
Systematic analysis identifies three-lncRNA signature as a potentially prognostic biomarker for lung squamous cell carcinoma using bioinformatics strategy
Introduction
Lung cancer (LC) with more than 2 million new cases and 1.7 million deaths in 2018, is the most commonly diagnosed malignancies and the top leading cause of cancer death worldwide (1). Non-small cell lung cancer (NSCLC) is the most common pathological type, and accounts for approximately 85% of all LCs (2). Among NSCLCs, lung squamous cell carcinoma (LUSC) with more than 400,000 new cases per year is the second most common type of NSCLCs, and accounts for 20–30% of NSCLCs (3,4). Due to the lack of sensibility to radiotherapy, surgery and chemotherapy are the main methods to combat LUSC (5). Despite advances in treatment methods of LUSC, the 5-year overall survival (OS) rate of LUSC patients with clinical I and II stages is about 40%, and that of LUSC patients with clinical III and IV stages less than poor 5% (6). A diagnosis for LUSC at an early stage contributes to offering a favorable prognosis, and the 5-year OS rate will significantly increase to 70–90% (7). Currently, disease stage and histological grade are still the basic methods for evaluating the diagnosis and prognosis of LUSC. However, it is limited to evaluate the predictive value in diagnosing early-stage LUSC by clinical and pathological symptoms, and it is difficult to predict the clinical outcomes due to the heterogeneity of LUSC. In addition, the molecular mechanisms underlying LUSC still remain unclear. Therefore, it is vital to further explore the molecular mechanisms, and identify potentially molecular diagnostic and prognostic markers and/or therapeutic targets to combat LUSC.
Smoking is the major risk factor for LUSC, which causes high number of mutations such as TP53, CDKN2A and PIK3CA (8). Although molecularly targeted therapies directly against those driver genes are an effective method to combat LUSC, development of molecularly targeted therapies has been less rapid for LUSC with respect to lung adenocarcinoma (LUAD), and molecularly targeted therapies are still not standard among LUSCs. Even though some genes such as DDR2, AKT and FGFR were considered as potentially molecular targets, few effective targeted therapies for LUSC are so far approved, and targeted therapies developed against LUAD are largely ineffective against LUSC (9,10). Recent efforts have focused on discovering novel, clinically actionable cancer driver genes by multi-omics analysis (10,11). However, due to the accumulation of large number of passenger mutations from prolonged exposure to carcinogens, identifying novel driver genes is challenging (8).
Long non-coding RNAs (lncRNAs) are an important class of non-coding RNAs (ncRNAs) with over 200 nucleotides long (12), and are widely reported to play roles as important regulatory components in tumor initiation and progression (13). However, little knowledge about the roles of lncRNAs is known in tumor biology. Nevertheless, due to the high tissue specificity of lncRNA expression, lncRNAs have been considered as potentially valuable diagnostic and prognostic biomarkers and/or therapeutic targets of some malignant tumors (14-17). Competitive endogenous RNA (ceRNA) hypothesis is so far considered as the foundation of playing the regulatory role for lncRNAs (18). LncRNA as ceRNA and mRNA can competitively bind microRNA (miRNA) by miRNA-response-elements (MREs) to play key roles in various biological processes such as tumor formation and metastasis (19). For instance, Fan’s team constructed lncRNA-miRNA-mRNA ceRNA network based on ceRNA theory to identify that a 4-lncRNA signature played roles in breast cancer (20). Liu and the colleagues revealed functional lncRNAs in gastric cancer by integrative analysis of dysregulated lncRNA-related ceRNA network (21). In addition, systematic analysis of lncRNA-related ceRNA network have been reported in many other cancers including thyroid carcinoma (22), glioblastoma (23), prostate cancer (24), and ovarian cancer (25). Collectively, the lncRNA-related ceRNA network findings revealed the lncRNA-associated regulatory mechanisms by miRNA-mediated lncRNA and mRNA interaction in cancer initiation and progression. For lncRNA-related ceRNA network of LUSC, several studies have reported some lncRNAs associated with LUSC development and clinical outcome (9,26,27). However, the findings reported from different studies are inconsistent due to differentially limited samples. The Cancer Genome Atlas (TCGA, 2018, the seventh edition of TNM staging criteria, https://cancergenome.nih.gov/) is a publically available cancer-related database which provides large-scale multi-dimensional molecular profile data related to over 30 cancer types. Using LUSC-related data with large sample size from TCGA database to identify LUSC-specific lncRNAs can increase statistical reliability of the current study. Another key issue is that most reported studies didn’t consider the co-expression characteristic of genes in ceRNA network, which may be difficult to target the key lncRNAs. Weighted gene co-expression network analysis (WGCNA) is a systematic biology method to describe the correlation patterns among genes across samples, and using WGCNA can find modules of highly correlated genes and identify key genes (28). Presently, WGCNA has been successfully applied in various biological contexts including cancer such as LC (29), bladder cancer (30), and colon cancer (31). Systematic analysis of lncRNA-miRNA-mRNA ceRNA network based on WGCNA contributes to insight into novel molecular mechanisms involved in LUSC. While, very little information is available on LUSC ceRNA based on WGCNA.
In this work, LUSC-related RNA-seq data were retrieved from the TCGA database, and were systematically integrated and analyzed based on bioinformatics methods including Differentially Expressed Gene Analysis (DEGA), WGCNA, Protein and Protein Interaction (PPI) network, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses, ceRNA network, and centrality analysis. Subsequently, survival analysis was performed to identify the key lncRNA signature in lncRNA-related ceRNA network as potentially valuable prognosticator associated with OS of LUSC patients. This study provides novel insights into molecular mechanisms underlying LUSC, and identifying novel lncRNAs may serve as potentially diagnostic and prognostic biomarkers and/or therapeutic targets to combat LUSC.
Methods
The flow chart of systematic bioinformatics analysis in the current study was showed in Figure 1.
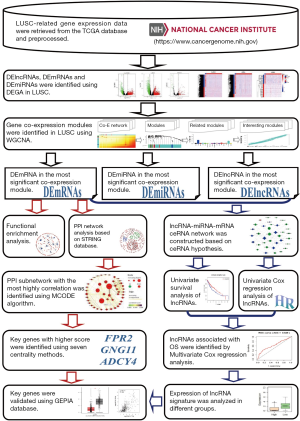
RNA-seq data and patient information collection
LUSC-related RNA-seq data were retrieved from TCGA database portal (2018, https://cancergenome.nih.gov/), and the inclusion criteria of RNA-seq data were included as follows: (I) histological diagnosis for LUSC; (II) except LUSC no other malignancy; (III) data with complete clinical information. A total of 475 LUSC tissues and 38 non-LUSC normal lung tissues RNA-seq data were retrieved. Among, 38 non-LUSC normal lung tissues were from the normal lung tissues nearby LUSC tissues of 38 LUSC patients. The clinical information of all LUSC samples patients was simultaneously obtained from TCGA database. Sample clustering was used to detect outlier, and 2 LUSC tissues samples were excluded. Finally, a total 473 LUSC-related and 38 non-LUSC normal lung tissues RNA-seq data was included. All RNA-seq data were approved by the Institutional Review Board of the relevant participating institutions and by the National Cancer Institute of NIH. No approval from the ethics committee was required. The present study meets the requirements of data usage and publishing from TCGA database. The clinical characteristics for LUSC patients were listed in Table 1.

Full table
RNA-seq data preprocessing and DEGA
All raw RNA-seq data were subjected to normalization using the trimmed mean of M-values method.
EdgeR package in Bioconductor project (version 3.8, http://www.bioconductor.org/) was used to screen the differentially expressed lncRNAs (DElncRNAs), differentially expressed miRNAs (DEmiRNAs) and differentially expressed mRNAs (DEmRNAs) between LUSC tissues and non-LUSC lung tissues samples (32). |Log (fold change)| (|logFC|) >2 and statistical P value <0.01 (P<0.01) were set as cut-off criteria. The volcano plots of RNA expression were visualized using the ggplot2 package (version 3.1.0, https://github.com/tidyverse/ggplot2). The heat map of differentially expressed RNAs (DERNAs) was plotted using the pheatmap package (version 1.0.10, https://CRAN.R-project.org/package=pheatmap).
WGCNA
WGCNA was used to construct the gene co-expression network among DElncRNAs, DEmiRNAs and DEmRNAs, and identify co-expression gene modules. WGCNA package (version 1.13) based on R was used to perform WGCNA (28). The expression matrix was firstly converted into an adjacency matrix, and an unsupervised co-expression relationship was constructed based on the adjacency matrix using Pearson correlation coefficients for gene pairs. The correlation adjacency matrix was strengthened by power β=10 (soft threshold), and the power parameter was selected based on scale-free topology criterion. Secondly, the adjacency matrix was converted into a topological matrix, and topological overlap measure (TOM) was used to measure the correlation of gene pairs. According to TOM-based dissimilarity, average linkage hierarchical clustering was performed to classify genes with coherent expression profiles into gene modules. Dynamic cutting algorithm was used to identify gene modules from the system cluster tree, and the modules with 95% similarity were merged into a module. Module eigengene (ME) was defined as the first principal components, and was the representative in module genes. Module membership (MM) was defined as the correlation between ME and gene module. Gene significance (GS) was indexed by log10 transformation of the p value of t-test measuring differential expression between LUSC and non-LUSC tissues. Module significance (MS) was defined as the average GS for all the genes in the module. More detailed description of WGCNA is presented by original article (28).
The genes in the most significant module were selected to further analyze.
PPI network construction and analysis
The interactive relationships among differentially expressed genes (DEGs) encoding proteins in the most significant gene co-expression module was analyzed by constructing a PPI network. The interactive information among DEGs encoding proteins was retrieved from online STRING database (version 11.0, https://string-db.org/) (33). The gene pairs with a combined score ≥ the highest confidence 0.9 were used to construct the PPI network. Cytoscape software (version 3.7.0, http://www.cytoscape.org/) was used to construct and visualize the interactive relationships among genes in whole PPI network (34). The subnetwork (highly correlated module) was extracted from whole PPI network using Molecular COmplex DEtection (MCODE) algorithm based on topological properties of whole PPI network, and a plugin MCODE (version 1.5.1) in Cytoscape was used to perform MCODE analysis (35). The threshold parameters were more severely set for Degree Cutoff=4, Node Score Cutoff=0.6, K-Core=4 and Max. Depth=100. The most significantly correlated module was used to next analyze.
Gene function analysis
GO and KEGG pathway enrichment analyses of DEmRNAs in PPI network were implemented using the clusterProfiler package (version 3.10.1), and an adjusted P<0.05 was considered to significantly enrich (36). GO analysis included Biological Process (BP), Cellular Component (CC) and Molecular Function (MF).
Identification of key genes and validation
Key genes in PPI network were identified using Centrality analyses. Centrality analyses including Subgraph Centrality, Degree Centrality, Eigenvector Centrality, Betweenness Centrality, Network Centrality, Information Centrality and Closeness Centrality were performed using a plugin CytoNCA (version 2.1.6) in Cytoscape (37). The genes with higher Centrality scores were identified as key gene, and essential genes were identified as intersecting genes of key genes obtained by seven Centrality methods. Gene Expression Profiling and Interactive Analyses (GEPIA, http://gepia.cancer-pku.cn) database is an interactive web server for analyzing gene expression data of tumors and normal tissues from TCGA and genotype-tissue expression database (38,39), and was used to validate the expression of essential genes in PPI network between LUSC and non-LUSC lung normal tissues and analyze the association with OS in LUSC patients. In GEPIA database, the expression analysis between LUSC and non-LUSC lung normal tissues was performed using log2(TPM+1), and survival analysis was perform using Log-Rank (LR) test (Mantel-Cox test)for hypothesis test. The LR P value, cox proportional hazard ratio (HR) and the 95% confidence interval (CI) information were calculated.
Construction of lncRNA-miRNA-mRNA ceRNA network
Co-expression genes in the most significant module were used to construct lncRNA-miRNA-mRNA ceRNA network, and ceRNA network was established based on ceRNA hypothesis that lncRNA directly bind miRNA by acting as miRNA sponge to indirectly regulate the function of mRNA (18). A ceRNA network was constructed according to the following steps: (I) three types of DERNAs in the most significant module were kept; (II) the potential target miRNAs of DElncRNAs, as well as lncRNAs and miRNAs interaction relationships were predicted using the online miRcode tool (miRcode 11, http://www.mircode.org/); (III) the potential target mRNAs of DEmiRNAs were predicted using the online tools including TargetScan (release 7.2, http://www.targetscan.org/), miRDB (version 5.0, http://mirdb.org/) and miRTarBase (release 7.0, http://mirtarbase.mbc.nctu.edu.tw/); (IV) based on ceRNA hypothesis, intersecting miRNAs negatively regulated by lncRNAs and mRNAs were selected to construct ceRNA network. A ceRNA network was constructed and visualized using an open-source Cytoscape software (34).
LUSC-specific prognostic lncRNA signatures identification
The associations between DElncRNAs in ceRNA network and OS in LUSC patients were evaluated using Kaplan-Meier (KM) estimate and Log-rank (LR) test in survival package (version 2.43-3, https://CRAN.R-project.org/package=survival) based on R. Group cut off of 50% was set, and LR P value, hazard ratio (HR) and 95% CI were computed. The statistical P<0.05 was considered as the significant association between DElncRNA and OS of LUSC patients. Further, univariate Cox proportional hazards regression model on the basis of survival package in R project was performed to evaluate the association between DElncRNAs in ceRNA network and OS of LUSC patients. The same characteristic parameters as LR method were computed and the same significant P value criterion was set. Overlapping DElncRNAs significantly associated with OS of LUSC patients in LR method and univariate Cox regression analysis was performed multivariate Cox hazards regression model with Stepwise method, and which was implemented using survival package in R project, and multivariate Cox hazards regression model was used to assess the prognostic value for LUSC. DElncRNA combination in the optimal Cox hazard regression model was used to further analyses. The hazards model was established as follows:
where “Exp” denoted the expression level of lncRNA, and “Coe” represented the regression coefficient from the multivariate Cox regression model (20). According to the median of above risk scores, LUSC patients were divided into high- and low-risk groups. Receiver operating characteristic (ROC) curve was constructed using survivalROC package (version 1.0.3, https://CRAN.R-project.org/package=survivalROC) based on R, and was used to measure the risk prediction rate of lncRNAs between high- and low-risk groups.
Statistical analysis
All statistical analyses were performed using R software. The comparison of gene expression in LUSC tissues and normal tissues was analyzed using Mean ± Standard Deviation. T-test was used to estimate the statistical difference, and P<0.05 was considered as significant difference between two groups in expression level.
Results
Identification of DElncRNA, DEmRNA and DEmiRNA
After all raw data were normalized, 8,809 lncRNAs, 18,275 mRNAs, and 821 miRNAs were extracted. Using DEGA based on EdgeR package, we identified DElncRNAs, DEmRNAs and DEmiRNAs between LUSC tissues samples and non-LUSC lung tissues samples. According to the |logFC|>2 and P<0.01 cut-off criteria, a total of 2,114 DElncRNAs (1,667 up- and 447 down-regulated, http://fp.amegroups.cn/cms/32d9bc8d31415aaab9827d0601d1fbbd/tlcr.2019.09.13-1.pdf), 3,485 DEmRNAs (2,324 up- and 1,161 down-regulated, http://fp.amegroups.cn/cms/a52c08dd78c153449baca4f523871775/tlcr.2019.09.13-2.pdf), and 175 DEmiRNAs (148 up- and 27 down-regulated, http://fp.amegroups.cn/cms/a63f404926063d02b2c7c606a1972a92/tlcr.2019.09.13-3.pdf) were identified. The distributions of DElncRNAs, DEmRNAs and DEmiRNAs were displayed using volcano plots in Figure 2A. The expression levels of DElncRNAs, DEmRNAs and DEmiRNAs between LUSC tissues and non-LUSC normal lung tissues were displayed using heat map in Figure 2B.
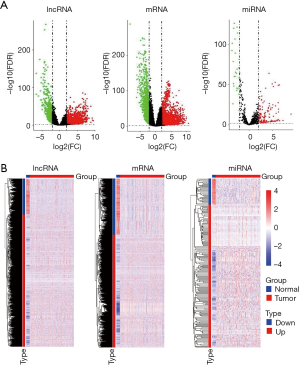
Co-expression module identification
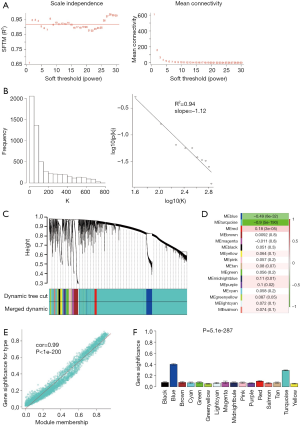
All DERNAs including 2,114 DElncRNAs, 3,485 DEmRNAs and 175 DEmiRNAs were used to reconstruct the transcriptional network using WGCNA algorithm. Pearson correlation matrix among genes were converted into a strengthened adjacency matrix by power β=10 based on scale-free topology criterion with R2=0.94 (Figure 3A,B). TOM of each gene pair was calculated, and 16 gene co-expression modules were identified by average linkage hierarchical clustering according to TOM-based dissimilarity measure (1-TOM) (Figure 3C). The correlation analysis between ME and LUSC showed that turquoise module (included 1,694 DElncRNAs and 2,654 DEmRNAs as well as 113 DEmiRNAs, http://fp.amegroups.cn/cms/23ad548482adf5ccf686ce250556033f/tlcr.2019.09.13-4.pdf) was the most significantly representative module (cor=−0.9 and P=5e-190) (Figure 3D). The MM in turquoise module possessed the most significant correlation in all modules (cor=0.99 and P<1e-200) (Figure 3E), and GS analysis across modules showed that turquoise module had the second highest gene significance with the lowest standard deviation (Figure 3F). Thus, the turquoise module was used for next analyses.
GO and KEGG enrichment analyses
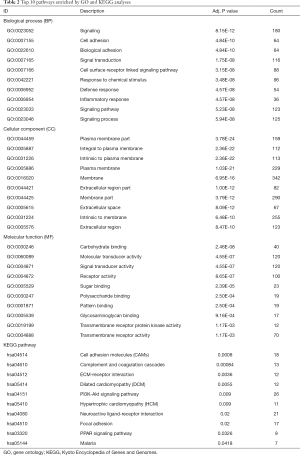
Based on MM, 660 MEs with |MM|>0.6 in turquoise module were selected to investigate the functions by GO and KEGG enrichment analyses. GO enrichment results showed that 216 BPs, 58 CCs and 33 MFs were significantly enriched. KEGG enrichment analysis showed that 11 KEGG pathways were significantly enriched. Top 10 GO terms and KEGG pathways were showed in Table 2, and the most significant BP, CC, MF and KEGG pathway were Signaling (GO:0023052, P=8.15E-12), Plasma membrane part (GO:0044459, P=3.78E-24), Carbohydrate binding (GO:0030246, P=2.46E-08) and Cell adhesion molecules (hsa04514, P=0.0008), separately. In order to elucidate the potentially biological complexities, pathway-gene network was constructed to show the relationships between pathways and genes (Figure 4A). PI3K-Akt signaling pathway contained the most significant genes in the network, which involved in many cellular functions such as transcription, translation, proliferation, growth, and survival.
Full table
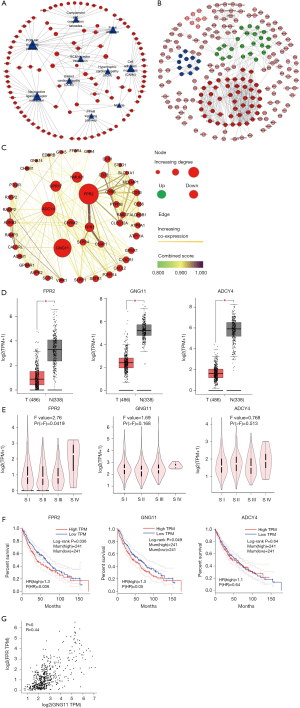
PPI network construction and key gene validation
DEmRNAs (MEs) in the most significant turquoise module were used to perform PPI network analysis. Based on MM, 660 MEs with |MM|>0.6 (653 positive correlation and 7 negative correlation) were selected to construct PPI network. At minimum required interaction score = the highest confidence 0.9, a total of 239 among 660 MEs was filtered into PPI network, and a PPI network with 239 nodes and 625 edges was established (Figure 4B). Highly correlated module analysis showed that 3 significantly correlated modules were identified in whole PPI network, and the most significant module (subnetwork 1) included 46 nodes (all genes down-regulated) and 326 edges (Score=14.489) (Figure 4C). Centrality analysis of genes in the subnetwork1 showed that top 3 genes among genes obtained by each Centrality method were FPR2 (logFC=−3.511, P=1.08e-74), GNG11 (logFC=−2.994, P=4.22e-110) and ADCY4 (logFC=−2.472, P=1.15e-99) (Table 3), and the 3 genes were identified as essential genes in subnetwork1. The expression analysis based on GEPIA database showed that the 3 genes were significantly down-regulated expressed in LUSC tissues (Figure 4D), and which were consistent with the results obtained in this study. The expression analysis between major pathological stagings showed that FPR2 (F=2.76 and P=0.0419) had significant difference between four stages, with higher expression in stage IV (Figure 4E). Survival analysis revealed that low mRNA expression of FPR2 and GNG11 resulted in a higher OS rate than high mRNA expression (P=0.036 and 0.049, respectively) (Figure 4F). Pearson correlation analysis showed that mRNA expression between FPR2 and GNG11 (R=0.44 and P=0) had more similar pattern in LUSC and normal lung tissues (Figure 4G), which demonstrated that FPR2 and GNG11 possessed the stronger co-expression in LUSC and normal lung tissues. The findings indicate that FPR2 and GNG11 may sever as potential prognosticators of OS in LUSC patients.
Full table
Construction of ceRNA network
The relationships among DElncRNA, DEmiRNA and DEmRNAs were required before lncRNA-miRNAs-mRNAs ceRNA network was constructed. Using online miRcode tool, the relationships among 1,694 DElncRNAs and 113 DEmiRNAs in the turquoise module were evaluated, and finally 121 LUSC-specific DElncRNAs (92 up- and 29 down-regulated, Table 4) and 18 LUSC-specific DEmiRNAs (12 up- and 6 down-regulated, Table 4) were putatively identified to interact. Using online tools including TargetScan, miRDB and miRTarBase, we predicted the relationships among DEmiRNAs and their target DEmRNAs. Finally, 3 LUSC-specific target DEmRNAs (1 up- and 2 down-regulated, Table 4) were putatively identified to interact with 18 LUSC-specific DEmiRNAs.
Full table
On the basis of the above data, lncRNA-miRNA-mRNA ceRNA network was constructed and visualized using Cytoscape software. Based on ceRNA hypothesis and the expression levels of DElncRNA, DEmiRNAs and DEmRNAs, two lncRNA-miRNA-mRNA ceRNA networks including over-expressed (Figure 5A) and under-expressed (Figure 5B) networks were constructed. In over-expressed ceRNA network, 65 up-regulated DElncRNAs, 6 down-regulated DEmiRNAs and 1 up-regulated DEmRNAs were included and 111 edges were contained. In under-expressed ceRNA network, 23 down-regulated DElncRNAs, 12 up-regulated DEmiRNAs and 1 down-regulated DEmRNAs were included and 78 edges were contained.
OS analysis of LUSC-specific lncRNA signature
The relationships of OS in LUSC patients and DElncRNAs in ceRNA network were determined using KM estimate and LR test. According to P<0.05 cut-off threshold, 8 DElncRNAs including AL391152.1 [P=3.834e-03, HR(high) =0.66346, 95% CI: 0.4967–0.8763], CACNA2D3-AS1 [P=8.581e-03, HR(high) =0.68679, 95% CI: 0.5149–0.9095], KCNQ5-IT1 [P=0.013, HR(high) =0.70386, 95% CI: 0.5266–0.9287], LNX1-AS1 [P=0.03775, HR(high) =1.34097, 95% CI: 1.016–1.79], MAGI2-AS3 [P=0.01855, HR(high) =1.39845, 95% CI: 1.0569–1.8601], POU6F2-AS2 [P=9.601e-03, HR(high) =0.69481, 95% CI: 0.5189–0.9147], SOX2-OT [P=0.03924, HR(high) =0.74685, 95% CI: 0.5612–0.9865] and TTTY16 [P=0.02715, HR(high) =1.3863, 95% CI: 1.0483–1.8538] were found to be related to OS of LUSC patients (Figure S1).
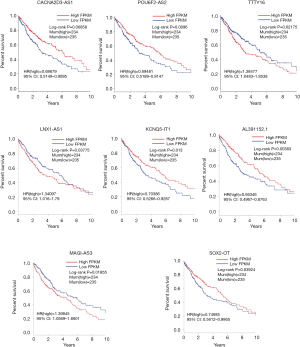
Establishment of lncRNA signature prognostic model
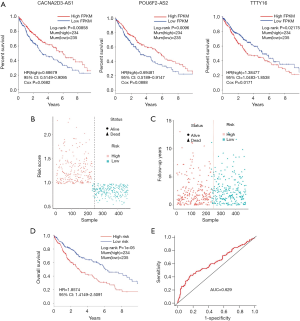
We used univariate regression analysis to identify DElncRNAs associated with the OS of LUSC patients. With P<0.05 cut-off threshold, a group of 10 DElncRNAs including TTTY16 [P=0.00583, HR(high) =1.155836, 95% CI: 1.04276–1.28118], ADAMTS9-AS2 [P=0.01317, HR(high) =1.15863, 95% CI: 1.03132–1.30165], AC006238.1 [P=0.014560, HR(high) =1.16733, 95% CI: 1.03107–1.32160], AC123595.1 [P=0.01636, HR(high) =1.16110, 95% CI: 1.02780–1.31168], CACNA2D3-AS1 [P=0.01905, HR(high) =0.87871, 95% CI: 0.78878–0.97903], KCNQ5-IT1 [P=0.02881, HR(high) =0.89105, 95% CI: 0.80350–0.98814], POU6F2-AS2 [P=0.03166, HR(high) =0.92865, 95% CI: 0.86801–0.99352], AL391152.1 [P=0.03411, HR(high) =0.90647, 95% CI: 0.82776–0.99267], CHODL-AS1 [P=0.03806, HR(high) =0.87980, 95% CI: 0.77952–0.99297] and LINC00462 [P=0.04096, HR(high) =0.93351, 95% CI: 0.87391–0.99718] was detected to have significantly prognostic value (http://fp.amegroups.cn/cms/23ad548482adf5ccf686ce250556033f/tlcr.2019.09.13-4.pdf). Furthermore, we found that 5 DElncRNAs (TTTY16, CACNA2D3-AS1, KCNQ5-IT1, POU6F2-AS2 and AL391152.1) were simultaneously identified to be related to OS in KM (LR test) and univariate regression analysis. The 5 DElncRNAs were fitted into the multivariate Cox regression model with Stepwise method. The result showed that a group of 3 DElncRNAs including TTTY16, POU6F2-AS2 and CACNA2D3-AS1 had significantly prognostic value with OS of LUSC patients, and 3-lncRNA prognostic model was established (P=0.002891). High lncRNA expression of POU6F2-AS2 and CACNA2D3-AS1, and low mRNA expression of TTTY16 resulted in a higher OS in patients (Figure 6A). With the risk score of each patient (http://fp.amegroups.cn/cms/d5fa0921ac865b7ce793c7183465233d/tlcr.2019.09.13-5.pdf), the LUSC patients were divided into high- and low-risk groups (Figure 6B,C). Comparing to low risk group, the mortality rate of patients in high-risk group was significantly higher [P=1e-05, HR(high) =1.8574, 95% CI: 1.4149–2.5091], and high-risk group was related with worse prognosis (Figure 6D). The ROC of 3-year survival correlation of the 3-lncRNA signature was analyzed, and the area under the curve (AUC) was computed. The AUC of 3-lncRNA signature was 0.629 (Figure 6E), which indicates its effectiveness as a prognostic biomarker in predicting OS of LUSC patients.
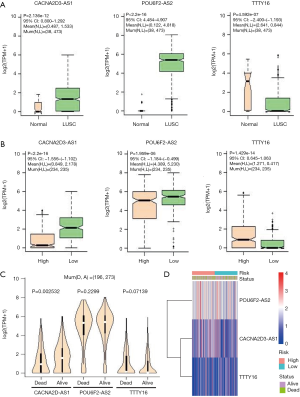
The expression levels of the 3 lncRNAs were analyzed between LUSC tissues and normal lung tissues and between high- and low- risk patient groups. CACNA2D3-AS1 and POU6F2-AS2 lncRNAs were significantly highly expressed in LUSC patients (P=2.136e-12 and <2.2e-16, separately, Figure 7A) with low risk scores (P<2.2e-16 and =1.959e-06, separately, Figure 7B). Whereas, TTTY16 lncRNA was significantly lowly expressed in LUSC patients (P=4.592e-07, Figure 7A) with high risk scores (P=1.429e-14, Figure 7B). Further, the expressions of three DElncRNAs between dead and alive groups were observed, and showed that the expression patterns of three DElncRNAs between dead and alive groups were separately consistent with that between LUSC and normal tissues and between low and high-risk groups (Figure 7C). The expression of CACNA2D-AS1 in alive group was significantly higher than that in dead group (P=0.002532, Figure 7C). The expressions of the 3 DElncRNAs in high- and risk-groups and survival status groups were showed in Figure 7D.
Discussion
LUSC is a common malignant cancer, and per year causes hundreds of thousands of deaths. Identifying specific diagnostic and prognostic biomarkers may contribute to improving survival rate among LUSC patients. To improve clinical outcomes, it is essential to identify LUSC-related prognostic signatures that predict those outcomes. Increasing evidence indicates that lncRNAs play key roles in many biological processes including cancer initiation and progression by ceRNA mechanism. In this study, we systematically analyzed the LUSC-related RNA-seq data from TCGA database by bioinformatics methods including WGCNA, GO and KEGG pathway analyses, PPI network construction, ceRNA network construction and survival analysis, and identified the key genes and key lncRNA signature associated with OS in LUSC patients. Finally, two genes including FPR2 and GNG11 in PPI network, and three lncRNA signature including CACNA2D3-AS1, POU6F2-AS2 and TTTY16 in ceRNA network were identified to associate with OS in LUSC patients.
FPR2 is an important member of 7 transmembrane G-protein-coupled FPR family, and which was originally identified to play critical roles in host defense (40). Recently, FPR2 has been reported to play a key role in human diseases such as cancers (41,42). For example, a study found that FPR2 promoted invasion and metastasis of gastric cancer cells, and negative FPR2 expression was associated with a higher OS of patients (41), which was consistent with our results. GNG11 is a member of guanine nucleotide-binding protein (G protein) gamma family, and play a role in transmembrane signaling system. Recent studies showed that GNG11 played roles in regulating cell growth and cellular senescence (43,44). So far, the functions of GNG11 is entirely unknown in cancer. No studies have so far reported any association of two genes including FPR2 and GNG11 with LUSC. This study is the first to show aberrant expressions of FPR2 and GNG11 were identified to associate with OS of LUSC patients, and the low mRNA expressions of FPR2 and GNG11 resulted in a higher OS rate in LUSC patients.
In recent years, some studies have investigated ceRNA in LUSC, and constructed ceRNA network to identify some lncRNAs associated with OS of LUSC patients (9,26,27). For example, Sui et al. constructed lcnRNA regulatory ceRNA by integrating analyzing RNA-seq data from TCGA database using DEGA and ceRNA network, and constructed a 22 lncRNAs multivariated Cox regression model and finally identified two lncRNAs including FMO6P and PRR26 had significantly prognostic value (26). In addition, Ning et al. also used LUSC-related from TCGA database to construct lncRNA-miRNA-mRNA ceRNA network, and identified six ceRNAs (PLAU, miR-31-5p, miR-455-3p, FAM83A-AS1, MIR31HG, and MIR99AHG) significantly correlated with survival by survival analysis based on KM method (9). These LUSC-related ceRNA studies provide novel knowledge for a better understanding the ceRNA interactive mechanism in LUSC biology, and contribute to improving the diagnosis and prognosis of LUSC patients. However, few lncRNA-related ceRNA network studies considered the co-expression among ceRNAs. In this study, we firstly used WGCNA to identify the co-expression modules in LUSC, and further constructed lncRNA-miRNA-mRNA ceRNA network based on co-expression genes in the most significant turquoise module according to ceRNA theory. Furthermore, KM (LR test) and univariate Cox regression analysis revealved that 5 DElncRNAs in ceRNA network (TTTY16, CACNA2D3-AS1, KCNQ5-IT1, POU6F2-AS2 and AL391152.1) were simultaneously identified to associate with OS of LUSC patients. Multivariate Cox regression analysis showed significant prognostic value of 3 of those lncRNAs (CACNA2D3-AS1, POU6F2-AS2 and TTTY16) in the OS of LUSC patients. A cumulative risk score of the 3-lncRNA showed that 3-lncRNA signature could independently predict OS in LUSC patients.
Among identified 3-lncRNA signature, POU6F2-AS2 lncRNA is an antisense lncRNA, and has so far been reported to play role in esophageal squamous cell carcinoma by modulating DNA repair (45). However, no studies reported the association between POU6F2-AS2 and OS of cancer-related patients. In our study, we found that POU6F2-AS2 was highly expressed in LUSC tissue, and LUSC patients with high expression possessed low risk scores. Similar to POU6F2-AS2, CACNA2D3-AS1 is an antisense lncRNA, and no studies so far has reported any association of CACNA2D3-AS1 with cancer. In our study, we noticed that CACNA2D3-AS1 in ceRNA network was up-regulated and competed with the down-regulated has-mir-140. Some previous studies showed that hsa-mir-140 often functioned as a tumor suppressor in cancer (46-48). Down-regulated hsa-mir-140 has been suggested to enhance cell proliferation and invasion in colorectal carcinoma and squamous cell LC (46,47), and low expression of hsa-mir-140 is associated with poor prognosis of spinal chordoma (49). This study firstly showed aberrant expression of CACNA2D3-AS1 in LUSC, and indicated that CACNA2D3-AS1 might sever as a potentially prognostic marker in LUSC. In addition, we observed that TTTY16 in ceRNA network was down-regulated in LUSC, and TTTY16 with low expression competed with up-regulated hsa-mir-183. TTTY16 has so far been reported to be associated with cancer (5), and a 14-gene expression signature including TTTY16 was related to the prognoses of LUSC patients (5). Previous studies also showed that hsa-mir-183 was highly expressed in cancer, and promoted tumor cell growth and migration (50,51), which indicated that the high expression of hsa-mir-183 was related to the poor prognosis. So far, no studies about TTTY16 regulating cancer initiation and progression by competing with has-mir-183 was reported. In this study, we are the first to report the role of TTTY16 by competing with hsa-mir-183 in LUSC, and low expression of TTTY16 was associated with good prognosis in LUSC. This study findings indicate that CACNA2D3-AS1 and POU6F2-AS2 may play roles as LUSC suppressors, and TTTY16 may function as LUSC promoter.
Despite the findings in clinical implications, some limitations must be noted. Firstly, the findings are obtained based on the TCGA database by pure bioinformatics methods. Although some findings have been validated by GEPIA database, the findings must be verified by experimental methods. Secondly, the 3 prognosticators of 3-lncRNA signature need be evaluated by a median follow-up duration (3 to 5 years).
Conclusions
Taken together, three lncRNAs were identified as a potentially prognostic lncRNA signature for LUSC patients. The findings provide novel insights into the lncRNA-related regulatory mechanism by ceRNA network in LUSC. Further, more functional studies are required to elucidate the lncRNA-related molecular mechanisms underlying LUSC.
Acknowledgments
Funding: This work was supported by National Natural Science Foundation of China (no 31660655), Yunnan Province Applied Basic Research Projects (nos 2016FB146, 2017FB042, 2018FE001) and Fund from Health and Family Planning Commission of Yunnan Province (no 2017NS261).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Blandin Knight S, Crosbie PA, Balata H, et al. Progress and prospects of early detection in lung cancer. Open Biol 2017;7. [Crossref] [PubMed]
- Ling DJ, Chen ZS, Liao QD, et al. Differential effects of MTSS1 on invasion and proliferation in subtypes of non-small cell lung cancer cells. Exp Ther Med 2016;12:1225-31. [Crossref] [PubMed]
- Gandara DR, Hammerman PS, Sos ML, et al. Squamous cell lung cancer: from tumor genomics to cancer therapeutics. Clin Cancer Res 2015;21:2236-43. [Crossref] [PubMed]
- Li J, Wang J, Chen Y, et al. A prognostic 4-gene expression signature for squamous cell lung carcinoma. J Cell Physiol 2017;232:3702-13. [Crossref] [PubMed]
- Tanoue LT, Detterbeck FC. New TNM classification for non-small-cell lung cancer. Expert Rev Anticancer Ther 2009;9:413-23. [Crossref] [PubMed]
- Nesbitt JC, Putnam JB Jr, Walsh GL, et al. Survival in early-stage non-small cell lung cancer. Ann Thorac Surg 1995;60:466-72. [Crossref] [PubMed]
- Campbell JD, Alexandrov A, Kim J, et al. Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. Nat Genet 2016;48:607-16. [Crossref] [PubMed]
- Ning P, Wu Z, Hu A, et al. Integrated genomic analyses of lung squamous cell carcinoma for identification of a possible competitive endogenous RNA network by means of TCGA datasets. PeerJ 2018;6:e4254. [Crossref] [PubMed]
- Cancer Genome Atlas Research N. Comprehensive genomic characterization of squamous cell lung cancers. Nature 2012;489:519-25. [Crossref] [PubMed]
- Cancer Genome Atlas Research N. Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014;511:543-50. [Crossref] [PubMed]
- Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet 2009;10:155-9. [Crossref] [PubMed]
- Fang Y, Fullwood MJ. Roles, Functions, and Mechanisms of Long Non-coding RNAs in Cancer. Genomics Proteomics Bioinformatics 2016;14:42-54. [Crossref] [PubMed]
- Arun G, Diermeier SD, Spector DL. Therapeutic Targeting of Long Non-Coding RNAs in Cancer. Trends Mol Med 2018;24:257-77. [Crossref] [PubMed]
- Chen L, Dzakah EE, Shan G. Targetable long non-coding RNAs in cancer treatments. Cancer Lett 2018;418:119-24. [Crossref] [PubMed]
- Wang L, Yu Z, Sun S, et al. Long non-coding RNAs: potential molecular biomarkers for gliomas diagnosis and prognosis. Rev Neurosci 2017;28:375-80. [Crossref] [PubMed]
- Deng H, Wang JM, Li M, et al. Long non-coding RNAs: New biomarkers for prognosis and diagnosis of colon cancer. Tumour Biol 2017;39:1010428317706332. [Crossref] [PubMed]
- Salmena L, Poliseno L, Tay Y, et al. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 2011;146:353-8. [Crossref] [PubMed]
- Conte F, Fiscon G, Chiara M, et al. Role of the long non-coding RNA PVT1 in the dysregulation of the ceRNA-ceRNA network in human breast cancer. PLoS One 2017;12:e0171661. [Crossref] [PubMed]
- Fan CN, Ma L, Liu N. Systematic analysis of lncRNA-miRNA-mRNA competing endogenous RNA network identifies four-lncRNA signature as a prognostic biomarker for breast cancer. J Transl Med 2018;16:264. [Crossref] [PubMed]
- Liu H, Zhang Z, Wu N, et al. Integrative Analysis of Dysregulated lncRNA-Associated ceRNA Network Reveals Functional lncRNAs in Gastric Cancer. Genes (Basel) 2018;9:E303. [Crossref] [PubMed]
- Lu M, Xu X, Xi B, et al. Molecular Network-Based Identification of Competing Endogenous RNAs in Thyroid Carcinoma. Genes (Basel) 2018;9:E44. [Crossref] [PubMed]
- Cao Y, Wang P, Ning S, et al. Identification of prognostic biomarkers in glioblastoma using a long non-coding RNA-mediated, competitive endogenous RNA network. Oncotarget 2016;7:41737-47. [Crossref] [PubMed]
- Du Z, Sun T, Hacisuleyman E, et al. Integrative analyses reveal a long noncoding RNA-mediated sponge regulatory network in prostate cancer. Nat Commun 2016;7:10982. [Crossref] [PubMed]
- Zhou M, Wang X, Shi H, et al. Characterization of long non-coding RNA-associated ceRNA network to reveal potential prognostic lncRNA biomarkers in human ovarian cancer. Oncotarget 2016;7:12598-611. [PubMed]
- Sui J, Xu SY, Han J, et al. Integrated analysis of competing endogenous RNA network revealing lncRNAs as potential prognostic biomarkers in human lung squamous cell carcinoma. Oncotarget 2017;8:65997-6018. [Crossref] [PubMed]
- Tang RX, Chen WJ, He RQ, et al. Identification of a RNA-Seq based prognostic signature with five lncRNAs for lung squamous cell carcinoma. Oncotarget 2017;8:50761-73. [PubMed]
- Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 2008;9:559. [Crossref] [PubMed]
- Ding M, Li F, Wang B, et al. A comprehensive analysis of WGCNA and serum metabolomics manifests the lung cancer-associated disordered glucose metabolism. J Cell Biochem 2019;120:10855-63. [Crossref] [PubMed]
- Di Y, Chen D, Yu W, et al. Bladder cancer stage-associated hub genes revealed by WGCNA co-expression network analysis. Hereditas 2019;156:7. [Crossref] [PubMed]
- Zhai X, Xue Q, Liu Q, et al. Colon cancer recurrenceassociated genes revealed by WGCNA coexpression network analysis. Mol Med Rep 2017;16:6499-505. [Crossref] [PubMed]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010;26:139-40. [Crossref] [PubMed]
- Szklarczyk D, Franceschini A, Wyder S, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 2015;43:D447-52. [Crossref] [PubMed]
- Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498-504. [Crossref] [PubMed]
- Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics 2003;4:2. [Crossref] [PubMed]
- Yu G, Wang LG, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 2012;16:284-7. [Crossref] [PubMed]
- Tang Y, Li M, Wang J, et al. CytoNCA: a cytoscape plugin for centrality analysis and evaluation of protein interaction networks. Biosystems 2015;127:67-72. [Crossref] [PubMed]
- Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 2017;45:W98-W102. [Crossref] [PubMed]
- Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017;19:649-58. [Crossref] [PubMed]
- Li L, Chen K, Xiang Y, et al. New development in studies of formyl-peptide receptors: critical roles in host defense. J Leukoc Biol 2016;99:425-35. [Crossref] [PubMed]
- Hou XL, Ji CD, Tang J, et al. FPR2 promotes invasion and metastasis of gastric cancer cells and predicts the prognosis of patients. Sci Rep 2017;7:3153. [Crossref] [PubMed]
- Xiang Y, Yao X, Chen K, et al. The G-protein coupled chemoattractant receptor FPR2 promotes malignant phenotype of human colon cancer cells. Am J Cancer Res 2016;6:2599-610. [PubMed]
- Hossain MN, Sakemura R, Fujii M, et al. G-protein gamma subunit GNG11 strongly regulates cellular senescence. Biochem Biophys Res Commun 2006;351:645-50. [Crossref] [PubMed]
- Takauji Y, Kudo I, En A, et al. GNG11 (G-protein subunit gamma 11) suppresses cell growth with induction of reactive oxygen species and abnormal nuclear morphology in human SUSM-1 cells. Biochem Cell Biol 2017;95:517-23. [Crossref] [PubMed]
- Liu J, Sun X, Zhu H, et al. Long noncoding RNA POU6F2-AS2 is associated with oesophageal squamous cell carcinoma. J Biochem 2016;160:195-204. [Crossref] [PubMed]
- Huang H, Wang Y, Li Q, et al. miR-140-3p functions as a tumor suppressor in squamous cell lung cancer by regulating BRD9. Cancer Lett 2019;446:81-9. [Crossref] [PubMed]
- Zhao Z, Liu W, Li J. miR-140-5p inhibits cell proliferation and invasion in colorectal carcinoma by targeting SOX4. Oncol Lett 2019;17:2215-20. [PubMed]
- Zhou Y, Wang B, Wang Y, et al. miR-140-3p inhibits breast cancer proliferation and migration by directly regulating the expression of tripartite motif 28. Oncol Lett 2019;17:3835-41. [PubMed]
- Zou MX, Huang W, Wang XB, et al. Identification of miR-140-3p as a marker associated with poor prognosis in spinal chordoma. Int J Clin Exp Pathol 2014;7:4877-85. [PubMed]
- Bi DP, Yin CH, Zhang XY, et al. MiR-183 functions as an oncogene by targeting ABCA1 in colon cancer. Oncol Rep 2016;35:2873-9. [Crossref] [PubMed]
- Zhang L, Quan H, Wang S, et al. MiR-183 promotes growth of non-small cell lung cancer cells through FoxO1 inhibition. Tumour Biol 2015;36:8121-6. [Crossref] [PubMed]


