
Role of liquid biopsy in oncogene-addicted non-small cell lung cancer
Introduction
The identification of molecular aberrations in key elements of signal transduction pathways involved in tumor growth and survival, so called oncogene-addicted tumors, has dramatically changed the treatment approach of non-small cell lung cancer (NSCLC). Mutations in the epidermal growth factor receptor (EGFR) and translocations involving the anaplastic lymphoma kinase (ALK) gene are the most frequent and clinically important targets. For patients carrying these alterations, a specific class of drugs, tyrosine kinase inhibitors (TKIs), has become the standard of treatment. EGFR TKIs gefitinib, erlotinib, afatinib and osimertinib, and the ALK inhibitors crizotinib, ceritinib, and alectinib have been approved in Europe and the United States for the treatment of advanced NSCLC (1). The majority of adenocarcinomas harbor at least one driver mutation that can potentially be a target for therapy and this has opened up for many new targeted therapies being studied in clinical trials (2). The clinical benefit of these targeted therapies is very high, with high response rates and improved quality-of-life for the patients. Objective response rates (ORR) of 60–70% and a disease control rate (DCR) of up to 80–90% are reported with these different TKIs (3). Other examples of oncogenic drivers for NSCLC include alterations in BRAF, HER2, MET, PIK3CA, ROS1, RET, AKT, DDR2, and KRAS (4). Most recently, targeted therapies for ROS1 and BRAF have been approved for the treatment of advanced NSCLC (5,6). Furthermore, new generations of EGFR and ALK TKIs have been developed, some of them also being active in treatment-refractory tumors where acquired resistance to first-generation TKIs has developed (7,8). The development in the field is rapid and an increasing number of targeted therapies can be expected to be approved for the treatment of advanced NSCLC in the coming years.
The diagnosis of metastatic NSCLC is usually made on small biopsies or on cytology specimens, and the scarcity of available tumor material is very frequent. Sometimes the available biological sample derives from a bone biopsy, with a consequent bad quality of the material that leads to unsuccessful molecular analysis. In addition, tumor could be heterogeneous and a biopsy done in a specific site could not necessarily represent the overall tumor molecular landscape. Liquid biopsy represents an optimal strategy to overcome these matters. Moreover, liquid biopsy can be used during patient treatment to monitor response and the onset of resistance mechanisms. Since tumor re-biopsy is an invasive procedure, and it is not feasible for all patients, the only chance is to characterize tumor alterations by liquid biopsy. Molecular analysis on liquid biopsy is usually quicker compared to that performed on tumor tissue biopsy, which requires procedures and evaluations performed by the pathologist, with a consequent shortening of the turnaround time.
In this review we summarized the utility of liquid biopsy in oncogene-addicted NSCLC, focusing the attention on the study of the different blood biomarkers, underlying pro and contra of each approach for molecular analysis.
Oncogene-addicted NSCLC: actionable oncogene alterations
EGFR mutation
A new era in NSCLC treatment has started in 2004, when mutations of the EGFR gene were discovered to be predictable of sensitivity to therapy with EGFR-TKIs (9,10). Initially, the IDEAL-1 and IDEAL-2 trials for gefitinib (11,12), and the TALENT and TRIBUTE for erlotinib (13,14), gave disappointing results, and then both drugs had a history of contrasting results until the success of studies where EGFR-TKIs were compared to chemotherapy in patients with activating EGFR mutations (7,15,16): both drugs prolong progression-free survival (PFS) but not overall survival (OS) compared with platinum-doublet chemotherapy (median PFS of 9.2–13.1 vs. 4.6–6.3 months, respectively).
Second-generation EGFR-TKI, afatinib, which inhibits irreversibly EGFR and other ErbB family receptor members, then demonstrated PFS benefit with respect to chemotherapy in 2 phase III trials, LUX-Lung 3 and LUX-Lung 6 (17,18). This drug demonstrated also significant improvement in terms of OS, in patients with tumor positive for EGFR exon 19 deletion (19).
Subsequently, third-generation EGFR-TKI osimertinib, already approved in the second line setting in patients at progression with first or second-generation TKIs who have developed the EGFR-T790M mutation (20), demonstrated a benefit in terms of PFS with respect to gefitinib or erlotinib, in previously untreated EGFR-mutated patients and was then approved in the first-line setting (21).
However, despite a clinical benefit is indubitably observed with the use of these drugs, resistance mechanisms appeared after about 12 months of treatment in almost all cases (7,15-18). The most common resistance mechanism to first and second generation TKIs is the gatekeeper T790M mutation occurring at exon 20 of EGFR. It is observed in about 50–70% of cases and patients developing this alteration become sensitive to osimertinib. For this reason, this drug was approved in the second line therapy of this setting of patients (7). This led to the necessity of tumor characterization after progression to first or second TKI treatment for T790M, which requires the availability of sufficient tumor tissue available for molecular analysis. As tumor re-biopsy is not always feasible as regards tumor localization and patient’s general health status, liquid biopsy has acquired more relevance for the detection of resistance mutations.
In contrast to first and second-generation TKIs, few information about acquired resistant mechanisms to osimertinib in the first-line setting has been known until now. In vitro studies and some case reports showed that secondary EGFR mutations (22,23), activation of AXL (24) and ERK (25) signaling, and small cell transformation can emerge as possible acquired resistance mechanisms in this setting. More data are available with regard to resistance mechanisms to osimertinib in the second line setting in T790M positive patients, for whom C797S mutation is observed in about 20–40% of cases, followed by the c-Met proto-oncogene (MET) amplification (14%), transformation into small-cell phenotype, amplification of the Erb-B2 Receptor Tyrosine Kinase 2 (HER2) or the Fibroblast Growth Factor Receptor 1 (FGFR1) genes, and mutations in the B-Raf Proto-Oncogene, Serine/Threonine Kinase (BRAF) (26,27).
ALK-translocation
In 2007, aberrant fusion of echinoderm microtubule associated protein-like 4 (EML4) anaplastic lymphoma kinase (ALK) was documented in NSCLC (28). Genetic rearrangement of ALK is detected in about 3–7% of NSCLC, and patients carrying such alteration are usually young, non-smokers and with adenocarcinoma (ADC) histology (29). The first drug developed against this alteration was crizotinib, an oral ATP-competitive selective inhibitor of the ALK, MET, and ROS1 tyrosine kinase. Two phase III trials, PROFILE 1007 (30) and PROFILE 1014 (31) showed a significant advantage of crizotinib with respect to chemotherapy in second- and first-line settings, respectively. Based on these results, the drug was approved for treatment of patients with ALK-rearranged advanced tumors, and became the first choice of treatment in untreated NSCLC patients carrying this alteration. However, the majority of patients undergo progression after about 1 year of treatment, with the development of both ALK-dependent and ALK-independent resistance mechanisms. The most common ALK secondary mutations observed are L1196M and G1269A (32-34), both interfering with crizotinib binding. Other mutant variants that reduce the ATP binding affinity of crizotinib include S1206Y, V1180L, and G1202R, while other secondary mutations, such as C1156Y, L1171T, L1152R, and L1198P, promote ATP binding and stabilize the ALK active conformation (35,36) (Figure 1). These mechanisms of resistance occur more frequently following treatment with second-generation ALK inhibitors, like ceritinib and alectinib, due to the greater potency and specificity of these agents (37). Moreover, these mutations are associated with resistance to both first- and second-generation ALK inhibitors (38), although brigatinib, another second-generation ALK inhibitor, is most active against secondary resistance mutation, with the exception of the G1202R (39). Lorlatinib, a third-generation ALK inhibitor, was then developed to overcome all ALK secondary resistance mutations, including G1202R, and there is potential indication for its use after failure to other ALK inhibitors (40). The identification of specific ALK mutations at progression during crizotinib or other TKIs is important to give the right drug to the right patient in the right time. As a consequence, liquid biopsy becomes essential to monitor the development of these mutations.
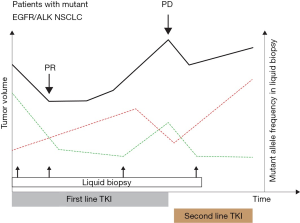
ROS1-rearrangement
Chromosomal rearrangements of the ROS1 gene were first described in NSCLC in 2007 (41). This alteration is present in about 1–2% of NSCLC patients, mainly in younger, never or light smokers and adenocarcinoma histology (42). ROS1-rearranged lung tumors are “addicted” to ROS1 for growth and survival (41). This finding implies a pharmacological sensitivity of the tumor to ROS1-directed TKIs (5,42). After preclinical evidences of crizotinib activity in ROS1 altered models, the phase I PROFILE 1001 study of crizotinib in ALK translocated patients was amended to include ROS1-rearranged patients, and it demonstrated an ORR and DCR to crizotinib of 72% and 90%, respectively (5). After these results, crizotinib was granted full approval by FDA in March 2016 for treatment of advanced ROS1-rearranged NSCLC.
As for EGFR-mutant and ALK-translocated patients, also for ROS1-rearranged tumors resistance mechanisms to crizotinib typically develop. These mechanisms are represented by secondary mutations in the ROS1 kinase domain (50–60% of cases), or by “off target” alterations in parallel pathways (43,44).
The most frequent ROS1 resistance mutation is the G2032R mutation, causing steric hindrance to drug binding, while not altering the oncogenic kinase activity (45). Other reported resistance mutations are the D2033N, S1986Y/F, L2026M and L1951R. Of all mutations, G2032R and L1951R confer the highest level of crizotinib-resistant phenotype in vitro (29). Less knowledge is available about “off target” mechanisms. Alterations of the KIT proto-oncogene receptor tyrosine kinase (KIT), KRAS proto-oncogene, GTPase (KRAS), EGFR, and BRAF genes have been demonstrated (46-48). Phenotypic changes, such as epithelial-to-mesenchymal transition (EMT), have also been demonstrated (44). Several other agents have shown activity in patients with ROS1-rearranged tumor, as well as in patients developing ROS1 resistance mutations. In particular, ceritinib and brigatinib showed comparable activity against resistance mutations, both showing activity against L2026M but not against G2032R, D2033N or L1951R (43). Lorlatinib showed a broader activity against different resistant mutations (48), however its activity against G2032R was limited in the clinic. Cabozantinib is one of the few agents showing activity against G2032R mutation, but its high toxicity limits its use in these patients (49).
Biomarkers from liquid biopsy
Cell-free DNA
The first evidence of the presence of cell-free DNA (cfDNA) in the body fluids was obtained in 1989 (50). Since then, a growing number of studies have demonstrated the usefulness of cfDNA analysis at all stages of cancer patient’s management, specifically in the detection of drug-resistance mutations to guide clinical decisions (51). The improvements of technologies based on next-generation sequencing (NGS) have enabled the use of cfDNA for the noninvasive interrogation of the tumor genome on the presence of multiple cancer-specific mutations with a single assay, along with chromosomal aberrations and gene rearrangements, particularly with the prospect to monitor, through serial sampling, the development of cancer clones in the course of therapy (52). Consequently, NGS-based analysis has become the gold standard for the analysis of alterations in common oncogenic drivers and to generate a picture of the overall mutational burden and tumor heterogeneity in NSCLC patients (53).
In physiological conditions, a large amount of cfDNA that has originated from different cell types, mostly through apoptosis and necrosis, can be found in the bloodstream (54). Sequencing data have identified a peak of fragment size around 166 bp, indicating that this cfDNA is normally wrapped around a nucleosome (55), and the epigenetic nucleosome footprint, along with the genetic modifications of the cfDNA molecule, reflects the genome of the cells of origin (56,57). However, longer cfDNA fragments, which normally escape the current extraction methods, can be found by the use of long-read sequencing techniques, and they associate mostly with extracellular vesicles (EVs), contributing to a consistent part of the cfDNA pool (58). Typically, half-life of cfDNA is of only a few hours (59), but its stability and permanence in the circulation can be increased through the inclusion inside EVs, which in turn may work as a vehicle to mediate horizontal transfer of functional DNA fragments and oncogenes (60,61).
A massive amount of circulating tumor DNA (ctDNA) can be released by the tumor mass, and its concentration correlates with tumor size and stage (55,62). Quantification of ctDNA in plasma can be used to assess tumor dynamics in patients undergoing treatment, reflecting responses and progression (63). Detection via blood tests of clinically relevant mutations in ctDNA may help predict the insurgence of cancer and define the anatomical site of the primary tumor of origin, facilitating the diagnosis of cancer at early stages and before the metastatic spread (64). At the moment, the most important use of ctDNA tests in clinical practice of NSCLC is related to the detection of the EGFR T790M resistance mutation and the monitoring of EGFR-sensitive mutations during the course of TKI treatment (65,66). However, despite an overall good specificity in the detection of ctDNA mutations (90–100%), sensitivity levels reached with the current methodologies are typically low (60–70%) (67-69). A reason for this suboptimal sensitivity in detecting cancer mutations in cfDNA lays on the fact that ctDNA is markedly diluted in the circulation by the higher abundance of background cfDNA released by normal cells (70). Further methods of analysis should integrate mutational information from ctDNA with data derived from the evaluation of the other blood biomarkers, which can improve sensitivity.
Other blood biomarkers
Although the analysis of ctDNA remains the gold standard for blood-based mutational assays, by providing clear advantages in term of simplicity and sensitivity, the need for specific biomarkers of immediate clinical utility leads the study of other tumor-derived components (71). Circulating tumor cells (CTCs), EVs, and cell-free microRNAs (cfmiRNAs) are constantly released from the tumor mass into the stream, but their use in liquid biopsy still lacks strong clinical validation to ensure standardization of the readout, while there is sufficient evidence that the time is mature to introduce the analysis of platelet RNA as a reliable diagnostic biomarker.
CTCs are extremely rare, usually one cell per milliliter of blood, in millions of background leukocytes (72). However, CTCs represent a most promising alternative to invasive biopsies for the comprehensive characterization of the tumor tissue at a single-cell level (73). CTC-based analysis of sequencing data can produce a complete overview of tumor alteration profile, complementing gene expression data with mutational status and genomic instability, which would not be achieved by cfDNA analysis alone (74). The ultimate progresses in the isolation of CTCs have enabled to draw a precise genetic signature of drug-resistance clones, with direct clinical relevance, being CTCs ultimately responsible for metastasization (75).
The functional relevance of EV release into the circulation is now well accepted, with their major role in cell-to-cell communication, mostly via delivering of active miRNAs, messenger RNAs (mRNAs), and proteins into the target cell (76,77). In parallel, the technical improvements in EV purification (78) have set the promise to establish the analysis of EV as a novel and reliable methodology for tumor diagnosis and progression (79). Several evidences indicate that EVs are directly involved in cancer resistance to therapy, and the information collected from tumor EVs could be used for early diagnosis of the metastatic propensity of a primary tumor (80). In fact, tumor-derived EVs can promote organ-specific metastasization of the cancer cell from which they are originated (81), by preparing the pre-metastatic niche through the engagement of the normal cell signaling in the surrounding microenvironment (82,83). In NSCLC EVs could serve as a robust predictive biomarker of tumor evolution during TKI treatment (84).
Traditionally, miRNAs, by accounting for over the 50% of all RNA species released into the circulation, are considered a direct readout of the altered tumor’s transcriptional program. Most of the cfmiRNAs found into the blood plasma are involved in cell-to-cell communication, and are delivered to target cells by selective loading within the EVs, although functional miRNAs can also be released directly into the bloodstream by dying cells (85). These cfmiRNAs have shown their diagnostic and prognostic relevance, in NSCLC, as in many other solid tumors (86).
Analysis of platelet RNA content has emerged as a highly sensitive and alternative method for blood-based diagnosis of cancer (87). Platelets are anucleate blood cells that can capture external RNAs released by other cells, via both EV-dependent or -independent mechanism (88). Normally, cancer cells can prepare their niche by secreting EVs able to transfer oncogenic RNAs into circulating platelets, which in turn release these RNAs at distant sites (89). The expression profile of these so-called tumor-educated platelets changes in response to contact with tumor EVs, reflecting the molecular signature of the cancer cell of origin, with important diagnostic consequences.
Methods for liquid biopsy analysis
In the routine clinical diagnostics, molecular analyses on liquid biopsy are mostly focused on the detection of targetable mutations of ctDNA of patients. To date, ctDNA is the unique circulating biomarker approved by Food and Drug Administration (FDA) for diagnosis of such mutations, while there are evidences that other circulating biomarkers could represent a diagnostic tool for primary diagnosis of lung cancer (90). Molecular testing on CTCs, EVs, and platelets remains challenging, and it seems they will enter the clinical practice in the near future.
In the last years, several works have focused the attention on which are the best practices to test liquid biopsy, from blood collection and handling to analysis methodologies (53,66).
Blood collection and handling
A key point for molecular analysis on liquid biopsy is the processing of the blood sample. Blood tubes containing ethylenediaminetratra-acetic acid (EDTA) are extensively used for the isolation of plasmatic ctDNA, mainly for the low cost and availability. On the other hand, blood collected in EDTA tubes is at a higher risk of cellular degradation, with the subsequent degradation of ctDNA, and contamination by genomic DNA released form blood cells (91,92). Hence, rapid isolation of the plasmatic components is necessary, up to the 6 hours from the blood collection (93). Time at blood processing could represent a logistic issue, especially if plasma processing is not possible immediately after blood withdrawal, as happens in large prospective clinical trials with centralized analyses. To solve this issue, the use of preservative tubes is an effective option to prevent ctDNA contamination by genomic DNA. Properly used, collection tubes such as Streck Cell-Free DNA BCT (La Vista, Nevada), cfDNA collection tubes (Roche Diagnostics, Germany), PAXgene tubes (Qiagen, Germany) are able to stabilize blood cells, preventing the release of genomic DNA, and to for at least one week at room temperature (92,94-96). The gold standard for centrifugation is to perform this step twice, with the second centrifugation that aims to eliminate debris. As reported, the two spins are at 800 and 2,000–16,000 g, respectively, for 10–20 minutes, while performing a third spin is still debated (97).
Extraction methods
The methodologies for ctDNA isolation represent a delicate step to consider when performing liquid biopsy analyses. To date, there is a need to standardize such methodologies between the different laboratories, as highlighted by a comparison study that demonstrated high variability between 56 laboratories, with DNA yields ranging from 2.87 to 224.02 ng/mL (98). Most of commercially available extraction kits are based on the binding of DNA molecules by silica-gel filter columns, magnetic beads, or phase isolation. In the last years, several automated extraction platforms have been developed (Promega Maxwell RSC, Promega; QIAsymphony ccfDNA, Qiagen; MagMax, ThermoFisher Scientific, MagNA Pure Compact, Roche Diagnostics), and comparative studies on manual and extraction methods have reported contrasting results in DNA yields (91,99-101). One thing worthy of attention is that extraction kits are selective for the fragment size of the eluted DNA, with the QIAamp blood mini kit and QIAamp Circulating Nucleic acid kit (Qiagen) enriching for DNA >200 bp and 166–200 bp, respectively. Depending on the chosen analysis methodology for molecular test, this could be relevant, because quantitative PCR (qPCR) primers preferentially amplify long DNA fragments (102). Another interesting study attempted to directly measure ctDNA through qPCR without previous extraction methods, reaching good DNA yields and demonstrating that a notable percentage of DNA is loss in the flow-throughs of column-based extraction kits (103). Given the high sensitivity of qPCR platforms, this could be a feasible and time-saving approach, even though extraction methods remain the gold standard for liquid biopsy molecular analyses.
Methodologies for molecular analysis
Even though ctDNA is present at higher concentrations in cancer patients with respect to healthy subjects (104), it represents less than 1% of total cfDNA (70), presenting technical challenges for sensitivity and specificity. To date, several approaches have been developed to discriminate ctDNA from cfDNA, and to diagnose clinically relevant mutations, with relative advantages and limitations (Table 1). Here we summarize the most common platforms for ctDNA testing.
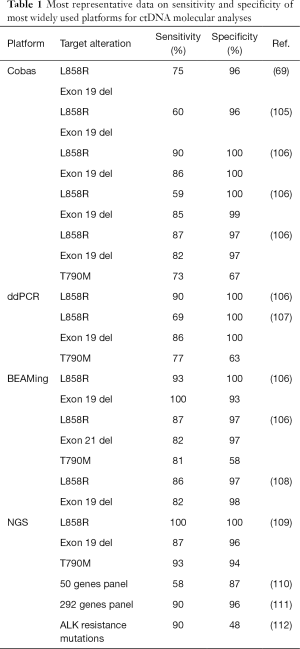
Full table
PCR-based technologies
PCR-based assays are focused on the detection of one or few mutations, but are able to be extremely sensitive. Platforms as Therascreen (Qiagen) and Cobas (Roche Diagnostics) are approved by FDA to test EGFR mutations (exon 19 deletion, L858R and T790M) on plasma samples of NSCLC patients when tissue analysis is not available. On the other hand, such platforms are validated only for allelic frequencies at >1%, but usually mutated alleles are found at lower frequencies (97). Nonetheless, Cobas platform have been used in a clinical study for the detection of T790M mutation, reaching the threshold of 0.02% of the allele fraction (106).
By qPCR is possible to detect allele frequencies at 10–20%, with high specificity. To enhance sensitivity, a number of technical modifications have been developed, such as the use of an oligo that blocks the 3'-end of the wild-type allele to avoid its amplification (i.e., Allele-Specific PCR, AS-PCR; Allele-Specific Non-Extendable Primer Blocker PCR, AS-NEPB-PCR; Peptide Nucleic Acid-Locked Nucleic Acid (PNA-LNA) PCR clamp) or modification of the PCR with the enrichment of allelic variants of normal and mutated DNA (co-amplification at lower denaturation temperature, COLD-PCR) (105,113).
Droplet digital PCR (ddPCR) is a robust technique to detect mutations at low frequencies, based on the generation of thousands to millions of droplets on a microfluidic chip, individually amplified, with high increase in sensitivity and specificity. In a study on early breast cancer patients, this technique reached 93.3% sensitivity and 100% specificity on ctDNA, compared to tumor tissue, for the detection of mutation of PIK3CA (114). For activating mutations in NSCLC patients, ddPCR demonstrated 100% specificity in detecting L858R mutation, and 79% specificity in detecting T790M mutation on ctDNA (107). Moreover, sensitivity could be further enhanced through suppressing the amplification of the wild-type allele fraction, reaching a threshold limit of 0.01% (115).
BEAMing (Beads, Emulsion, Amplification, Magnetics) is a modified ddPCR that uses oil emulsion to generate water droplets, analyzed with fluorescent terminators in flow cytometry to detect mutant alleles. BEAMing has reached high rates of sensitivity and specificity for EGFR activating mutations (106), even though producing some discordance with tissue testing, possibly for tumor heterogeneity (108).
NGS
NGS is a high throughput technology and it is becoming the routine methodology for tumor tissue profiling, especially for its large applicability to different pathologies. Several massively parallel sequencing platforms have been developed, that can be classified on the basis of the sequencing method used: pyrosequencing (Roche 454), sequencing-by-synthesis (Illumina), ion semiconductor-based (Ion-torrent) (66). NGS is able to interrogate a high quantity of DNA bases in relatively short turnaround times, allowing to perform analyses on the entire genome (Whole Genome Sequencing, WGS), coding sequences (Whole Exome Sequencing, WES), or targeted gene panels; an additional characteristic is that NGS is capable to simultaneously detect mutations, insertions, deletions, copy number variations and genomic rearrangements. Nonetheless, high sensitivity and specificity are accompanied by random errors at rates ranging from 0.1% to 1% (116), but recent improvements have demonstrated that sensitivity and specificity could not represent a technical issue in the future (109,117,118). On the other hand, sensitivity is influenced by the choice to perform WGS, WES or targeted panels, given that enlarging the number of target regions will inevitably reduce the coverage, and WGS and WES would not detect allele frequencies less than 5%, presenting a clinical issue in identifying early resistance mutations at low frequencies in the ctDNA (97). On the other hand, targeted panels ensure consistently deeper coverage (>10,000×) (119,120). Oncomine panel for Ion torrent (ThermoFischer Scientific) has reached 77% sensitivity in detecting clinically relevant mutations in ctDNA of NSCLC patients (121), while the validation of a multiplex PCR-NGS panel on ctDNA of early stage NSCLC reached >99% sensitivity and 99.6% specificity, with the promising to long-term monitoring of disease (122). Generally, WGS and WES track a molecular profile of the tumor, while targeted panels are more useful to identify sensitizing mutations. The nature of these platforms generates random error rates that become more consistent when testing ctDNA, where sensitizing mutations are often present at low frequencies. To address this issue, methodologies using molecular barcoding and “selectors” have been implemented, such as the construction of algorithms, with the final result to dramatically improve sensitivity (52,97,111,113). One advantage of NGS platforms is the capability to detect a large number of somatic mutations, also with unknown function and in tumor suppressor genes, adding simultaneous information from the same sample. Moreover, the possibility to analyze gene copy variations and chromosomal rearrangements is an added value for these platforms. This is very useful to detect and monitor clinically relevant rearrangements, as NGS demonstrated high concordance rates with IHC for ALK fusions diagnosis (123,124). NGS is the ideal methodology to scan ctDNA for the presence of multiple resistance mutations of ALK that could develop after TKI treatment (112).
On the other hand, NGS present limitations that should be highlighted: NGS is more expensive than other methodologies able to detect mutations in ctDNA, and usually larger amounts of input material are needed for ctDNA molecular testing, especially for the degradation of the specimen. Moreover, NGS analyses require a bioinformatic additional analyses, and the lack of standardized mathematical model has generated the existence of several algorithms with the same aim.
Application of liquid biopsy in the clinical practice
Baseline identification of oncogenic drivers
Liquid biopsy can provide an advantage for treatment-naive patients because the diagnostic tissue can be spared for immunohistochemistry necessary for immunotherapy. However, not always a tumor shed sufficient amount of DNA for detection in peripheral blood. In advanced stage disease approximately 85% sensitivity is the highest achieved. Slow-growing tumors before treatment are associated with false-negative results in plasma more frequently than more disseminated tumors. Variant allele frequency (VAF) in peripheral blood can drastically change as a consequence of therapy. For this reason, blood samples for cfDNA analysis should be drawn before any therapy, because one or two weeks of treatment are sufficient to make undetectable a previously plasma-positive patient (117). To select treatment-naive patients for EGFR detection on ctDNA the same criteria of DNA detection from tumor tissue should be used: advanced or metastatic nonsquamous NSCLC or squamous NSCLC in never smokers and/or younger patients. Liquid biopsy is possible at diagnosis in all patients to be tested for EGFR mutations. This approach is recommended if tumor tissue is scarce or unavailable, or even when more than 2 weeks are necessary for detection in the tumor tissue and invasive procedures for biopsy may be risky or contraindicated. A particular condition is represented by bone biopsies, which can be sufficient for a histological diagnosis, but decalcification can damage tumor DNA limiting molecular testing. If the detection of an actionable mutation in ctDNA gives a positive result, this finding is sufficient for starting a targeted treatment. Conversely, a negative result should be considered inconclusive and a secondary test is needed (53).
Conversely, the search for ALK translocation in ctDNA from treatment-naive patients is not supported by prospective studies. Some retrospective studies showed the limitation of qPCR to achieve the detection of this alteration. Droplet digital PCR seems to be more effective, but this approach has not been validated yet. An acceptable sensitivity with optimal specificity is provided by NGS, but the available data are not specific for ALK rearrangement (110,125,126).
Monitoring of treatment efficacy
When resistance occurs during first- and second-generation EGFR-TKIs as first-line setting in NSCLC patients with EGFR actionable mutations, the detection of T790M must be prompt to allow starting the third-generation EGFR-TKI osimertinib (20). Perhaps this scenario will change with the use of osimertinib as first-line treatment on the basis of the results from the FLAURA trial (21). However, liquid biopsy might continue to be useful to understand the resistance mechanisms to osimertinib. Testing of EGFR alterations with an EGFR assay of sufficient sensitivity is recommended for patients progressing, either clinically or radiologically, during treatment with first- or second-generation EGFR TKIs. If on liquid biopsy T790M mutation is absent, this result should be considered inconclusive and a further assessment is recommended with a more sensitive and/or more comprehensive test on cfDNA or DNA from a tumor re-biopsy. An example of comprehensive analysis is NGS multiplex panel, which detects not only the common resistance mechanism T790M but it can detect a spectrum of alterations. Conversely, a positive result for EGFR T790M is considered adequate to start osimertinib as second-line treatment after progression on a first- or second-generation EGFR-TKI. If the T790M mutation is also absent in tissue biopsy, the NGS analysis may help finding other resistance mechanisms to address patients to a clinical trial or an expanded access program. If the primary EGFR mutation is found, a negative T790M mutation result is more credible. If both primary EGFR mutation and T790M mutation are negative, it is probable that the tumor is not shedding sufficient levels of DNA for its detection. In this case further investigation through liquid biopsy can be performed later (53).
The detection of ALK resistance mutations at disease progression during ALK TKIs can have a role in clinical management in the future. Currently, this approach cannot guide the choice of further line ALK TKI at progression. Some studies highlighted the ability of ddPCR and NGS in detecting ALK resistance mutations in cfDNA (127,128). However, NGS should be considered the optimal approach for the detection of these mutations because of the wide range of resistance alterations known to date. Moreover, the concordance of ALK mutations from tissue biopsy and liquid biopsy is 100% (129).
Similarly, cfDNA analysis could be applied to ROS1-positive patients treated with crizotinib, as also for these patients resistance mutations could emerge and drive subsequent targeted treatment (117,118,130). However, also in this case, no clinical indication is present to indicate the ROS1 test on cfDNA.
Conclusions
The identification of gene alterations in oncogene-addicted NSCLC is essential to draw a target treatment. Baseline EGFR mutations and ALK or ROS1 rearrangements are the only actually actionable with specific TKIs available for clinical practice. Likewise, resistance mechanisms have to be detected to change treatment at disease progression during first-line TKI. The main challenges for the detection of these alterations in tumor tissue are represented by the scarcity of tumor cells from tissue biopsy and the limitations of tissue re-biopsy in pretreated patients.
Liquid biopsy, intended as the search for oncogene alterations in blood samples, has been currently applied for EGFR-mutant NSCLC and it is under validation for ALK and ROS1 positive NSCLC. Various methods are available for both targeted detection of single alterations or broad-spectrum analyses in ctDNA. In the meantime, other circulating material (CTCs, EVs, cfmiRNAs) from tumor cells is under investigation. However, data and technology available until now make us suppose that the substitution of tissue biopsy with liquid biopsy is far from an actual clinical application. To date liquid biopsy holds just a complementary role in addition to tissue biopsy.
Acknowledgments
None.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
References
- Tsakonas G, Ekman S. Oncogene-addicted non-small cell lung cancer and immunotherapy. J Thorac Dis 2018;10:S1547-55. [Crossref] [PubMed]
- Imielinski M, Berger AH, Hammerman PS, et al. Mapping the Hallmarks of Lung Adenocarcinoma with Massively Parallel Sequencing. Cell 2012;150:1107-20. [Crossref] [PubMed]
- Holleman MS, van Tinteren H, Groen HJM, et al. First-line tyrosine kinase inhibitors in EGFR mutation-positive non-small-cell lung cancer: a network meta-analysis. Onco Targets Ther 2019;12:1413-21. [Crossref] [PubMed]
- Schulze AB, Evers G, Kerkhoff A, et al. Future Options of Molecular-Targeted Therapy in Small Cell Lung Cancer. Cancers 2019;11:690. [Crossref] [PubMed]
- Shaw AT, Ou SHI, Bang YJ, et al. Crizotinib in ROS1 -Rearranged Non-Small-Cell Lung Cancer. N Engl J Med 2014;371:1963-71. [Crossref] [PubMed]
- Baik CS, Myall NJ, Wakelee HA. Targeting BRAF ‐Mutant Non‐Small Cell Lung Cancer: From Molecular Profiling to Rationally Designed Therapy. Oncologist 2017;22:786-96. [Crossref] [PubMed]
- Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or Carboplatin-Paclitaxel in Pulmonary Adenocarcinoma. N Engl J Med 2009;361:947-57. [Crossref] [PubMed]
- Novello S, Mazières J, Oh IJ, et al. Alectinib versus chemotherapy in crizotinibpretreated anaplastic lymphoma kinase (ALK)-positive non-small-cell lung cancer: Results from the phase III ALUR study. Ann Oncol 2018;29:1409-16. [Crossref] [PubMed]
- Lynch TJ, Bell DW, Sordella R, et al. Activating Mutations in the Epidermal Growth Factor Receptor Underlying Responsiveness of Non-Small-Cell Lung Cancer to Gefitinib. N Engl J Med 2004;350:2129-39. [Crossref] [PubMed]
- Paez JG, Jänne PA, Lee JC, et al. EGFR Mutations in Lung Cancer: Correlation with Clinical Response to Gefitinib Therapy. Science 2004;304:1497-500. [Crossref] [PubMed]
- Fukuoka M, Yano S, Giaccone G, et al. Multi-Institutional Randomized Phase II Trial of Gefitinib for Previously Treated Patients With Advanced Non-Small-Cell Lung Cancer. J Clin Oncol 2003;21:2237-46. [Crossref] [PubMed]
- Kris MG, Natale RB, Herbst RS, et al. Efficacy of Gefitinib, an Inhibitor of the Epidermal Growth Factor Receptor Tyrosine Kinase, in Symptomatic Patients With Non-Small Cell Lung Cancer. JAMA 2003;290:2149-58. [Crossref] [PubMed]
- Herbst RS, Prager D, Hermann R, et al. TRIBUTE: A Phase III Trial of Erlotinib Hydrochloride (OSI-774) Combined With Carboplatin and Paclitaxel Chemotherapy in Advanced Non-Small-Cell Lung Cancer. J Clin Oncol 2005;23:5892-9. [Crossref] [PubMed]
- Gatzemeier U, Pluzanska A, Szczesna A, et al. Phase III Study of Erlotinib in Combination With Cisplatin and Gemcitabine in Advanced Non-Small-Cell Lung Cancer: The Tarceva Lung Cancer Investigation Trial. J Clin Oncol 2007;25:1545-52. [Crossref] [PubMed]
- Rosell R, Carcereny E, Gervais R, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol 2012;13:239-46. [Crossref] [PubMed]
- Maemondo M, Inoue A, Kobayashi K, et al. Gefitinib or Chemotherapy for Non-Small-Cell Lung Cancer with Mutated EGFR. N Engl J Med 2010;362:2380-8. [Crossref] [PubMed]
- Sequist LV, Yang JCH, Yamamoto N, et al. Phase III Study of Afatinib or Cisplatin Plus Pemetrexed in Patients With Metastatic Lung Adenocarcinoma With EGFR Mutations. J Clin Oncol 2013;31:3327-34. [Crossref] [PubMed]
- Wu YL, Zhou C, Hu CP, et al. Afatinib versus cisplatin plus gemcitabine for first-line treatment of Asian patients with advanced non-small-cell lung cancer harbouring EGFR mutations (LUX-Lung 6): an open-label, randomised phase 3 trial. Lancet Oncol 2014;15:213-22. [Crossref] [PubMed]
- Yang JCH, Sequist LV, Geater SL, et al. Clinical activity of afatinib in patients with advanced non-small-cell lung cancer harbouring uncommon EGFR mutations: a combined post-hoc analysis of LUX-Lung 2, LUX-Lung 3, and LUX-Lung 6. Lancet Oncol 2015;16:830-8. [Crossref] [PubMed]
- Mok TS, Wu YL, Ahn MJ, et al. Osimertinib or Platinum-Pemetrexed in EGFR T790M-Positive Lung Cancer. N Engl J Med 2017;376:629-40. [Crossref] [PubMed]
- Soria JC, Ohe Y, Vansteenkiste J, et al. Osimertinib in Untreated EGFR-Mutated Advanced Non-Small-Cell Lung Cancer. N Engl J Med 2018;378:113-25. [Crossref] [PubMed]
- Nishino M, Suda K, Kobayashi Y, et al. Effects of secondary EGFR mutations on resistance against upfront osimertinib in cells with EGFR-activating mutations in vitro. Lung Cancer 2018;126:149-55. [Crossref] [PubMed]
- Uchibori K, Inase N, Nishio M, et al. Identification of Mutation Accumulation as Resistance Mechanism Emerging in First-Line Osimertinib Treatment. J Thorac Oncol 2018;13:915-25. [Crossref] [PubMed]
- Namba K, Shien K, Takahashi Y, et al. Activation of AXL as a Preclinical Acquired Resistance Mechanism Against Osimertinib Treatment in EGFR -Mutant Non-Small Cell Lung Cancer Cells. Mol Cancer Res 2019;17:499-507. [Crossref] [PubMed]
- Ku BM, Choi MK, Sun JM, et al. Acquired resistance to AZD9291 as an upfront treatment is dependent on ERK signaling in a preclinical model. PLoS One 2018;13:e0194730. [Crossref] [PubMed]
- Le X, Puri S, Negrao M V, et al. Landscape of EGFR-Dependent and -Independent Resistance Mechanisms to Osimertinib and Continuation Therapy Beyond Progression in EGFR -Mutant NSCLC. Clin Cancer Res 2018;24:6195-203. [Crossref] [PubMed]
- Oxnard GR, Hu Y, Mileham KF, et al. Assessment of Resistance Mechanisms and Clinical Implications in Patients With EGFR T790M-Positive Lung Cancer and Acquired Resistance to Osimertinib. JAMA Oncol 2018;4:1527-34. [Crossref] [PubMed]
- Soda M, Choi YL, Enomoto M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature 2007;448:561-6. [Crossref] [PubMed]
- Katayama R, Lovly CM, Shaw AT. Therapeutic Targeting of Anaplastic Lymphoma Kinase in Lung Cancer: A Paradigm for Precision Cancer Medicine. Clin Cancer Res 2015;21:2227-35. [Crossref] [PubMed]
- Kwak EL, Bang YJ, Camidge DR, et al. Anaplastic Lymphoma Kinase Inhibition in Non-Small-Cell Lung Cancer. N Engl J Med 2010;363:1693-703. [Crossref] [PubMed]
- Solomon BJ, Mok T, Kim DW, et al. First-Line Crizotinib versus Chemotherapy in ALK -Positive Lung Cancer. N Engl J Med 2014;371:2167-77. [Crossref] [PubMed]
- Shaw AT, Engelman JA. ALK in Lung Cancer: Past, Present, and Future. J Clin Oncol 2013;31:1105-11. [Crossref] [PubMed]
- Katayama R, Friboulet L, Koike S, et al. Two Novel ALK Mutations Mediate Acquired Resistance to the Next-Generation ALK Inhibitor Alectinib. Clin Cancer Res 2014;20:5686-96. [Crossref] [PubMed]
- Doebele RC, Pilling AB, Aisner DL, et al. Mechanisms of Resistance to Crizotinib in Patients with ALK Gene Rearranged Non-Small Cell Lung Cancer. Clin Cancer Res 2012;18:1472-82. [Crossref] [PubMed]
- Katayama R, Shaw AT, Khan TM, et al. Mechanisms of Acquired Crizotinib Resistance in ALK-Rearranged Lung Cancers. Sci Transl Med 2012;4:120ra17. [Crossref] [PubMed]
- Toyokawa G, Hirai F, Inamasu E, et al. Secondary Mutations at I1171 in the ALK Gene Confer Resistance to Both Crizotinib and Alectinib. J Thorac Oncol 2014;9:e86-7. [Crossref] [PubMed]
- Gainor JF, Dardaei L, Yoda S, et al. Molecular Mechanisms of Resistance to First- and Second-Generation ALK Inhibitors in ALK-Rearranged Lung Cancer. Cancer Discov 2016;6:1118-33. [Crossref] [PubMed]
- Fontana D, Ceccon M, Gambacorti-Passerini C, et al. Activity of second-generation ALK inhibitors against crizotinib-resistant mutants in an NPM-ALK model compared to EML4-ALK. Cancer Med 2015;4:953-65. [Crossref] [PubMed]
- Zhang S, Anjum R, Squillace R, et al. The Potent ALK Inhibitor Brigatinib (AP26113) Overcomes Mechanisms of Resistance to First- and Second-Generation ALK Inhibitors in Preclinical Models. Clin Cancer Res 2016;22:5527-38. [Crossref] [PubMed]
- Shaw AT, Felip E, Bauer TM, et al. Lorlatinib in non-small-cell lung cancer with ALK or ROS1 rearrangement: an international, multicentre, open-label, single-arm first-in-man phase 1 trial. Lancet Oncol 2017;18:1590-9. [Crossref] [PubMed]
- Rikova K, Guo A, Zeng Q, et al. Global Survey of Phosphotyrosine Signaling Identifies Oncogenic Kinases in Lung Cancer. Cell 2007;131:1190-203. [Crossref] [PubMed]
- Bergethon K, Shaw AT, Ignatius Ou SH, et al. ROS1 Rearrangements Define a Unique Molecular Class of Lung Cancers. J Clin Oncol 2012;30:863-70. [Crossref] [PubMed]
- Gainor JF, Tseng D, Yoda S, et al. Patterns of Metastatic Spread and Mechanisms of Resistance to Crizotinib in ROS1 -Positive Non-Small-Cell Lung Cancer. JCO Precis Oncol 2017. doi: 10.1200/PO.17.00063. [Crossref]
- Song A, Kim TM, Kim DW, et al. Molecular Changes Associated with Acquired Resistance to Crizotinib in ROS1-Rearranged Non-Small Cell Lung Cancer. Clin Cancer Res 2015;21:2379-87. [Crossref] [PubMed]
- Awad MM, Katayama R, McTigue M, et al. Acquired resistance to crizotinib from a mutation in CD74-ROS1. N Engl J Med 2013;368:2395-401. [Crossref] [PubMed]
- Dziadziuszko R, Le AT, Wrona A, et al. An Activating KIT Mutation Induces Crizotinib Resistance in ROS1-Positive Lung Cancer. J Thorac Oncol 2016;11:1273-81. [Crossref] [PubMed]
- Vaishnavi A, Schubert L, Rix U, et al. EGFR Mediates Responses to Small-Molecule Drugs Targeting Oncogenic Fusion Kinases. Cancer Res 2017;77:3551-63. [Crossref] [PubMed]
- Cargnelutti M, Corso S, Pergolizzi M, et al. Activation of RAS family members confers resistance to ROS1 targeting drugs. Oncotarget 2015;6:5182-94. [Crossref] [PubMed]
- Drilon A, Rekhtman N, Arcila M, et al. Cabozantinib in patients with advanced RET -rearranged non-small-cell lung cancer: an open-label, single-centre, phase 2, single-arm trial. Lancet Oncol 2016;17:1653-60. [Crossref] [PubMed]
- Stroun M, Anker P, Maurice P, et al. Neoplastic Characteristics of the DNA Found in the Plasma of Cancer Patients. Oncology 1989;46:318-22. [Crossref] [PubMed]
- Wan JCM, Massie C, Garcia-Corbacho J, et al. Liquid biopsies come of age: Towards implementation of circulating tumour DNA. Nat Rev Cancer 2017;17:223-38. [Crossref] [PubMed]
- Forshew T, Murtaza M, Parkinson C, et al. Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Sci Transl Med 2012;4:136ra68. [Crossref] [PubMed]
- Rolfo C, Mack PC, Scagliotti G V, et al. Liquid Biopsy for Advanced Non-Small Cell Lung Cancer (NSCLC): A Statement Paper from the IASLC. J Thorac Oncol 2018;13:1248-68. [Crossref] [PubMed]
- Stroun M, Lyautey J, Lederrey C, et al. About the possible origin and mechanism of circulating DNA. Clin Chim Acta 2001;313:139-42. [Crossref] [PubMed]
- Thierry AR, Mouliere F, Gongora C, et al. Origin and quantification of circulating DNA in mice with human colorectal cancer xenografts. Nucleic Acids Res 2010;38:6159-75. [Crossref] [PubMed]
- Rizvi NA, Hellmann MD, Snyder A, et al. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 2015;348:124-8. [Crossref] [PubMed]
- Sun K, Jiang P, Chan KCA, et al. Plasma DNA tissue mapping by genome-wide methylation sequencing for noninvasive prenatal, cancer, and transplantation assessments. Proc Natl Acad Sci 2015;112:E5503-12. [Crossref] [PubMed]
- Thakur BK, Zhang H, Becker A, et al. Double-stranded DNA in exosomes: a novel biomarker in cancer detection. Cell Res 2014;24:766-9. [Crossref] [PubMed]
- Yao W, Mei C, Nan X, et al. Evaluation and comparison of in vitro degradation kinetics of DNA in serum, urine and saliva: A qualitative study. Gene 2016;590:142-8. [Crossref] [PubMed]
- Kahlert C, Melo SA, Protopopov A, et al. Identification of double-stranded genomic DNA spanning all chromosomes with mutated KRAS and p53 DNA in the serum exosomes of patients with pancreatic cancer. J Biol Chem 2014;289:3869-75. [Crossref] [PubMed]
- Dvořáková M, Karafiát V, Pajer P, et al. DNA released by leukemic cells contributes to the disruption of the bone marrow microenvironment. Oncogene 2013;32:5201-9. [Crossref] [PubMed]
- Bettegowda C, Sausen M, Leary RJ, et al. Detection of Circulating Tumor DNA in Early- and Late-Stage Human Malignancies. Sci Transl Med 2014;6:224ra24. [Crossref] [PubMed]
- Diehl F, Schmidt K, Choti MA, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med 2008;14:985-90. [Crossref] [PubMed]
- Cohen JD, Li L, Wang Y, et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science 2018;359:926-30. [Crossref] [PubMed]
- Ulivi P. Non-Invasive Methods to Monitor Mechanisms of Resistance to Tyrosine Kinase Inhibitors in Non-Small-Cell Lung Cancer: Where Do We Stand? Int J Mol Sci 2016;17:1186. [Crossref] [PubMed]
- Lim M, Kim CJ, Sunkara V, et al. Liquid Biopsy in Lung Cancer: Clinical Applications of Circulating Biomarkers (CTCs and ctDNA). Micromachines 2018;9:100. [Crossref] [PubMed]
- Oxnard GR, Paweletz CP, Kuang Y, et al. Noninvasive Detection of Response and Resistance in EGFR-Mutant Lung Cancer Using Quantitative Next-Generation Genotyping of Cell-Free Plasma DNA. Clin Cancer Res 2014;20:1698-705. [Crossref] [PubMed]
- Douillard JY, Ostoros G, Cobo M, et al. Gefitinib Treatment in EGFR Mutated Caucasian NSCLC: Circulating-Free Tumor DNA as a Surrogate for Determination of EGFR Status. J Thorac Oncol 2014;9:1345-53. [Crossref] [PubMed]
- Mok T, Wu YL, Lee JS, et al. Detection and Dynamic Changes of EGFR Mutations from Circulating Tumor DNA as a Predictor of Survival Outcomes in NSCLC Patients Treated with First-line Intercalated Erlotinib and Chemotherapy. Clin Cancer Res 2015;21:3196-203. [Crossref] [PubMed]
- Diaz LA, Bardelli A. Liquid Biopsies: Genotyping Circulating Tumor DNA. J Clin Oncol 2014;32:579-86. [Crossref] [PubMed]
- Murtaza M, Dawson SJ, Tsui DWY, et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature 2013;497:108-12. [Crossref] [PubMed]
- Alix-Panabières C, Pantel K. Challenges in circulating tumour cell research. Nat Rev Cancer 2014;14:623-31. [Crossref] [PubMed]
- Krebs MG, Sloane R, Priest L, et al. Evaluation and prognostic significance of circulating tumor cells in patients with non-small-cell lung cancer. J Clin Oncol 2011;29:1556-63. [Crossref] [PubMed]
- Neumann MHD, Bender S, Krahn T, et al. ctDNA and CTCs in Liquid Biopsy - Current Status and Where We Need to Progress. Comput Struct Biotechnol J 2018;16:190-5. [Crossref] [PubMed]
- Massagué J, Obenauf AC. Metastatic colonization by circulating tumour cells. Nature 2016;529:298-306. [Crossref] [PubMed]
- Valadi H, Ekström K, Bossios A, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 2007;9:654-9. [Crossref] [PubMed]
- van Niel G, D’Angelo G, Raposo G. Shedding light on the cell biology of extracellular vesicles. Nat Rev Mol Cell Biol 2018;19:213-28. [Crossref] [PubMed]
- Boriachek K, Islam MN, Möller A, et al. Biological Functions and Current Advances in Isolation and Detection Strategies for Exosome Nanovesicles. Small 2018. [Crossref] [PubMed]
- EL Andaloussi S, Mäger I, Breakefield XO, et al. . Extracellular vesicles: Biology and emerging therapeutic opportunities. Nat Rev Drug Discov 2013;12:347-57. [Crossref] [PubMed]
- Syn N, Wang L, Sethi G, et al. Exosome-Mediated Metastasis: From Epithelial-Mesenchymal Transition to Escape from Immunosurveillance. Trends Pharmacol Sci 2016;37:606-17. [Crossref] [PubMed]
- Hoshino A, Costa-Silva B, Shen TL, et al. Tumour exosome integrins determine organotropic metastasis. Nature 2015;527:329-35. [Crossref] [PubMed]
- Costa-Silva B, Aiello NM, Ocean AJ, et al. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat Cell Biol 2015;17:816-26. [Crossref] [PubMed]
- Ortiz A, Gui J, Zahedi F, et al. An Interferon-Driven Oxysterol-Based Defense against Tumor-Derived Extracellular Vesicles. Cancer Cell 2019;35:33-45.e6. [Crossref] [PubMed]
- van der Mijn JC, Sol N, Mellema W, et al. Analysis of AKT and ERK1/2 protein kinases in extracellular vesicles isolated from blood of patients with cancer. J Extracell Vesicles 2014;3:25657. [Crossref] [PubMed]
- Godoy PM, Bhakta NR, Barczak AJ, et al. Large Differences in Small RNA Composition Between Human Biofluids. Cell Rep 2018;25:1346-58. [Crossref] [PubMed]
- Schwarzenbach H, Nishida N, Calin GA, et al. Clinical relevance of circulating cell-free microRNAs in cancer. Nat Rev Clin Oncol 2014;11:145-56. [Crossref] [PubMed]
- van der Meijden PEJ, Heemskerk JWM. Platelet biology and functions: new concepts and clinical perspectives. Nature Reviews Cardiology 2019;16:166-79. [Crossref] [PubMed]
- Gay LJ, Felding-Habermann B. Contribution of platelets to tumour metastasis. Nat Rev Cancer 2011;11:123-34. [Crossref] [PubMed]
- Best MGG, Sol N, Kooi I, et al. RNA-Seq of Tumor-Educated Platelets Enables Blood-Based Pan-Cancer, Multiclass, and Molecular Pathway Cancer Diagnostics. Cancer Cell 2015;28:666-76. [Crossref] [PubMed]
- Wu Z, Yang Z, Dai Y, et al. Update on liquid biopsy in clinical management of non-small cell lung cancer. Onco Targets Ther 2019;12:5097-109. [Crossref] [PubMed]
- van Dessel LF, Beije N, Helmijr JCA, et al. Application of circulating tumor DNA in prospective clinical oncology trials - standardization of preanalytical conditions. Mol Oncol 2017;11:295-304. [Crossref] [PubMed]
- Nikolaev S, Lemmens L, Koessler T, et al. Circulating tumoral DNA: Preanalytical validation and quality control in a diagnostic laboratory. Anal Biochem 2018;542:34-9. [Crossref] [PubMed]
- Lam NYM, Rainer TH, Chiu RWK, et al. EDTA Is a Better Anticoagulant than Heparin or Citrate for Delayed Blood Processing for Plasma DNA Analysis. Clin Chem 2004;50:256-7. [Crossref] [PubMed]
- Rothwell DG, Smith N, Morris D, et al. Genetic profiling of tumours using both circulating free DNA and circulating tumour cells isolated from the same preserved whole blood sample. Mol Oncol 2016;10:566-74. [Crossref] [PubMed]
- Parpart-Li S, Bartlett B, Popoli M, et al. The effect of preservative and temperature on the analysis of circulating tumor DNA. Clin Cancer Res 2017;23:2471-7. [Crossref] [PubMed]
- Kang Q, Henry NL, Paoletti C, et al. Comparative analysis of circulating tumor DNA stability In K3EDTA, Streck, and CellSave blood collection tubes. Clin Biochem 2016;49:1354-60. [Crossref] [PubMed]
- Volckmar AL, Sültmann H, Riediger A, et al. A field guide for cancer diagnostics using cell-free DNA: From principles to practice and clinical applications. Genes Chromosom Cancer 2018;57:123-39. [Crossref] [PubMed]
- Malentacchi F, Pizzamiglio S, Verderio P, et al. Influence of storage conditions and extraction methods on the quantity and quality of circulating cell-free DNA (ccfDNA): the SPIDIA-DNAplas External Quality Assessment experience. Clin Chem Lab Med 2015;53:1935-42. [Crossref] [PubMed]
- Wolf A, Beller K, Groemminger S, et al. Purification of Circulating Cell-Free DNA from Plasma and Urine Using the Automated Large-Volume Extraction on the QIAsymphony(R) SP Instrument. Adv Exp Med Biol 2016;924:179-85. [Crossref] [PubMed]
- Pérez-Barrios C, Nieto-Alcolado I, Torrente M, et al. Comparison of methods for circulating cell-free DNA isolation using blood from cancer patients: Impact on biomarker testing. Transl Lung Cancer Res 2016;5:665-72. [Crossref] [PubMed]
- Page K, Guttery DS, Zahra N, et al. Influence of Plasma Processing on Recovery and Analysis of Circulating Nucleic Acids. PLoS One 2013;8:e77963. [Crossref] [PubMed]
- Lu JL, Liang ZY. Circulating free DNA in the era of precision oncology: Pre- and post-analytical concerns. Chronic Dis Transl Med 2016;2:223-30. [Crossref] [PubMed]
- Breitbach S, Tug S, Helmig S, et al. Direct Quantification of Cell-Free, Circulating DNA from Unpurified Plasma. PLoS One 2014;9:e87838. [Crossref] [PubMed]
- Sozzi G, Conte D, Leon M, et al. Quantification of free circulating DNA as a diagnostic marker in lung cancer. J Clin Oncol 2003;21:3902-8. [Crossref] [PubMed]
- Weber B, Meldgaard P, Hager H, et al. Detection of EGFR mutations in plasma and biopsies from non-small cell lung cancer patients by allele-specific PCR assays. BMC Cancer 2014;14:294. [Crossref] [PubMed]
- Thress KS, Brant R, Carr TH, et al. EGFR mutation detection in ctDNA from NSCLC patient plasma: A cross-platform comparison of leading technologies to support the clinical development of AZD9291. Lung Cancer 2015;90:509-15. [Crossref] [PubMed]
- Sacher AG, Paweletz C, et al. Prospective validation of rapid plasma genotyping as a sensitive and specific tool for guiding lung cancer care. JAMA Oncol 2016;2:1014-22. [Crossref] [PubMed]
- Oxnard GR, Thress KS, Alden RS, et al. Association Between Plasma Genotyping and Outcomes of Treatment With Osimertinib (AZD9291) in Advanced Non-Small-Cell Lung Cancer. J Clin Oncol 2016;34:3375-82. [Crossref] [PubMed]
- Reckamp KL, Melnikova VO, Karlovich C, et al. A Highly Sensitive and Quantitative Test Platform for Detection of NSCLC EGFR Mutations in Urine and Plasma. J Thorac Oncol 2016;11:1690-700. [Crossref] [PubMed]
- Couraud S, Vaca-Paniagua F, Villar S, et al. Noninvasive diagnosis of actionable mutations by deep sequencing of circulating free DNA in lung cancer from never-smokers: a proof-of-concept study from BioCAST/IFCT-1002. Clin Cancer Res 2014;20:4613-24. [Crossref] [PubMed]
- Newman AM, Lovejoy AF, Klass DM, et al. Integrated digital error suppression for improved detection of circulating tumor DNA. Nat Biotechnol 2016;34:547-55. [Crossref] [PubMed]
- Dagogo-Jack I, Rooney MM, Lin JJ, et al. Treatment with Next-Generation ALK Inhibitors Fuels Plasma ALK Mutation Diversity. Clin Cancer Res 2019. [Epub ahead of print]. [Crossref] [PubMed]
- Elazezy M, Joosse SA. Techniques of using circulating tumor DNA as a liquid biopsy component in cancer management. Comput Struct Biotechnol J 2018;16:370-8. [Crossref] [PubMed]
- Beaver JA, Jelovac D, Balukrishna S, et al. Detection of cancer DNA in plasma of patients with early-stage breast cancer. Clin Cancer Res 2014;20:2643-50. [Crossref] [PubMed]
- Zhang S, Chen Z, Huang C. at al. Ultrasensitive and quantitative detection of EGFR mutations in plasma samples from patients with non-small-cell lung cancer using a dual PNA clamping-mediated LNA-PNA PCR clamp. Analyst 2019;144:1718-24. [Crossref] [PubMed]
- Glenn TC. Field guide to next-generation DNA sequencers. Mol Ecol Resour 2011;11:759-69. [Crossref] [PubMed]
- Plagnol V, Woodhouse S, Howarth K, et al. Analytical validation of a next generation sequencing liquid biopsy assay for high sensitivity broad molecular profiling. PLoS One 2018;13:e0193802. [Crossref] [PubMed]
- Leighl NB, Page RD, Raymond VM, et al. Clinical Utility of Comprehensive Cell-free DNA Analysis to Identify Genomic Biomarkers in Patients with Newly Diagnosed Metastatic Non-small Cell Lung Cancer. Clin Cancer Res 2019;25:4691-700. [Crossref] [PubMed]
- Uchida J, Kato K, Kukita Y, et al. Diagnostic Accuracy of Noninvasive Genotyping of EGFR in Lung Cancer Patients by Deep Sequencing of Plasma Cell-Free DNA. Clin Chem 2015;61:1191-6. [Crossref] [PubMed]
- Narayan A, Carriero NJ, Gettinger SN, et al. Ultrasensitive measurement of hotspot mutations in tumor DNA in blood using error-suppressed multiplexed deep sequencing. Cancer Res 2012;72:3492-8. [Crossref] [PubMed]
- Rachiglio AM, Abate RE, Sacco A, et al. Limits and potential of targeted sequencing analysis of liquid biopsy in patients with lung and colon carcinoma. Oncotarget 2016;7:66595-605. [Crossref] [PubMed]
- Abbosh C, Birkbak NJ, Wilson GA, et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature 2017;545:446-51. [Crossref] [PubMed]
- Lin C, Shi X, Yang S, et al. Comparison of ALK detection by FISH, IHC and NGS to predict benefit from crizotinib in advanced non-small-cell lung cancer. Lung Cancer 2019;131:62-8. [Crossref] [PubMed]
- Scattone A, Catino A, Schirosi L, et al. Discordance between FISH, IHC, and NGS Analysis of ALK Status in Advanced Non-Small Cell Lung Cancer (NSCLC): a Brief Report of 7 Cases. Transl Oncol 2019;12:389-95. [Crossref] [PubMed]
- Nilsson RJA, Karachaliou N, Berenguer J, et al. Rearranged EML4-ALK fusion transcripts sequester in circulating blood platelets and enable blood-based crizotinib response monitoring in non-small-cell lung cancer. Oncotarget 2016;7:1066-75. [Crossref] [PubMed]
- Dagogo-Jack I, Brannon AR, Ferris LA, et al. Tracking the Evolution of Resistance to ALK Tyrosine Kinase Inhibitors through Longitudinal Analysis of Circulating Tumor DNA. JCO Precis Oncol 2018. doi: 10.1200/PO.17.00160. [Crossref]
- Combaret V, Iacono I, Bellini A, et al. Detection of tumor ALK status in neuroblastoma patients using peripheral blood. Cancer Med 2015;4:540-50. [Crossref] [PubMed]
- Bordi P, Tiseo M, Rofi E, et al. Detection of ALK and KRAS Mutations in Circulating Tumor DNA of Patients With Advanced ALK-Positive NSCLC With Disease Progression During Crizotinib Treatment. Clin Lung Cancer 2017;18:692-7. [Crossref] [PubMed]
- Page K, Powles T, Slade MJ, et al. The importance of careful blood processing in isolation of cell-free DNA. Ann N Y Acad Sci 2006;1075:313-7. [Crossref] [PubMed]
- Dagogo-Jack I, Rooney M, Nagy RJ, et al. Molecular Analysis of Plasma From Patients With ROS1-Positive NSCLC. J Thorac Oncol 2019;14:816-24. [Crossref] [PubMed]


