
Sensitive molecular testing methods can demonstrate NSCLC driver mutations in malignant pleural effusion despite non-malignant cytology
Introduction
Cytology is currently the standard method of diagnosing malignant pleural effusion (MPE), however the sensitivity is approximately 60–70%, and is dependent upon tumour load, volume of fluid assessed and cytopathologist experience (1,2). Lung adenocarcinoma is the commonest cause of MPE and following cytologic diagnosis, molecular testing of MPE cell block specimens is feasible (3-5).
The recent introduction of highly sensitive molecular testing techniques, such as amplicon-based parallel sequencing (APS) (6) and improved and complete enrichment CO-amplification at lower denaturation temperature PCR (ICECOLD PCR) (7-9) is rapidly changing the application of DNA sequencing for lung cancer patients. These techniques are currently applied only to specimens demonstrated to be malignant by cytologic/histologic examination. The sensitivity of these methods suggests molecular testing could also provide an alternative approach for detection of MPE.
We investigated whether the sensitive molecular testing methods of APS and ICECOLD PCR could detect tumour-derived mutations in cytologically non-MPEs from patients with non-small cell lung cancer (NSCLC).
Methods
This project was approved by the Human Research Ethics Committee of the Royal Melbourne Hospital (LNR/15/MH/289).
Patient selection
Retrospective review of institutional records identified 61 patients with a final diagnosis of MPE in 2014 (1). In five of these patients, initial pleural centesis specimens demonstrated non-malignant results on cytologic examination (including both morphologic examination and immunohistochemical examination with TTF-1 and calretinin). Subsequent clinical investigation in these patients confirmed malignant pleural involvement by NSCLC (Figure 1). These five patients (Table 1) form the basis of this report.
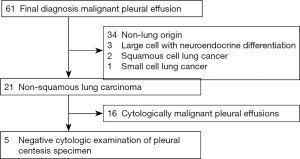

Full table
Mutation testing—reference specimens
Tumour mutation status was established by amplicon parallel sequencing of separate diagnostic clinical specimens (see Table 1).
Cell blocks and DNA extraction
Archival formalin-fixed paraffin embedded (FFPE) cell-blocks of the non-malignant pleural fluid samples were obtained. Five unstained sections of 10-micron thickness were cut onto glass slides, dewaxed, then macro-dissected. DNA was extracted using the QIAamp DNA FFPE Tissue Kit (Qiagen, Hilden, Germany).
Amplicon parallel sequencing
All non-malignant pleural fluid samples were analysed in triplicate using APS. Targeted regions of EGFR exons 18, 19, 20, 21, KRAS and NRAS exons 2, 3, 4, BRAF exon 15, and PIK3CA exons 9 and 20 were amplified by multiplex polymerase chain reaction (PCR) and sequenced on a MiSeq Next Generation Sequencer (Illumina, SanDiego, USA). Mutations were detected using MiSeq Reporter Software (Illumina, San Diego, USA). Only mutations at >1% frequency in at least two out of three assays were recorded.
ICECOLD PCR
The more sensitive ICECOLD PCR method was used for samples in which a patient’s known mutation was not detected by APS. After the multiplex reaction the samples were Pico-tested for double-stranded DNA content, with those having concentrations over 30 ng/µL considered to have passed primary amplification, followed by dilution to a uniform 4 ng. Transgenomic ICE-me kit (Transgenomic, Omaha, USA) reagents were used for the nested ICE COLD enrichment reaction, with the manufacturers thermal cycler protocols. Verity thermal cyclers used were stepped down to a 38.4% ramp rate (1.5 degrees/second). Samples underwent BigDye 3.1 cycle-sequencing on a 3730xl DNA analyzer. Sequencing traces were analysed on Sequencher 3.5 software. Variant detection was determined by dual comparison—wild-type control (WTC) and sample-peak under peak (PuP) levels. Variants were deemed genuine if they were;
- Higher by >3× than any PuP in the WTC at the same nucleotide location, and,
- Higher by >2× than any PuP in the surrounding region of the tested sample. WTC DNA was derived from FFPE material.
Results
The histo-cytological findings and corresponding molecular results of reference specimens are presented in Table 1, alongside cytologic and molecular findings from non-malignant pleural fluid specimens.
Molecular testing results—reference specimens
Two patients had known lung adenocarcinoma diagnosed by core-biopsy, with KRAS c.34G>T G12C mutation detected on molecular testing of the clinical biopsy specimens. One patient had pleomorphic carcinoma with a known KRAS c.34G>T G12C mutation detected in a pleural biopsy specimen. The remaining two patients demonstrated wild type EGFR, KRAS, NRAS, BRAF and PIK3CA on testing of cytologically malignant pleural fluid samples.
Molecular testing results—cytologically non-malignant pleural fluid specimens
In all three patients with known KRAS mutations detected in reference diagnostic specimens, the expected mutation was detected by the highly sensitive molecular testing techniques in cytologically negative pleural effusion specimens.
For the two adenocarcinoma patients with KRAS c.34G>T G12C mutation, APS testing of the cytologically non-malignant pleural fluid specimens demonstrated the same KRAS mutation at approximately 2% mutant allele frequency.
In the patient with pleural biopsy demonstrating KRAS c.34G>T G12C mutation, APS testing of cytologically non-malignant pleural fluid did not demonstrate a mutation. However, further molecular analysis of the same pleural fluid with the more sensitive method of ICECOLD PCR detected the KRAS variant at a mutant allele frequency of 0.7% (Figure 2).
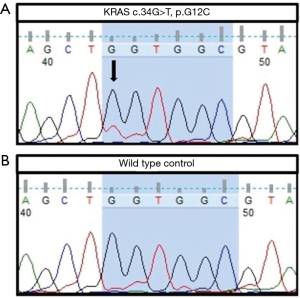
There was no mutation detected by APS on the cytologically non-malignant pleural fluid samples from the two patients with wild-type findings from the reference samples (specificity 100%).
Discussion
We demonstrate that highly sensitive mutation detection methods can detect driver mutations in cytologically non-malignant pleural fluids. Using APS and ICECOLD-PCR we detected mutations in all three cytologically non-malignant pleural fluids for which a separate diagnostic clinical reference specimen demonstrated a mutation. In each case the detected mutation was identical to that reported in the reference sample. Importantly, there were no false-positive mutations detected in any samples, including two samples with known wild type tumours. Our findings demonstrate the feasibility of molecular diagnostic techniques for improvement of diagnostic assessment of pleural effusions in patients with lung cancer.
Critically, the three patients in this cohort where KRAS mutations were detected on cytologically non-malignant specimens were subsequently demonstrated clinically to have MPE, indicating their molecular findings were not false-positive results. Carcinoma cells are recognised in cytology samples on their morphology, and therefore must be intact and well preserved to be recognized on cytological examination (10). We hypothesise that the mutation-positive but cytologically non-MPE samples had small numbers of poorly visualised, degenerate, fragmented, or entirely lysed tumour cells, as the source of tumour-derived DNA.
There is significant potential for sensitivity of cytologic detection of MPE to be improved by molecular diagnostic methods such as those used in the current study. Benefits may include obviating need for more invasive procedures, and avoiding delayed diagnoses. Patients with known early stage lung cancer who develop a cytologically negative pleural effusion may benefit from molecular testing prior to radical-intent treatment to avoid potentially futile therapy.
The ability of sensitive PCR-based techniques to detect a range of driver mutations in NSCLC within cytologically malignant pleural fluid specimens is well established (11,12). However, to our knowledge only two prior studies have applied molecular diagnostic methods to cytologically negative pleural fluids in patients with confirmed driver mutations. Buttitta et al. used next-generation sequencing to detect activating EGFR mutations in patients with known EGFR mutated cancers. An activating EGFR mutation was detected in only one of five pleural fluid specimens examined (13). As part of a larger study on pleural fluids that were mostly cytologically malignant, Akamatsu et al. used a combination of a combination of methods to identify mutations in three of 17 (18%) cytologically non-malignant pleural fluids from patients with lung carcinoma (14). However, they did not report which mutations were found in these particular samples, the mutant allele frequency, which method was used, or whether the same mutations were confirmed in another tumour sample. It therefore remains unclear whether the detected variants were from pleural spread of known tumour, from a benign proliferative process in the pleura, or potential artefacts.
The high sensitivity for detection of mutations in our study (100%) likely reflects the higher analytical sensitivity of the methods we used. The APS method we used has a limit of detection of 1% mutant allele frequency, and ICECOLD PCR has a reported limit of detection of 0.05% (9,15), therefore our sequential testing method should have detected mutations to the level of 0.05%. There are more sensitive methods of mutation detection now available, with analytical sensitivity below this level, such as BEAMing, digital droplet PCR, and SAFE-SeqS (16), and these could be even more effective at detecting tumour derived mutations, although false positives could arise from contamination with circulating tumour DNA or circulating tumour cells.
Limitations
This is a small retrospective study. Larger studies using various high sensitivity mutation detection methods are required. The clinical significance of cytological negative/mutation-positive effusion also requires further consideration. Other approaches to enhancing detection of MPE may complement PCR-based detection of tumour markers. Metabolic-based assays for rapid detection of rare metabolically active tumor cells in cytologically non-malignant pleural fluid specimens are reported (17). These demonstrate “live” tumour cells, though the clinical predictive significance of this finding, similar to that for detection of circulating tumour cells in early stage NSCLC, remains unclear. Detection of driver mutations is likely to be highly significant/specific, though negative predictive value is not established.
Recent animal studies suggest KRAS mutations may promote development of MPE (18), suggesting such patients are at higher risk of development of MPE. Future studies including patients with other driver mutations are required to confirm findings.
Use of highly sensitive molecular techniques may also be of diagnostic utility in other lung cancer specimen types, such as bronchoscopic specimens (19). Such specimens may be non-diagnostic or not suitable for molecular testing (20,21), due to small specimen volume, or due to presence of a significant proportion of non-malignant cells (22). Molecular testing even on a single bronchial brushing specimen may significantly enhance diagnostic sensitivity of minimally invasive investigation (23,24), though this remains to be examined in future studies.
Conclusions
The sensitive molecular methods Amplicon parallel sequencing and ICECOLD PCR may detect cancer-driver mutations in cytologically non-malignant pleural fluids from patients with lung adenocarcinoma. Molecular testing of pleural effusions demonstrates significant potential utility in improved diagnosis of pleural effusions in NSCLC patients.
Acknowledgments
This work was supported by RMH Home Lottery Grant-in-Aid.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This project was approved by the Human Research Ethics Committee of the Royal Melbourne Hospital (LNR/15/MH/289).
References
- Loveland P, Christie M, Hammerschlag G, et al. Diagnostic yield of pleural fluid cytology in malignant effusions: an Australian tertiary centre experience. Intern Med J 2018;48:1318-24. [Crossref] [PubMed]
- Rivera MP, Mehta AC, Wahidi MM. Establishing the diagnosis of lung cancer: Diagnosis and management of lung cancer, 3rd ed: American College of Chest Physicians evidence-based clinical practice guidelines. Chest 2013;143:e142S-65S.
- Yang J, Lee OJ, Son SM, et al. EGFR Mutation Status in Lung Adenocarcinoma-Associated Malignant Pleural Effusion and Efficacy of EGFR Tyrosine Kinase Inhibitors. Cancer Res Treat 2018;50:908-16. [Crossref] [PubMed]
- Chen YL, Lee CT, Lu CC, et al. Epidermal Growth Factor Receptor Mutation and Anaplastic Lymphoma Kinase Gene Fusion: Detection in Malignant Pleural Effusion by RNA or PNA Analysis. PLoS One 2016;11:e0158125. [Crossref] [PubMed]
- Li W, Zhang Z, Guo L, et al. Assessment of cytology based molecular analysis to guide targeted therapy in advanced non-small-cell lung cancer. Oncotarget 2016;7:8332-40. [PubMed]
- König K, Peifer M, Fassunke J, et al. Implementation of Amplicon Parallel Sequencing Leads to Improvement of Diagnosis and Therapy of Lung Cancer Patients. J Thorac Oncol 2015;10:1049-57. [Crossref] [PubMed]
- How-Kit A, Lebbe C, Bousard A, et al. Ultrasensitive detection and identification of BRAF V600 mutations in fresh frozen, FFPE, and plasma samples of melanoma patients by E-ice-COLD-PCR. Anal Bioanal Chem 2014;406:5513-20. [Crossref] [PubMed]
- Luke JJ, Oxnard GR, Paweletz CP, et al. Realizing the potential of plasma genotyping in an age of genotype-directed therapies. J Natl Cancer Inst 2014;106. [Crossref] [PubMed]
- Tran HT, Legendre BL, Kim ES, et al. The use of improved and complete enrichment co-amplification at lower denaturation temperature (ICE COLD-PCR) method for the detection of EGFR and KRAS mutations from cell-free plasma DNA of non-small cell lung cancer (NSCLC) patients. J Clin Oncol 2014;32:abstr 8058.
- Pereira TC, Saad RS, Liu Y, et al. The diagnosis of malignancy in effusion cytology: a pattern recognition approach. Adv Anat Pathol 2006;13:174-84. [Crossref] [PubMed]
- DeMaio A, Clarke JM, Dash R, et al. Yield of Malignant Pleural Effusion for Detection of Oncogenic Driver Mutations in Lung Adenocarcinoma. J Bronchology Interv Pulmonol 2019;26:96-101. [Crossref] [PubMed]
- Wu SG, Liu YN, Yu CJ, et al. Driver mutations of young lung adenocarcinoma patients with malignant pleural effusion. Genes Chromosomes Cancer 2018;57:513-21. [Crossref] [PubMed]
- Buttitta F, Felicioni L, Del Grammastro M, et al. Effective assessment of egfr mutation status in bronchoalveolar lavage and pleural fluids by next-generation sequencing. Clin Cancer Res 2013;19:691-8. [Crossref] [PubMed]
- Akamatsu H, Koh Y, Kenmotsu H, et al. Multiplexed molecular profiling of lung cancer using pleural effusion. J Thorac Oncol 2014;9:1048-52. [Crossref] [PubMed]
- How-Kit A, Daunay A, Buhard O, et al. Major improvement in the detection of microsatellite instability in colorectal cancer using HSP110 T17 E-ice-COLD-PCR. Hum Mutat 2018;39:441-53. [Crossref] [PubMed]
- Siravegna G, Marsoni S, Siena S, et al. Integrating liquid biopsies into the management of cancer. Nat Rev Clin Oncol 2017;14:531-48. [Crossref] [PubMed]
- Tang Y., Wang Z., Li Z, et al. High-throughput screening of rare metabolically active tumor cells in pleural effusion and peripheral blood of lung cancer patients. Proc Natl Acad Sci U S A 2017;114:2544-9. [Crossref] [PubMed]
- Agalioti T, Giannou AD, Krontira AC, et al. Mutant KRAS promotes malignant pleural effusion formation. Nat Commun 2017;8:15205. [Crossref] [PubMed]
- Steinfort DP, Bonney A, See K, et al. Sequential multimodality bronchoscopic investigation of peripheral pulmonary lesions. Eur Respir J 2016;47:607-14. [Crossref] [PubMed]
- Bonney A, Beaty A, See K, et al. Diagnostic Utility of Bronchial Brush-Tip Washings for the Immunohistochemical Assessment of Peripheral Lung Lesions. Acta Cytol 2016;60:74-8. [Crossref] [PubMed]
- Bonney A, Christie M, Beaty A, et al. The feasibility of molecular testing on cell blocks created from brush tip washings in the assessment of peripheral lung lesions. J Thorac Dis 2016;8:2551-5. [Crossref] [PubMed]
- Ilie M, Hofman V, Ortholan C, et al. Predictive clinical outcome of the intratumoral CD66b-positive neutrophil-to-CD8-positive T-cell ratio in patients with resectable nonsmall cell lung cancer. Cancer 2012;118:1726-37. [Crossref] [PubMed]
- Vannitamby A, Hendry S, Makadia T, et al. A Novel Approach to Detect Programed Death Ligand 1 (PD-L1) Status and Multiple Tumor Mutations Using a Single Non-Small-Cell Lung Cancer (NSCLC) Bronchoscopy Specimen. J Mol Diagn 2019;21:186-97. [Crossref] [PubMed]
- Vannitamby A, Hendry S, Irving L, et al. Novel multiplex droplet digital PCR assay for scoring PD-L1 in non-small cell lung cancer biopsy specimens. Lung Cancer 2019;134:233-7. [Crossref] [PubMed]


