
Immunocytochemistry of cytology specimens for predictive biomarkers in lung cancer
Predictive biomarkers are markers of therapeutic efficacy, the results of which are essential for determining patient care. With a growing number of predictive biomarkers that have emerged in non-small cell lung carcinoma (NSCLC), there has been a paradigm shift in the management of these patients. Targeted therapies with tyrosine kinase inhibitors (TKIs) in patients with sensitizing mutations in EGFR or ALK and ROS1 rearranged tumors, and immune checkpoint inhibitor therapy in patients expressing PD-L1 are just some of the examples of how predictive biomarker testing has become an integral part of NSCLC standard of care (1).
Of the various predictive biomarker testing methods, immunohistochemistry (IHC) is common, cost-effective, and easily available in most laboratories, has a rapid turn-around time, can be performed on relatively fewer number of tumor cells and poses the least amount of technical challenges when compared to other molecular and cytogenetics methods (2,3). While most predictive IHC assays are validated primarily on formalin-fixed paraffin-embedded (FFPE) histologic tissue samples, a large fraction of NSCLC patients are diagnosed on cytology samples, resulting in an increasing demand for predictive biomarker testing on cytologic specimens.
Most laboratories follow the recommendations from the College of American Pathologists (CAP) guideline for IHC assay validation on FFPE tissue samples; however cytologic specimens pose a greater challenge for validation (4). This is in part due to the wide variety of cytologic specimen preparations that comprise multiple preanalytic variables including a variety of collection media, preservatives, fixatives, storage conditions, processing techniques, and stains among others (2). Therefore, implementing IHC assays that are standardized and validated on FFPE histologic tissue samples on these cytologic specimens require a thorough and rigorous validation process. While several professional organizations have issued recommendations for the use of cytologic specimens for ancillary testing in NSCLC samples (5,6), specific guidelines for assay validation for immunocytochemistry (ICC) on cytology samples (for instance number of samples, selection of markers etc.) are largely lacking and are typically at the discretion of the individual laboratory medical director.
Preanalytical factors in cytologic specimens
A recent article from the International Association for the Study of Lung Cancer (IASLC) Pathology Committee states that “all cytologic preparations, including cell blocks, ethanol fixed, and air-dried slides” can be used for ICC (7). Of the various cytologic specimen preparations, cell blocks are the most widely used. This is in part due to the easy availability, the ease of getting multiple sections for a panel of markers, and the ease of validation as standardized protocols for FFPE histologic tissue can be easily implemented on cytology cell blocks for automated immunostainers (2). The wide acceptance of cell block sections for ICC uses the premise that all cell block protocols (irrespective of preparation protocol) use 10% neutral buffered formalin as the final fixation step prior to processing into an FFPE block. However, there is no standardized protocol for the type of collection media, prefixation, and processing technique and there is a wide variation amongst cytopathology laboratories. While the varied processing methodologies do not significantly impact diagnostic yield, several recent studies have highlighted issues with immunostaining of specific markers that demonstrate reduced antigenicity and false negative results mostly related to ethanol or methanol-based fixatives used prior to cell block preparation (8-11).
Non-cell block cytologic preparations including air-dried and alcohol fixed direct smears, cytospins and liquid based cytology (LBC) preparations pose an even greater challenge for ICC validation. Of these, immunostaining of ethanol-fixed smears or cytospins are used more frequently, with prior Papanicolau staining that can identify areas or cells of interest, or air-dried unfixed extra slides that can be used for ICC usually after some sort of post-fixation step involving formalin or acetone (12-14). While there are studies in the literature that suggest some fixatives can alter the antigenicity and results of ICC in cytologic samples, a report from the United Kingdom National External Quality Assessment Service (UK NEQAS) suggests that all non-formalin fixatives, with the exception of acetone, yield a quality of immunostaining comparable to that of formalin fixation alone (15).
Given the implications of biomarker reporting in NSCLC, where the results of ICC are used to guide patient care, it is critical that a rigorous protocol validation with possible optimization of existing FFPE protocols for histologic tissue, together with careful quality control (QC) measures, prior to any clinical implementation is required for all cytologic cell block and non-cell block specimens. Analytic validation of an ICC assay requires positive and negative control slides to be processed in the same way as the test samples (4). However, due to the difficulty of finding standardized control slides, most laboratories resort to using FFPE histologic sections as controls for ICC, a practice that is inadequate for non-FFPE cytologic specimen preparations (16). Commercially available cell lines with specific target antigen expression may be used as a positive control for ICC assay, but not all antigens are available as cell lines, in addition to posing a significant resource commitment. This underscores the need for adequate quality assurance measures with internal QC checks and participating in external QC programs as part of the continued QC metrics for ICC biomarker assays (2,15).
Predictive biomarkers in lung cancer cytology
ICC for ALK rearrangements
The current College of American Pathologist/ International Association for the Study of Lung Cancer/ Association for Molecular Pathology (CAP/IASLC/AMP) guidelines recommend the use of ALK IHC using D5F3 (Cell Signaling Technology, Danvers, MA) and 5A4 (Novocastra, Leica Biosystems; Newcastle Upon Tyne, UK) clones as an alternative to testing for ALK rearrangements using a fluorescence in situ hybridization (FISH) assay (17). The ALK D5F3 clone uses a Ventana automated immunoassay (Ventana ALK D5F3 CDx Assay, Ventana Medical Systems, Tucson, AZ) that has been approved by the United States Food and Drug Administration (FDA) as a companion diagnostic kit for the use of crizotinib in patients with ALK rearrangements (17,18).
Most cytology studies using ALK ICC have been performed using the D5F3 or 5A4 clones on cell block preparations and demonstrated 100% sensitivity with specificities ranging between 83% and 100% (19-24). Studies using non-cell block preparations including smears, cytospins and LBC preparations have shown slightly lower sensitivity (66–100%), suggesting that ICC on non-FFPE substrates may require some additional optimization (25-28). Table 1 summarizes some of the studies in the published literature that report ALK ICC in cytology specimens.
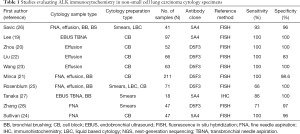
Full table
ICC for ROS1 rearrangements
ROS1 IHC using the highly sensitive but relatively less specific D4D6 (Cell Signaling Technology)clone has been recommended by the CAP/IASLC/AMP as a screening only tool where any cases that are positive by the IHC assay need molecular or cytogenetic confirmation (17). Unlike ALK, ROS1 IHC using D4D6 has not been FDA approved to be used as a companion diagnostic in patients with ROS1 rearrangements. Relatively few studies in cytology using ROS1 D4D6 ICC have been published; immunostaining using cell blocks as well as non-cell block smear and cytospin preparations have demonstrated sensitivities of 88–100% and specificities of 92–98% (29,30) (Table 2). More recently, a Ventana ROS1 antibody (SP384) has been developed with high sensitivity and specificity; however, there is very limited data on cytology samples (31-33).
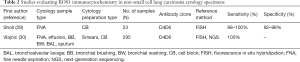
Full table
ICC for PD-L1 expression
A number of PD-L1 IHC assays have been developed as separate predictive biomarker assays that are associated with separate checkpoint inhibitors in separate clinical trials, making the validation and implementation of PD-L1 IHC challenging. The Dako 22C3 pharmDx and Ventana SP263 assay have been FDA approved as companion diagnostic tests with specific tumor proportion scores (TPS) to determine eligibility for immune checkpoint inhibitors (2,3). Part of the challenge of evaluating and interpreting PD-L1 ICC in cytology is the fact that none of the clinical trials involving checkpoint inhibitors included cytologic specimens. In addition, PD-L1 expression is known to have spatial heterogeneity, making the assessment of PD-L1 TPS on cytology specimens challenging (34-37). Further issues involve evaluating PD-L1 clones such as SP143 that require evaluation of both tumor and immune cell components, which may be difficult to perform on a cytologic specimen that lacks architecture, especially in sites such as a lymph node with metastatic tumor (3,38).
Notwithstanding, several studies have analyzed the concordance of PD-L1 expression in comparison to histologic tissue samples, showing that PD-L1 can be reliably assessed on cytologic samples (39-43). Most published studies were performed using cytology cell block preparations; however a few studies have reported ICC on cytologic smears with good concordance with histologic specimens (41,44-46). One study also reported clinical response to check point inhibitor therapy in patients with high PD-L1 expression in cytology samples, providing clinical evidence of a predictive role of PD-L1 ICC in cytology samples (47).
ICC for other predictive markers
Other predictive markers that may have some use in the setting on NSCLC include EGFR mutation specific antibodies, L858R (43B2) and the 15 base pair E746-A750 deletion in exon 19 (6B6). Both these antibodies are highly specific (96–99%) and have been shown to be suitable on cytologic preparations including cell blocks, smears, cytospin and LBC preparations (48-52). However, since they only detect a subset of EGFR mutations, current guidelines do not recommend the use of EGFR mutation specific antibodies for selecting patients due to the low sensitivity (ranging from 47% to 92%) (17).
A small subset of NSCLC patients may have a BRAF mutation (2–4%). The BRAF V600E mutation specific antibody (VE1) can be used as a surrogate for detecting BRAF V600E mutations in these patients, similar to its use in melanoma and colon cancer patients (53-56). However, 40-50% of BRAF mutations in NSCLC patients are non-V600E mutations and therefore limits the use of this antibody for screening NSCLC patients using the VE1 antibody (57). A few studies have evaluated the use of BRAF V600E (VE1) antibody in histologic specimens, but published literature on BRAF V600E VE1 ICC in cytology samples is largely lacking (53,54).
There is growing evidence on the usefulness of detecting MET alterations (MET exon 14 skipping mutations and MET amplifications) in NSCLC patients; however evaluating MET protein expression using IHC (clone SP44) does not seem to be provide an efficient screening method (3,58,59).
NTRK fusions may be detected in a very small subset of NSCLC patients (60). Screening using NTRK antibody (EPR17341) may be an option; however currently there is limited literature demonstrating the usefulness of using the NTRK antibody as a predictive marker in NSCLC patients (61). A recent study reported a sensitivity of 87.5% and specificity of 100% for detection of NTRK fusions in lung carcinomas using the NTRK antibody (EPR17341) (62).
Predictive ICC in cytology: opportunities and limitations
The versatility of specimen preparations in cytology provides a variety of options for the application of predictive ICC. Most ICC assays can be performed on very limited number of tumor cells and results are cost-effective (compared to other molecular/cytogenetic options) with rapid turn-around time. The main difficulty of implementing predictive ICC on cytology specimens is the need for optimization and additional validation to ensure rigorous QC which is frequently challenging due to the lack of standardized processing protocols for cytologic specimens. Therefore, cytology remains an underutilized resource for predictive ICC in NSCLC patients.
In addition, interpretation of predictive ICC on non-cell block preparations may sometimes pose difficulty. For example PD-L1 expression is defined as partial or complete membrane staining of tumors cells, and nuclear or cytoplasmic staining is considered non-specific. However, in non-cell block preparations with the three-dimensionality of cell clusters and overlapping intact cells, membranous staining may often appear as cytoplasmic staining (2) (Figure 1). Also, non-specific staining of background cells such as histiocytes and inflammatory cells can frequently lead to overestimation of PD-L1 TPS (2,44) (Figure 2). The absence of a corresponding section (as available in cell blocks) with a confirmatory immunostain such as TTF-1 that can help identify the tumor cells for accurate PD-L1 estimation, adds to the challenge of interpreting predictive ICC on non-cell block preparations.
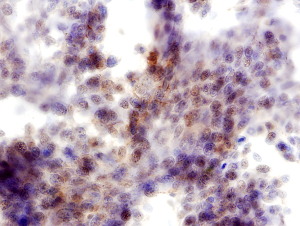
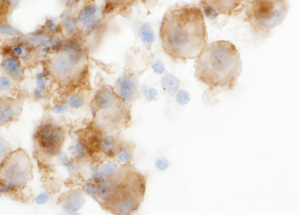
Conclusions
With the increasing number of predictive biomarkers available for treating NSCLC patients, the need to provide rapid, reliable, cost-effective results from NSCLC specimens is very important. IHC is a widely available and less technically challenging assay than molecular testing that can be performed effectively on most FFPE tissue. Translating these predictive biomarker assays into cytology specimens provide a few challenges. However, with proper optimization and rigorous validation, ICC can be performed on all cytologic specimens. Being able to implement predictive ICC assays in routine cytology practice can provide a rapid and cost-effective option for evaluating predictive biomarkers, especially in cases with limited tumor that precludes molecular testing.
Acknowledgments
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editor (Sanja Dacic) for the series “Selected Highlights of the 2019 Pulmonary Pathology Society Biennial Meeting” published in Translational Lung Cancer Research. The article was sent for external peer review organized by the Guest Editor and the editorial office.
Conflicts of Interest: The author has completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tlcr.2019.12.31). The series “Selected Highlights of the 2019 Pulmonary Pathology Society Biennial Meeting” was commissioned by the editorial office without any sponsorship or funding. The author has no conflicts of interest to declare.
Ethical Statement: The author is accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Hanna N, Johnson D, Temin S, et al. Systemic Therapy for Stage IV Non-Small-Cell Lung Cancer: American Society of Clinical Oncology Clinical Practice Guideline Update Summary. J Oncol Pract 2017;13:832-7. [Crossref] [PubMed]
- Jain D, Nambirajan A, Borczuk A, et al. Immunocytochemistry for predictive biomarker testing in lung cancer cytology. Cancer Cytopathol 2019;127:325-39. [Crossref] [PubMed]
- Mino-Kenudson M. Immunohistochemistry for predictive biomarkers in non-small cell lung cancer. Transl Lung Cancer Res 2017;6:570-87. [Crossref] [PubMed]
- Fitzgibbons PL, Bradley LA, Fatheree LA, et al. Principles of analytic validation of immunohistochemical assays: Guideline from the College of American Pathologists Pathology and Laboratory Quality Center. Arch Pathol Lab Med 2014;138:1432-43. [Crossref] [PubMed]
- Roy-Chowdhuri S, Aisner DL, Allen TC, et al. Biomarker Testing in Lung Carcinoma Cytology Specimens: A Perspective From Members of the Pulmonary Pathology Society. Arch Pathol Lab Med. 2016;140:1267-72. [Crossref] [PubMed]
- Layfield LJ, Roy-Chowdhuri S, Baloch Z, et al. Utilization of ancillary studies in the cytologic diagnosis of respiratory lesions: The papanicolaou society of cytopathology consensus recommendations for respiratory cytology. Diagn Cytopathol 2016;44:1000-9. [Crossref] [PubMed]
- Yatabe Y, Dacic S, Borczuk AC, et al. Best Practices Recommendations for Diagnostic Immunohistochemistry in Lung Cancer. J Thorac Oncol 2019;14:377-407. [Crossref] [PubMed]
- Sauter JL, Grogg KL, Vrana JA, et al. Young investigator challenge: Validation and optimization of immunohistochemistry protocols for use on cellient cell block specimens. Cancer Cytopathol 2016;124:89-100. [Crossref] [PubMed]
- Gruchy JR, Barnes PJ, Dakin Hache KA. CytoLyt(R) fixation and decalcification pretreatments alter antigenicity in normal tissues compared with standard formalin fixation. Appl Immunohistochem Mol Morphol 2015;23:297-302. [Crossref] [PubMed]
- Wagner DG, Russell DK, Benson JM, et al. Cellient automated cell block versus traditional cell block preparation: a comparison of morphologic features and immunohistochemical staining. Diagn Cytopathol 2011;39:730-6. [Crossref] [PubMed]
- Gorman BK, Kosarac O, Chakraborty S, et al. Comparison of breast carcinoma prognostic/predictive biomarkers on cell blocks obtained by various methods: Cellient, formalin and thrombin. Acta Cytol 2012;56:289-96. [Crossref] [PubMed]
- Skoog L, Tani E. Immunocytochemistry: an indispensable technique in routine cytology. Cytopathology 2011;22:215-29. [Crossref] [PubMed]
- Shidham VB, Chang CC, Rao RN, et al. Immunostaining of cytology smears: a comparative study to identify the most suitable method of smear preparation and fixation with reference to commonly used immunomarkers. Diagn Cytopathol 2003;29:217-21. [Crossref] [PubMed]
- Roh MH, Schmidt L, Placido J, et al. The application and diagnostic utility of immunocytochemistry on direct smears in the diagnosis of pulmonary adenocarcinoma and squamous cell carcinoma. Diagn Cytopathol 2012;40:949-55. [Crossref] [PubMed]
- Kirbis IS, Maxwell P, Flezar MS, et al. External quality control for immunocytochemistry on cytology samples: a review of UK NEQAS ICC (cytology module) results. Cytopathology 2011;22:230-7. [Crossref] [PubMed]
- Fischer AH, Schwartz MR, Moriarty AT, et al. Immunohistochemistry practices of cytopathology laboratories: a survey of participants in the College of American Pathologists Nongynecologic Cytopathology Education Program. Arch Pathol Lab Med 2014;138:1167-72. [Crossref] [PubMed]
- Lindeman NI, Cagle PT, Aisner DL, et al. Updated Molecular Testing Guideline for the Selection of Lung Cancer Patients for Treatment With Targeted Tyrosine Kinase Inhibitors: Guideline From the College of American Pathologists, the International Association for the Study of Lung Cancer, and the Association for Molecular Pathology. Arch Pathol Lab Med 2018;142:321-46. [Crossref] [PubMed]
- Available online: https://www.accessdata.fda.gov/scripts/cdrh/cfdocs/cfpma/pma.cfm?id=P140025
- Lee K, Um SW, Jeong BH, et al. Triple Gene Analysis Using Samples Obtained by Endobronchial Ultrasound-guided Transbronchial Needle Aspiration. Intern Med 2016;55:3105-11. [Crossref] [PubMed]
- Zhou J, Yao H, Zhao J, et al. Cell block samples from malignant pleural effusion might be valid alternative samples for anaplastic lymphoma kinase detection in patients with advanced non-small-cell lung cancer. Histopathology 2015;66:949-54. [Crossref] [PubMed]
- Minca EC, Lanigan CP, Reynolds JP, et al. ALK status testing in non-small-cell lung carcinoma by FISH on ThinPrep slides with cytology material. J Thorac Oncol 2014;9:464-8. [Crossref] [PubMed]
- Liu L, Zhan P, Zhou X, S, et al. Detection of EML4-ALK in lung adenocarcinoma using pleural effusion with FISH, IHC, and RT-PCR methods. PLoS One 2015;10:e0117032. [Crossref] [PubMed]
- Wang W, Tang Y, Li J, et al. Detection of ALK rearrangements in malignant pleural effusion cell blocks from patients with advanced non-small cell lung cancer: a comparison of Ventana immunohistochemistry and fluorescence in situ hybridization. Cancer Cytopathol 2015;123:117-22. [Crossref] [PubMed]
- Sullivan HC, Fisher KE, Hoffa AL, et al. The role of immunohistochemical analysis in the evaluation of EML4-ALK gene rearrangement in lung cancer. Appl Immunohistochem Mol Morphol 2015;23:239-44. [Crossref] [PubMed]
- Rosenblum F, Hutchinson LM, Garver J, et al. Cytology specimens offer an effective alternative to formalin-fixed tissue as demonstrated by novel automated detection for ALK break-apart FISH testing and immunohistochemistry in lung adenocarcinoma. Cancer Cytopathol 2014;122:810-21. [Crossref] [PubMed]
- Savic S, Bode B, Diebold J, et al. Detection of ALK-positive non-small-cell lung cancers on cytological specimens: high accuracy of immunocytochemistry with the 5A4 clone. J Thorac Oncol 2013;8:1004-11. [Crossref] [PubMed]
- Tanaka H, Tone K, Hayashi A, et al. Clinical application of immunocytochemical detection of ALK rearrangement on cytology slides for detection or screening of lung adenocarcinoma. Lung Cancer 2013;80:289-92. [Crossref] [PubMed]
- Zhang C, Randolph ML, Jones KJ, et al. Anaplastic Lymphoma Kinase Immunocytochemistry on Cell-Transferred Cytologic Smears of Lung Adenocarcinoma. Acta Cytol 2015;59:213-8. [Crossref] [PubMed]
- Sholl LM, Sun H, Butaney M, et al. ROS1 immunohistochemistry for detection of ROS1-rearranged lung adenocarcinomas. Am J Surg Pathol 2013;37:1441-9. [Crossref] [PubMed]
- Vlajnic T, Savic S, Barascud A, et al. Detection of ROS1-positive non-small cell lung cancer on cytological specimens using immunocytochemistry. Cancer Cytopathol 2018;126:421-9. [Crossref] [PubMed]
- Conde E, Hernandez S, Martinez R, et al. Assessment of a New ROS1 Immunohistochemistry Clone (SP384) for the Identification of ROS1 Rearrangements in Patients with Non-Small Cell Lung Carcinoma: the ROSING Study. J Thorac Oncol 2019;14:2120-32. [Crossref] [PubMed]
- Hofman V, Rouquette I, Long-Mira E, et al. Multicenter Evaluation of a Novel ROS1 Immunohistochemistry Assay (SP384) for Detection of ROS1 Rearrangements in a Large Cohort of Lung Adenocarcinoma Patients. J Thorac Oncol 2019;14:1204-12. [Crossref] [PubMed]
- Huang RSP, Smith D, Le CH, et al. Correlation of ROS1 Immunohistochemistry With ROS1 Fusion Status Determined by Fluorescence In Situ Hybridization. Arch Pathol Lab Med 2020;144:735-41. [Crossref] [PubMed]
- Büttner R, Gosney JR, Skov BG, et al. Programmed Death-Ligand 1 Immunohistochemistry Testing: A Review of Analytical Assays and Clinical Implementation in Non-Small-Cell Lung Cancer. J Clin Oncol 2017;35:3867-76. [Crossref] [PubMed]
- Thunnissen E, Kerr KM, Herth FJ, et al. The challenge of NSCLC diagnosis and predictive analysis on small samples. Practical approach of a working group. Lung Cancer 2012;76:1-18. [Crossref] [PubMed]
- Munari E, Zamboni G, Lunardi G, et al. PD-L1 Expression Heterogeneity in Non-Small Cell Lung Cancer: Defining Criteria for Harmonization between Biopsy Specimens and Whole Sections. J Thorac Oncol 2018;13:1113-20. [Crossref] [PubMed]
- Munari E, Zamboni G, Marconi M, et al. PD-L1 expression heterogeneity in non-small cell lung cancer: evaluation of small biopsies reliability. Oncotarget 2017;8:90123-31. [Crossref] [PubMed]
- Tsao MS, Kerr KM, Dacic S, et al. IASLC Atlas of PD-L1 Immunohistochemistry Testing in Lung Cancer, 1st Ed. International Association for the Study of Lung Cancer: IASLC, 2017.
- Heymann JJ, Bulman WA, Swinarski D, et al. Programmed death-ligand 1 expression in non-small cell lung carcinoma: Comparison among cytology, small biopsy, and surgical resection specimens. Cancer Cytopathol 2017.125896-907.
- Skov BG, Skov T. Paired Comparison of PD-L1 Expression on Cytologic and Histologic Specimens From Malignancies in the Lung Assessed With PD-L1 IHC 28-8pharmDx and PD-L1 IHC 22C3pharmDx. Appl Immunohistochem Mol Morphol 2017;25:453-9. [Crossref] [PubMed]
- Munari E, Zamboni G, Sighele G, et al. Expression of programmed cell death ligand 1 in non-small cell lung cancer: Comparison between cytologic smears, core biopsies, and whole sections using the SP263 assay. Cancer Cytopathol 2019;127:52-61. [Crossref] [PubMed]
- Ilie M, Juco J, Huang L, et al. Use of the 22C3 anti-programmed death-ligand 1 antibody to determine programmed death-ligand 1 expression in cytology samples obtained from non-small cell lung cancer patients. Cancer Cytopathol 2018;126:264-74. [Crossref] [PubMed]
- Russell-Goldman E, Kravets S, Dahlberg SE, et al. Cytologic-histologic correlation of programmed death-ligand 1 immunohistochemistry in lung carcinomas. Cancer Cytopathol 2018;126:253-63. [Crossref] [PubMed]
- Noll B, Wang WL, Gong Y, et al. Programmed death ligand 1 testing in non-small cell lung carcinoma cytology cell block and aspirate smear preparations. Cancer Cytopathol 2018;126:342-52. [Crossref] [PubMed]
- Jain D, Sukumar S, Mohan A, et al. Programmed death-ligand 1 immunoexpression in matched biopsy and liquid-based cytology samples of advanced stage non-small cell lung carcinomas. Cytopathology 2018;29:550-7. [Crossref] [PubMed]
- Lozano MD, Abengozar-Muela M, Echeveste JI, et al. Programmed death-ligand 1 expression on direct Pap-stained cytology smears from non-small cell lung cancer: Comparison with cell blocks and surgical resection specimens. Cancer Cytopathol 2019;127:470-80. [Crossref] [PubMed]
- Torous VF, Rangachari D, Gallant BP, et al. PD-L1 testing using the clone 22C3 pharmDx kit for selection of patients with non-small cell lung cancer to receive immune checkpoint inhibitor therapy: are cytology cell blocks a viable option? J Am Soc Cytopathol 2018;7:133-41. [Crossref] [PubMed]
- Kawahara A, Azuma K, Sumi A, et al. Identification of non-small-cell lung cancer with activating EGFR mutations in malignant effusion and cerebrospinal fluid: rapid and sensitive detection of exon 19 deletion E746-A750 and exon 21 L858R mutation by immunocytochemistry. Lung Cancer 2011;74:35-40. [Crossref] [PubMed]
- Kawahara A, Taira T, Abe H, et al. Fixation effect of SurePath preservative fluids using epidermal growth factor receptor mutation-specific antibodies for immunocytochemistry. Cancer Cytopathol 2014;122:145-52. [Crossref] [PubMed]
- Yoshida M, Nagatomo T, Ohnishi T, et al. Detection of epidermal growth factor receptor mutations in lung adenocarcinoma cytological specimens by immunocytochemistry. Mol Clin Oncol 2017;7:981-7. [PubMed]
- Hasanovic A, Ang D, Moreira AL, et al. Use of mutation specific antibodies to detect EGFR status in small biopsy and cytology specimens of lung adenocarcinoma. Lung Cancer 2012;77:299-305. [Crossref] [PubMed]
- Bellevicine C, Bianco A, Malapelle U, et al. Performance of EGFR mutant-specific antibodies in different cytological preparations: a validation study. Cytopathology 2015;26:99-105. [Crossref] [PubMed]
- Sasaki H, Shimizu S, Tani Y, et al. Usefulness of immunohistochemistry for the detection of the BRAF V600E mutation in Japanese lung adenocarcinoma. Lung Cancer 2013;82:51-4. [Crossref] [PubMed]
- Ilie M, Long E, Hofman V, et al. Diagnostic value of immunohistochemistry for the detection of the BRAFV600E mutation in primary lung adenocarcinoma Caucasian patients. Ann Oncol 2013;24:742-8. [Crossref] [PubMed]
- Karbel HAE, Ejam SS, Naji AZ. Immunohistochemical Study Using Monoclonal VE1 Antibody Can Substitute the Molecular Tests for Apprehension of BRAF V600E Mutation in Patients with Non-small-Cell Lung Carcinoma. Anal Cell Pathol (Amst) 2019;2019:2315673.
- Gow CH, Hsieh MS, Lin YT, et al. Validation of Immunohistochemistry for the Detection of BRAF V600E-Mutated Lung Adenocarcinomas. Cancers 2019;11:866. [Crossref] [PubMed]
- Nguyen-Ngoc T, Bouchaab H, Adjei AA, et al. BRAF Alterations as Therapeutic Targets in Non-Small-Cell Lung Cancer. J Thorac Oncol 2015;10:1396-403. [Crossref] [PubMed]
- Lindeman NI, Cagle PT, Aisner DL, et al. Updated Molecular Testing Guideline for the Selection of Lung Cancer Patients for Treatment With Targeted Tyrosine Kinase Inhibitors: Guideline From the College of American Pathologists, the International Association for the Study of Lung Cancer, and the Association for Molecular Pathology. J Mol Diagn 2018;20:129-59. [Crossref] [PubMed]
- Guo R, Berry LD, Aisner DL, et al. MET IHC Is a Poor Screen for MET Amplification or MET Exon 14 Mutations in Lung Adenocarcinomas: Data from a Tri-Institutional Cohort of the Lung Cancer Mutation Consortium. J Thorac Oncol 2019;14:1666-71. [Crossref] [PubMed]
- Ricciuti B, Brambilla M, Metro G, et al. Targeting NTRK fusion in non-small cell lung cancer: rationale and clinical evidence. Med Oncol 2017;34:105. [Crossref] [PubMed]
- Hechtman JF, Benayed R, Hyman DM, et al. Pan-Trk Immunohistochemistry Is an Efficient and Reliable Screen for the Detection of NTRK Fusions. Am J Surg Pathol 2017;41:1547-51. [Crossref] [PubMed]
- Solomon JP, Linkov I, Rosado A, et al. NTRK fusion detection across multiple assays and 33,997 cases: diagnostic implications and pitfalls. Mod Pathol 2020;33:38-46. [Crossref] [PubMed]


