CT-based radiomics signature for the stratification of N2 disease risk in clinical stage I lung adenocarcinoma
Introduction
As the N2 status in non-small cell lung cancer (NSCLC) dictates whether surgical treatment or neoadjuvant chemotherapy is conducted as the optimal intervention, accurate preoperative mediastinal lymph node staging is crucial for treatment decision in patients with resectable NSCLC (1).
Invasive staging modalities such as mediastinoscopy and endobronchial ultrasound transbronchial needle aspiration (EBUS-TBNA) are currently considered as the gold standards for N2 disease diagnosis. These are well-established techniques that can be used to obtain tissue diagnosis confirmation of N2 involvement. However, because of the related complications, risk stratification of N2 involvement is necessary before undertaking of any invasive staging procedure (2). Compared to computed tomography (CT), positron emission tomography (PET) demonstrates superior performance with sensitivity of 81.3% and specificity of 79.4% in mediastinal staging of resectable lung cancer, thus sparing more patients from invasive staging procedures (3,4). However, PET’s false negative and positive rates in lymph node staging cannot be overlooked. It was reported that the performance of PET depended on lymph node’s station, and the sensitivity of PET for N2 disease at the subcarinal lymph node station was only 29% (5,6). In addition, due to its high expense and relatively low incidence of N2 disease, routine PET use in clinical stage I NSCLC [lung cancer stage I (T1/T2aN0M0): tumor less than 4 cm in greatest dimension (T1/T2a), no regional lymph node metastasis (N0), no distant metastasis (M0)] is controversial and often precluded by economic considerations in many countries (3,7-9).
Radiomics is a novel approach to quantitatively decode radiographic information from lung nodules, which has demonstrated promising performance in diagnosing malignant from benign lesions (10), predicting the status of epidermal growth factor receptor (EGFR) mutations (11), discriminating among the histomorphological lung adenocarcinoma subtypes (12), and identifying invasive from preinvasive lesions (13). However, the relationship between radiomics and N2 disease in clinical stage I NSCLC has not been fully investigated. Previous studies (14-16) have reported that radiomics analysis can predict lymph node disease in breast, bladder, and colorectal cancers. Furthermore, the occurrence of N2 metastasis in clinical stage I lung adenocarcinoma has already been proven to correlate with qualitative CT features, such as tumor diameter, location, the ratio of solid component, etc. (8,17,18).
Therefore, we hypothesized that CT-based radiomics could more accurately predict N2 disease than traditional CT features. This study aimed to develop a predictive model to stratify N2 disease risk for patients with clinical stage I lung adenocarcinoma based on radiomics analysis.
Methods
Study population
The Institutional Review Board of Shanghai Pulmonary Hospital approved this retrospective study with a waiver of informed consent (No. k19-134Y). Patients with clinical stage I lung adenocarcinoma, who received curative surgery in Shanghai Pulmonary Hospital, were evaluated for their eligibility. Patients admitted between January 2014 and December 2014 served as training cohort; those from January 2015 to June 2015 were treated as validation cohort. Participants were included in the study with the following criteria: (I) lung nodule less than 4 cm on CT scan; (II) maximum short-axis diameter of mediastinal lymph node less than 1 cm on contrast CT scan; (III) pathologically-confirmed primary lung adenocarcinoma; and (IV) conduction of systemic mediastinal lymph node dissection if pathology indicated invasive adenocarcinoma. Patients with multiple lesions or undergoing neoadjuvant chemotherapy or radiotherapy were excluded. Demographic and clinical characteristics were collected from medical records.
Image acquisition and nodule segmentation
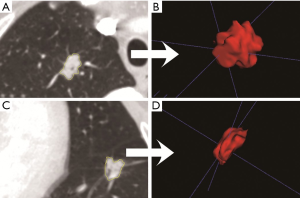
Chest high resolution CT scans were obtained during patient’s full inspiration using the Somatom Definition AS (Siemens Medical Systems, Germany) or Brilliance 40 (Philips Medical Systems, Netherlands) at 120 KVp tube energy and 200 mAs effective dose. All CT images were reconstructed using a medium sharp reconstruction algorithm with a slice thickness of 1 mm with 0.7 mm increment. And the size of CT images is 512×512 pixels. The identified CT scans were downloaded from the Picture Archiving and Communication Systems. As manual delineation was time-consuming and prone to inter-observer variability, nodule segmentation was performed using semi-automatic GrowCut segmentation algorithm in 3D slicer (an open-sourced software, www.slicer.org, Figure S1). Compared to manual segmentation, semi-automatic segmentation was more robust and with increased accuracy of nodule delineation, thus allowing for reproducible radiomics feature extraction (19). Two researchers with more than three years of experience in lung nodule screening performed nodule segmentation, which was confirmed by a radiologist with twenty years of experience in thoracic imaging.
Radiomics feature extraction
Radiomics features were extracted using PyRadiomics in Python, which is an open-source platform that sets the reference standard for obtaining radiomics data from medical images. The PyRadiomics platform was recently developed to standardize the calculation of radiomics feature algorithms and ease the feature extraction process in order to improve reproducibility of the findings (20). In this study, 1,078 radiomics features were extracted consisting of five classes: (I) first order statistics (n=266); (II) shape (n=14); (III) gray level co-occurrence matrix (n=350); (4) gray level run length matrix (n=224); and (5) gray level size zone matrix (n=224). The extracted radiomics features were listed in the Supplementary materials (Table S1). The computing algorithms can be found at www.radiomics.io and the image biomarker standardization initiative (IBSI) presented a document to standardize the nomenclature and definitions of radiomic feature (21).
Full table
Radiomics feature selection and model construction
Because of high dimensionality and multicollinearity, radiomics feature selection was conducted by sequentially applying minimum redundancy maximum relevance (mRMR) (22) and least absolute shrinkage and selection operator (LASSO) (23) techniques. Prior to feature selection and model building, no data transformation or standardization was conducted. Using the training dataset, we first applied the mRMR method to rank each radiomics feature according to its relevance with the N2 status and redundancy with other radiomics features. The top 100 most significant features were selected as candidates for LASSO analysis. By introducing a tuning parameter to penalize the coefficient of variables that entered into the regression model, LASSO aimed to reduce the possibility of overfitting. With the increase in the tuning parameter (λ), the absolute values of variable coefficients were reduced toward zero, and less variables were then selected. The area under the curve (AUC) was used as the criteria of model performance, and the model with maximum AUC was selected. The computing algorithms for feature selection and model building were listed in the Supplementary material.
Based on the results of LASSO regression of the training dataset, a radiomics signature was established, and its predictive performance was assessed in the training and validation datasets. The differentiation performance of the radiomics signature was quantified via receiver operator characteristics (ROC) analysis, and the calibration performance was evaluated using a calibration curve. Due to the existence of imbalanced classes, AUC value may not completely reflect the actual model performance, and the precision and recall were also calculated for model assessment. Additionally, a logistic regression model incorporating preoperative clinical factors was also constructed to predict N2 status, and its predictive performance was compared with the radiomics signature.
Statistical analysis
All statistical analyses were performed with R, and the “mRMRe” and “glmnet” packages were used to perform mRMR analysis and LASSO regression, respectively. Continuous and categorical variables were compared using the t and Fisher’s exact tests, respectively. For logistic analysis, variables with P value less than 0.1 in univariate analysis were entered into the model using a forward stepwise method. A two-sided P value was computed, and a difference was considered significant at P<0.05.
Results
Baseline characteristics of the patients
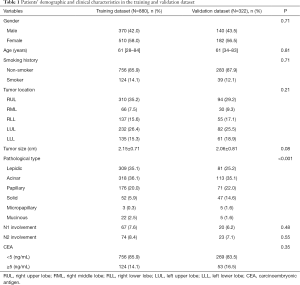
The study included 880 and 332 patients in the training and validation datasets, respectively. Demographic and clinical features of the patients were summarized in Table 1. In the training dataset, 58.0% of patients were female with 14.1% having a smoking history, and the median age was 61 years (range, 28–84 years). In the validation dataset, most patients were female (56.5%) and non-smokers (87.9%), and the median age was 61 years (range, 34–83 years). Mean tumor diameters were 2.15±0.71 and 2.06±0.81 cm in the training and validation datasets, respectively. The incidences of N2 involvement were 8.4% and 7.1% in the training and validation datasets, respectively.
Full table
Radiomics feature and its correlation with tumor characteristics
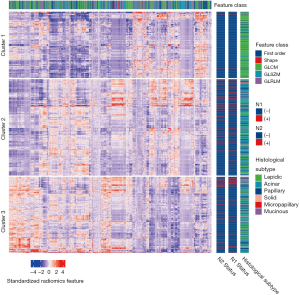
After Z-score standardization of the radiomics features, unsupervised K-means clustering was conducted for the 880 patients in the training dataset (Figure 1), which displayed three clusters of patients with similar radiomics features. The clinical parameters were compared between the three clusters, and the N1 and N2 disease and pathological subtypes (P=4.67×10−6, P=9.58×10−6, and P=2.20×10−16, respectively) were significantly different.
Radiomics feature selection and model construction
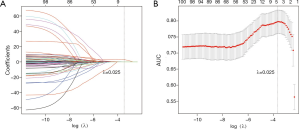
By applying mRMR analysis, the 1,078 radiomics features were ranked based on their relevance with N2 disease and redundancy with other features. The 100 highly ranked radiomics features (Figures S2,S3) were selected as candidates for LASSO regression analysis. In the LASSO regression, λ was selected through 10-fold cross-validation. As the log(λ) changed from −10 to 0, the number of variables that entered into the model was reduced, and the absolute values of the coefficients of the variables declined towards zero (Figure 2A). When λ was equal to 0.025 with a log λ of −3.38, the LASSO regression model demonstrated the best predictive performance with maximum AUC (Figure 2B). The optimal radiomics feature subset was determined at this point, and five radiomics features with nonzero coefficients in LASSO regression were selected (Table 2). Based on the five selected radiomics features, a radiomics signature was constructed to predict the N2 status, and the coefficients for radiomics score calculation were also showed in Table 2.
Full table
Model performance and validation
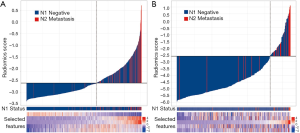
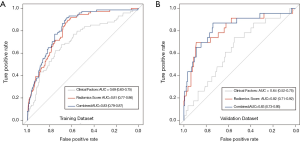
The radiomics signature was calculated for each patient in the training and validation datasets, and its relationship with N2/N1 status was presented in a waterfall plot (Figure 3). The radiomics signature showed good discriminatory performance with an AUC of 0.81 [95% confidence interval (CI), 0.77–0.86] and 0.82 (95% CI, 0.71–0.92) in the training and validation datasets, respectively.
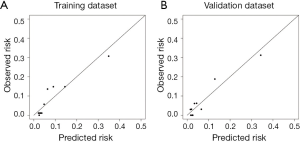
The calibration curve suggested that the predicted probabilities of N2 metastasis correlated well with the observed probabilities (Figure S4). And, the precision and recall were 0.92 and 0.21 in the training dataset and 0.92 and 0.22 in the validation dataset.
To compare the predictive performance of the radiomics signature to that of clinical factors, a multivariate logistic regression model incorporating preoperative clinical factors was established to predict N2 status (Tables S2,S3). In the logistic model, tumor size and carcinoembryonic antigen (CEA) [odds ratio (OR), 1.82; 95% CI, 1.30–2.55 and OR, 3.83; 95% CI, 2.24–6.46, respectively] were independent predictors of N2 disease. And, the precision and recall of the clinical model were 0.62 and 0.17, respectively. The independent clinical predictors and radiomics signature were both incorporated in order to construct build a combined model for predicting N2 disease (Table S4). As shown in Figure 4, radiomics signature showed better predictive performances than clinical factors (P<0.001), and the difference of predictive performance between radiomics signature and combined model was not statistically significant (P=0.23).
Full table

Full table

Full table
Discussion
Accurate mediastinal lymph node staging is vital for guiding a patient’s treatment decision and estimating prognosis in surgically resectable NSCLC (24). In this study, a predictive model incorporating radiomics information was developed in order to stratify N2 disease risk, which demonstrated a better predictive performance than conventional methods (8,17).
Currently, lung adenocarcinoma accounts for more than 80% of early-staged NSCLC and was more likely to be associated with N2 metastasis than other pathological types (8). It has been reported that the incidence of N2 disease in clinical stage I NSCLC was 4.0–16.8% (6,8,18,25). Risk stratification of N2 disease is crucial before performing invasive mediastinal lymph node staging techniques such as mediastinoscopy and EBUS-TBNA (2,26). Contrast-enhanced thoracic CT is currently the most widely used modality for mediastinal lymph node staging in NSCLC (3). And, the size of lymph node on CT scan serves as the only criterion for diagnosing malignant N2 disease, and lymph nodes larger than 1 cm are generally treated as N2 involvement. However, the sensitivity and specificity of CT scan in mediastinal lymph node staging is very poor. Cerfolio et al. (5) reported that the correlation between lymph node size and malignant probability is weak, and a malignant mediastinal lymph node is not significantly larger than a benign one. According to this study’s results, 15% of mediastinal lymph nodes <1 cm proved to be malignant, and 43% of mediastinal lymph nodes >1 cm were actually negative. Therefore, preoperative mediastinal lymph node staging based only on CT scans is highly inaccurate, which also explains the relatively high N2 disease incidence in our study (8,17).
PET detects lymph nodes with abnormally high functional activity and plays an important role in lung cancer staging. Compared with chest CT imaging, PET displays improved sensitivity and specificity in mediastinal staging (3). The maximum standardized uptake value (SUV) in PET measures the lesion’s metabolic activity, which is used to differentiate malignant from benign lesions. Although the optimal threshold of maximum SUV in mediastinal lymph node staging is still debatable, a maximum SUV of >2.5 was generally designated for diagnosing N2 disease (27). Despite its advantages, PET scan has several limitations in mediastinal lymph node staging. The diagnostic accuracy of PET varies depending upon different lymph nodes’ stations. Regarding mediastinal lymph node stations 6–8, PET sensitivity was poor (29–40%) (5,6). A high lesion SUV is associated with abnormal metabolic activity and is not specific to malignant disease. Therefore, false positive results often occur. It was reported that as high as 76% of N2 disease observed on PET scans were false positives after checking them with mediastinoscopy (5,9). And, the high cost of PET scan is also an obstacle to its routine use in mediastinal staging (3,8).
In previous studies (8,15,28), several models utilizing clinical and radiological characteristics have been developed to predict patient-specific probability of N2 metastasis in order to make cost-effective mediastinal staging strategy. In our study, 8.4% of patients with clinical stage I lung adenocarcinoma demonstrated N2 positive results. In agreement with other studies (8,18), we found that tumor sizes and CEA levels significantly correlated with N2 status. In addition, the predictive performance of the model incorporating clinical factors gave an AUC of about 0.7, which was similar to that observed in previously reported models (8,17).
Radiomics, also termed quantitative image analysis, is a newly developed field, which has the ability to maximize the information obtained from medical images and provide more accurate and comprehensive lesion characterization (26). Medical imaging, as the most commonly used, non-invasive modality, has offered much more information for lesion characterization than we could have perceived. By computerized image analysis, radiomics enables us to make the best use of medical images. It has been established that radiomics features significantly correlate with tumor stage, histology, and prognosis (29,30). An integrated analysis (31) reported that CT-based radiomics phenotypes also correlated with the driver mutation in lung adenocarcinoma and can predict EGFR and K-ras mutation status. Song et al. (12) also found that radiomics features have the potential to predict micropapillary component in invasive lung adenocarcinoma. In this study, a CT-based radiomics signature was established in order to predict N2 disease, and it demonstrated more accurate predictive performance than clinical factors. The energy feature quantifies the size and density of the tumor, and the other selected features measure the extent of heterogeneity by analyzing the asymmetry distribution of pixels or the texture of tumor presented in CT images.
Most patients in this study did not undergo PET scans for preoperative mediastinal lymph node staging. Thus, it is impossible to compare the diagnostic performance of radiomics signature and maximum SUV in stratifying N2 disease risk. And, the study only included patients with invasive adenocarcinoma and mediastinal lymph node less than 1 cm, which may compromise the generalizability of the results. Further studies are required in order to externally validate the signature. As a result of easy accessibility of open-sourced software for standardized radiomics analyses and advanced automatic segmentation techniques, it is much easier to obtain patient’s radiomics information. Therefore, the radiomics signature can be easily utilized for calculating patient-specific probability of N2 involvement. Based on the stratified N2 disease risk, customized mediastinal staging strategy can then be implemented.
In conclusion, the established CT-based radiomics signature could more accurately stratify the risk of N2 disease in clinical stage IA lung adenocarcinoma, which could help clinicians create patient-specific mediastinal staging strategies.
Supplementary
Radiomics feature extraction
Radiomics features were extracted using PyRadiomics package in Python (www.radiomics.io). Features implemented in the PyRadiomics were used for feature extraction, which consisted of five classes: first order statistics (n=19), shape (n=14), gray level cooccurrence matrix (n=25), gray level run length matrix (n=16) and gray level size zone matrix (n=16).
Using wavelet and LoG (Laplacian of Gaussian) filters for image processing, radiomics features were extracted both from the original CT images and filtered images. Regarding the wavelet filtering, the built-in stationary wavelet transformation was used through a high band-pass or lower band-pass filter in X, Y and Z directions, which created eight different pre-processed CT-images. For the LoG filtering, CT images were pre-processed using 3D LoG filter, and five different filtered images were created by changing σ to 5.0, 4.0, 3.0, 2.0 and 1.0 mm. As the shape radiomics features can only be extracted from the original images, in total, 1,078 radiomics features were obtained. The extracted features were listed in Table S1 and the computing algorithm can be found at www.radiomics.io.
Selected radiomics features and radiomics signature calculation
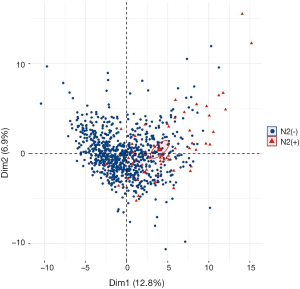
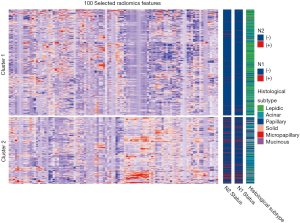
The top 100 most significant radiomics features were selected based on its relevance with the N2 status and redundancy with other radiomics features by applying mRMR analysis. Unsupervised cluster analysis of these 100 features in training cohort produced two clusters of patients with similar radiomics pattern, and the N2/N1 positive rate and the distribution of histological subtype were significantly different between the two clusters (Figure S2). In principal component analysis of the 100 radiomics features, the first two dimensions (PC1, PC2) respectively account for 12.8% and 6.9% of variance and patients with N2 metastasis demonstrated significantly different pattern in the scatterplot of PC1 and PC2 than N2 negative patients (Figure S3).
Afterwards, five radiomics features were further selected from the 100 candidate features for model development by using LASSO analysis (performed using glmnet package in R platform). LASSO is a powerful method for regression with high dimensional features. In our study, the LASSO method was combined with binary logistic regression analysis. We used the LASSO model to select the most important prognostic features from the training dataset. The formula of this method is:
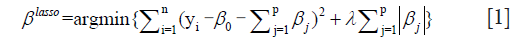
where, λ is the harmonic parameter. Benefited from the absolute constraint (λ), LASSO method shrinks coefficients and change some coefficients to zero. Therefore, LASSO method can be used for feature selection.
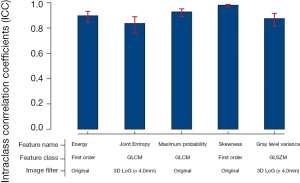
Selected radiomics features and the correspond coefficients were listed in Table 2. The radiomics signature for each patient was calculated as feature1 * coefficient1 + feature2 * coefficient2… featurei * coefficienti + constant value (−3.6). In order to confirm the stability of the five selected features, 100 randomly selected nodules were semi-automatically segmented by the two researchers; the intraclass coefficients (ICC) of the selected radiomics features were all larger than 0.8 (Figure S5).
N2 disease prediction using preoperative clinical factors
In training dataset, 74 patients were pathologically confirmed to be N2 positive. And, there was no significant difference between patients with N2 disease and those without in terms of gender, age, smoking history and lobar location (Table S2). Compared with nodules without N2 disease, the lung nodules with N2 involvement were significantly larger (OR, 1.82; 95% CI, 1.30–2.55) and more likely with elevated CEA (OR, 3.83; 95% CI, 2.24–6.46).
However, when the radiomics signature and the above independent clinical factor were all incorporated into the logistic model, the effect of nodule size faded away, and the contribution of CEA (OR, 2.64; 95% CI, 1.47–4.64) to the model was also reduced (Table S3).
Acknowledgments
Funding: This study was supported by Clinical Research Foundation of Shanghai Pulmonary Hospital (FK1943, FK1936, FK1944, FK1941), Shanghai Municipal Health Commission (2018ZHYL0102, 2019SY072, 201940018), Medicine and Public Health Scientific Projects in Zhejiang Province (2020KY270), and Huamei Key Research Foundation (2019HMZD05).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The Institutional Review Board of Shanghai Pulmonary Hospital approved this retrospective study with a waiver of informed consent (No. k19-134Y).
References
- Martins RG, D’Amico TA, Loo BW, et al. The Management of Patients With Stage IIIA Non–Small Cell Lung Cancer With N2 Mediastinal Node Involvement. J Natl Compr Canc Netw 2012;10:599-613. [Crossref] [PubMed]
- Decaluwé H, Dooms C, D'Journo XB, et al. Mediastinal staging by videomediastinoscopy in clinical N1 non-small cell lung cancer: a prospective multicentre study. Eur Respir J 2017. [Crossref] [PubMed]
- Whitson BA, Groth SS, Maddaus MA. Recommendations for optimal use of imaging studies to clinically stage mediastinal lymph nodes in non-small-cell lung cancer patients. Lung Cancer 2008;61:177-85. [Crossref] [PubMed]
- Schmidt-Hansen M, Baldwin DR, Hasler E, et al. PET-CT for assessing mediastinal lymph node involvement in patients with suspected resectable non-small cell lung cancer. Cochrane Database Syst Rev 2014.CD009519. [PubMed]
- Jie Y, Meng X, Gu A, et al. Metabolic volume parameters based on different thresholds with baseline 18F-FDG PET/CT as prognostic factors for survival in stage III non-small cell lung cancer. Transl Cancer Res 2017;6:732-45. [Crossref]
- Lin JT, Yang XN, Zhong WZ, et al. Association of maximum standardized uptake value with occult mediastinal lymph node metastases in cN0 non-small cell lung cancer. Eur J Cardiothorac Surg 2016;50:914-9. [Crossref] [PubMed]
- Amin MB, Edge S, Greene F, et al. American Joint Committee on Cancer Staging Manual. 8th ed. New York: Springer; 2017.
- Zhang Y, Sun Y, Xiang J, et al. A prediction model for N2 disease in T1 non-small cell lung cancer. J Thorac Cardiovasc Surg 2012;144:1360-4. [Crossref] [PubMed]
- Kozower BD, Meyers BF, Reed CE, et al. Does positron emission tomography prevent nontherapeutic pulmonary resections for clinical stage IA lung cancer? Ann Thorac Surg 2008;85:1166-9; discussion 1169-70. [Crossref] [PubMed]
- Hawkins S, Wang H, Liu Y, et al. Predicting Malignant Nodules from Screening CT Scans. J Thorac Oncol 2016;11:2120-8. [Crossref] [PubMed]
- Liu Y, Kim J, Balagurunathan Y, et al. Radiomic Features Are Associated With EGFR Mutation Status in Lung Adenocarcinomas. Clin Lung Cancer 2016;17:441-8.e6. [Crossref] [PubMed]
- Song SH, Park H, Lee G, et al. Imaging Phenotyping Using Radiomics to Predict Micropapillary Pattern within Lung Adenocarcinoma. J Thorac Oncol 2017;12:624-32. [Crossref] [PubMed]
- Chae HD, Park CM, Park SJ, et al. Computerized texture analysis of persistent part-solid ground-glass nodules: differentiation of preinvasive lesions from invasive pulmonary adenocarcinomas. Radiology 2014;273:285-93. [Crossref] [PubMed]
- Huang YQ, Liang CH, He L, et al. Development and Validation of a Radiomics Nomogram for Preoperative Prediction of Lymph Node Metastasis in Colorectal Cancer. J Clin Oncol 2016;34:2157-64. [Crossref] [PubMed]
- Wu S, Zheng J, Li Y, et al. A Radiomics Nomogram for the Preoperative Prediction of Lymph Node Metastasis in Bladder Cancer. Clin Cancer Res 2017;23:6904-11. [Crossref] [PubMed]
- Dong Y, Feng Q, Yang W, et al. Preoperative prediction of sentinel lymph node metastasis in breast cancer based on radiomics of T2-weighted fat-suppression and diffusion-weighted MRI. Eur Radiol 2018;28:582-91. [Crossref] [PubMed]
- Shafazand S, Gould MK. A clinical prediction rule to estimate the probability of mediastinal metastasis in patients with non-small cell lung cancer. J Thorac Oncol 2006;1:953-9. [Crossref] [PubMed]
- Ye B, Cheng M, Li W, et al. Predictive factors for lymph node metastasis in clinical stage IA lung adenocarcinoma. Ann Thorac Surg 2014;98:217-23. [Crossref] [PubMed]
- Parmar C, Rios Velazquez E, Leijenaar R, et al. Robust Radiomics feature quantification using semiautomatic volumetric segmentation. PLoS One 2014;9:e102107. [Crossref] [PubMed]
- van Griethuysen JJM, Fedorov A, Parmar C, et al. Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Res 2017;77:e104-7. [Crossref] [PubMed]
- Zwanenburg A, Leger S, Vallières M, et al. Image biomarker standardisation initiative. 2019. (arXiv:1612.07003).
- De Jay N, Papillon-Cavanagh S, Olsen C, et al. mRMRe: an R package for parallelized mRMR ensemble feature selection. Bioinformatics 2013;29:2365-8. [Crossref] [PubMed]
- McNeish DM. Using Lasso for Predictor Selection and to Assuage Overfitting: A Method Long Overlooked in Behavioral Sciences. Multivariate Behav Res 2015;50:471-84. [Crossref] [PubMed]
- De Leyn P, Dooms C, Kuzdzal J, et al. Revised ESTS guidelines for preoperative mediastinal lymph node staging for non-small-cell lung cancer. Eur J Cardiothorac Surg 2014;45:787-98. [Crossref] [PubMed]
- Defranchi SA, Cassivi SD, Nichols FC, et al. N2 disease in T1 non-small cell lung cancer. Ann Thorac Surg 2009;88:924-8. [Crossref] [PubMed]
- Dooms C, Tournoy KG, Schuurbiers O, et al. Endosonography for mediastinal nodal staging of clinical N1 non-small cell lung cancer: a prospective multicenter study. Chest 2015;147:209-15. [Crossref] [PubMed]
- Bryant AS, Cerfolio RJ, Klemm KM, et al. Maximum standard uptake value of mediastinal lymph nodes on integrated FDG-PET-CT predicts pathology in patients with non-small cell lung cancer. Ann Thorac Surg 2006;82:417-22; discussion 422-3. [Crossref] [PubMed]
- Chen K, Yang F, Jiang G, et al. Development and validation of a clinical prediction model for N2 lymph node metastasis in non-small cell lung cancer. Ann Thorac Surg 2013;96:1761-8. [Crossref] [PubMed]
- Lee G, Lee HY, Park H, et al. Radiomics and its emerging role in lung cancer research, imaging biomarkers and clinical management: State of the art. Eur J Radiol 2017;86:297-307. [Crossref] [PubMed]
- Aerts HJ, Velazquez ER, Leijenaar RT, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun 2014;5:4006. [Crossref] [PubMed]
- Rios Velazquez E, Parmar C, Liu Y, et al. Somatic Mutations Drive Distinct Imaging Phenotypes in Lung Cancer. Cancer Res 2017;77:3922-30. [Crossref] [PubMed]