and the application of circulating tumor cells.
Acknowledgments
The authors would like to thank Rui-Ting Li and Meng Sun for their technical and scientific advice.
Funding: This work was supported by the National Natural Science Foundation of China (81802808), the National Natural Science Foundation of China (81701304), and the Key Research and Development Plan of Shaanxi Province 2017SF-176.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was approved by the 211th Hospital of Chinese People’s Liberation Army (Ethical number: KY-2014-006). All included patients were informed about the nature of the study and gave their written informed consent.
References
- Siegel R, Ma J, Zou Z, et al. Cancer statistics, 2014. CA Cancer J Clin 2014;64:9-29. [Crossref] [PubMed]
- Jemal A, Siegel R, Xu J, et al. Cancer statistics, 2010. CA Cancer J Clin 2010;60:277-300. [Crossref] [PubMed]
- Ries LA. Influence of extent of disease, histology, and demographic factors on lung cancer survival in the SEER population-based data. Semin Surg Oncol 1994;10:21-30. [Crossref] [PubMed]
- Aberle DR, Adams AM, Berg CD, et al. Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med 2011;365:395-409. [Crossref] [PubMed]
- Chen D, Dai C, Kadeer X, et al. New horizons in surgical treatment of ground-glass nodules of the lung: experience and controversies. Ther Clin Risk Manag 2018;14:203-11. [Crossref] [PubMed]
- Ciriaco P, Muriana P, Negri G. Pulmonary nodules and mini-invasive lung resection: do we have the right “tool” for their intraoperative localization? J Thorac Dis 2017;9:4216-8. [Crossref] [PubMed]
- Dai J, Yu G, Yu J. Can CT imaging features of ground-glass opacity predict invasiveness? A meta-analysis. Thorac Cancer 2018;9:452-8. [Crossref] [PubMed]
- Lee HJ, Lee CH, Jeong YJ, et al. IASLC/ATS/ERS International Multidisciplinary Classification of Lung Adenocarcinoma: novel concepts and radiologic implications. J Thorac Imaging 2012;27:340-53. [Crossref] [PubMed]
- Collins J, Stern EJ. Ground-glass opacity at CT: the ABCs. AJR Am J Roentgenol 1997;169:355-67. [Crossref] [PubMed]
- Ghosh D, Poisson LM. "Omics" data and levels of evidence for biomarker discovery. Genomics 2009;93:13-6. [Crossref] [PubMed]
- Claudino WM, Goncalves PH, di Leo A, et al. Metabolomics in cancer: a bench-to-bedside intersection. Crit Rev Oncol Hematol 2012;84:1-7. [Crossref] [PubMed]
- Spratlin JL, Serkova NJ, Eckhardt SG. Clinical applications of metabolomics in oncology: a review. Clin Cancer Res 2009;15:431-40. [Crossref] [PubMed]
- Want EJ, Nordstrom A, Morita H, et al. From exogenous to endogenous: the inevitable imprint of mass spectrometry in metabolomics. J Proteome Res 2007;6:459-68. [Crossref] [PubMed]
- Dettmer K, Aronov PA, Hammock BD. Mass spectrometry-based metabolomics. Mass Spectrom Rev 2007;26:51-78. [Crossref] [PubMed]
- Ristimaa J, Gergov M, Pelander A, et al. Broad-spectrum drug screening of meco-nium by liquid chromatography with tandem mass spectrometry and time-of-flight mass spectrometry. Anal Bioanal Chem 2010;398:925-35. [Crossref] [PubMed]
- Pelander A, Ristimaa J, Ojanpera I. Vitreous humor as an alternative matrix for comprehensive drug screening in postmortem toxicology by liquid chromatography-time-of-flight mass spectrometry. J Anal Toxicol 2010;34:312-8. [Crossref] [PubMed]
- Wikoff WR, Hanash S, DeFelice B, et al. Diacetylspermine Is a Novel Prediagnostic Serum Biomarker for Non-Small-Cell Lung Cancer and Has Additive Performance With Pro-Surfactant Protein B. J Clin Oncol 2015;33:3880-6. [Crossref] [PubMed]
- Fahrmann JF, Kim K, DeFelice BC, et al. Investigation of metabolomic blood bi-omarkers for detection of adenocarcinoma lung cancer. Cancer Epidemiol Biomarkers Prev 2015;24:1716-23. [Crossref] [PubMed]
- Kuhl C, Tautenhahn R, Bottcher C, et al. CAMERA: an integrated strategy for com-pound spectra extraction and annotation of liquid chromatography/mass spectrometry data sets. Anal Chem 2012;84:283-9. [Crossref] [PubMed]
- Trygg J, Holmes E, Lundstedt T. Chemometrics in metabonomics. J Proteome Res 2007;6:469-79. [Crossref] [PubMed]
- van Velzen EJ, Westerhuis JA, van Duynhoven JP, et al. Multilevel data analysis of a crossover designed human nutritional intervention study. J Proteome Res 2008;7:4483-91. [Crossref] [PubMed]
- Fong MY, McDunn J, Kakar SS. Identification of metabolites in the normal ovary and their transformation in primary and metastatic ovarian cancer. PLoS One 2011;6:e19963. [Crossref] [PubMed]
- Ke C, Hou Y, Zhang H, et al. Plasma Metabolic Profiles in Women are Menopause Dependent. PLoS One 2015;10:e0141743. [Crossref] [PubMed]
- Zhang T, Wu X, Yin M, et al. Discrimination between malignant and benign ovarian tumors by plasma metabolomic profiling using ultra performance liquid chromatography/mass spectrometry. Clin Chim Acta 2012;413:861-8. [Crossref] [PubMed]
- Wishart DS, Jewison T, Guo AC, et al. HMDB 3.0--The Human Metabolome Data-base in 2013. Nucleic Acids Res 2013;41:D801-7. [Crossref] [PubMed]
- Kanehisa M, Goto S, Sato Y, et al. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res 2014;42:D199-205. [Crossref] [PubMed]
- Louis E, Adriaensens P, Guedens W, et al. Detection of Lung Cancer through Metabolic Changes Measured in Blood Plasma. J Thorac Oncol 2016;11:516-23. [Crossref] [PubMed]
- Hori S, Nishiumi S, Kobayashi K, et al. A metabolomic approach to lung cancer. Lung Cancer 2011;74:284-92. [Crossref] [PubMed]
- Barzilai N, Huffman DM, Muzumdar RH, et al. The critical role of metabolic path-ways in aging. Diabetes 2012;61:1315-22. [Crossref] [PubMed]
- Fahrmann JF, Grapov D, DeFelice BC, et al. Serum phosphatidylethanolamine levels distinguish benign from malignant solitary pulmonary nodules and represent a potential diagnostic biomarker for lung cancer. Cancer Biomark 2016;16:609-17. [PubMed]
- Huang LJ, Chen SX, Luo WJ, et al. Proteomic analysis of secreted proteins of non-small cell lung cancer. Ai Zheng 2006;25:1361-7. [PubMed]
- Wang X, Li N, Liu B, et al. A novel human phosphatidylethanolamine-binding protein resists tumor necrosis factor alpha-induced apoptosis by inhibiting mitogen-activated protein kinase pathway activation and phosphatidylethanolamine externalization. J Biol Chem 2004;279:45855-64. [Crossref] [PubMed]
- Yu GP, Chen GQ, Wu S, et al. The expression of PEBP4 protein in lung squamous cell carcinoma. Tumour Biol 2011;32:1257-63. [Crossref] [PubMed]
- Marien E, Meister M, Muley T, et al. Non-small cell lung cancer is characterized by dramatic changes in phospholipid profiles. Int J Cancer 2015;137:1539-48. [Crossref] [PubMed]
- Zinrajh D, Horl G, Jurgens G, et al. Increased phosphatidylethanolamine N-methyltransferase gene expression in non-small-cell lung cancer tissue predicts shorter patient survival. Oncol Lett 2014;7:2175-9. [Crossref] [PubMed]
- Sreekumar A, Poisson LM, Rajendiran TM, et al. Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature 2009;457:910-4. [Crossref] [PubMed]
- Yu L, Aa J, Xu J, et al. Metabolomic phenotype of gastric cancer and precancerous stages based on gas chromatography time-of-flight mass spectrometry. J Gastroenterol Hepatol 2011;26:1290-7. [Crossref] [PubMed]
- Cascino A, Muscaritoli M, Cangiano C, et al. Plasma amino acid imbalance in patients with lung and breast cancer. Anticancer Res 1995;15:507-10. [PubMed]


