
Next-generation sequencing to dynamically detect mechanisms of resistance to ALK inhibitors in ALK-positive NSCLC patients: a case report
Introduction
Approximately 3–7% of non-small cell lung cancer (NSCLC) patients have a genomic rearrangement of the anaplastic lymphoma kinase (ALK) gene (1), the most prevalent fusion partner being the echinoderm microtubule-associated protein-like 4 (EML4-ALK) (2). The development of molecular targeted therapies such as tyrosine kinase inhibitors (TKIs) of the ALK locus has dramatically changed the quality of life and prognosis of ALK-positive NSCLC patients, with significantly better survival than that provided by chemotherapy [median OS approximately 50 and 12 months, respectively (3)]. However, despite the efficacy of ALK inhibitors, patients ultimately relapse within 2 to 3 years while receiving therapy (4). In order to detect molecular resistance mechanisms, tumor re-biopsy is required, although this is seldom feasible in routine clinical practice. Conversely, the analysis of liquid biopsies, can potentially overcome the aforementioned limitation, being a promising approach for identifying resistance mechanisms arising during disease progression. Resistance mutations in the ALK locus have been reported to occur in around 20% of EML4-ALK NSCLC patients treated with ALK-TKIs (5). Unlike patients with NSCLC who harbor an EGFR mutation, where the mechanism of resistance to first-line TKI treatment is mainly due to the p.Thr790Met (c.2369C>T) mutation (6), multiple resistance mutations to ALK inhibitors have been described in the ALK locus (7,8). On the other hand, distinct mutations in the ALK locus confer different sensitivities on a variety of ALK-TKIs (9,10); for example, pre-clinical evidence suggests that the p.Ile1171Asn (c.3512T>A) mutation confers resistance to crizotinib and alectinib, but not to ceritinib (5). Furthermore, the therapeutic arsenal against ALK-positive tumors has increased in recent years, with several ALK-TKIs recently being approved by the FDA (4). In routine clinical practice, these treatments are prescribed empirically upon first disease progression, rather than being based on the tumor molecular profile.
Digital PCR (dPCR), although an adequate approach for detecting and quantifying somatic mutations using liquid biopsies, can only detect a few already-known mutations at a given time, which means the method is of limited use for monitoring ALK-positive NSCLC patients. On the other hand, monitoring tumor burden by tracking single somatic mutations could be limited by tumor heterogeneity, especially in advanced stages (11). This limitation can be overcome by using novel NGS approaches that allow for simultaneous low-frequency variant calling of multiple mutations (12). We present the following case report in accordance with the CARE Guideline (13).
Case presentation
A 52-year-old Spanish man, a former smoker (10 cigarettes/day), with no significant past medical and family history, who developed cough, dysphonia and significant weight loss, with no relevant findings on physical examination, was diagnosed in June 2014 as having a poorly differentiated lung adenocarcinoma stage IV (cT2N3M1c TNM 7ª ed. with bone metastasis) by a positron emission computed tomography (CT) scan and a pathological examination. Clinico-pathological characteristics of the patient are described in Table 1.

Full table
A mutational study of the tumor biopsy was not performed in the hospital where the patient was initially diagnosed due to lack of material available for molecular analysis. Plasma samples were obtained in the Medical Oncology Department of Hospital Puerta de Hierro-Majadahonda. The patient was a participant in a research study that aimed to evaluate the clinical utility of liquid biopsies in NSCLC patients. The study protocol was approved by the Hospital Puerta de Hierro Ethics Committee (internal code PIE14/00064). The patient signed the appropriate informed consent on 9 October 2015.
The patient was initially treated with four cycles of cisplatin (75 mg/m2) plus pemetrexed (500 mg/m2) chemotherapy in August 2014 and he continued maintenance therapy with pemetrexed (500 mg/m2). Unfortunately, after 12 months of chemotherapy a CT scan revealed progressive disease (PD), manifesting as new vertebral lesions.
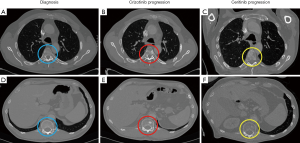
A second biopsy was performed using a Vysis LSI ALK Dual Color Break Apart FISH probe kit (Vysis, Downers Grove, IL), which revealed an EML4-ALK rearrangement. Based on this new result, the patient received 250 mg crizotinib daily from October to December 2015. In December 2015, despite good treatment tolerance, a CT scan showed a new blastic metastasis in the axial skeleton and a slight increase in the size of the lung lesion (Figure 1), and the patient was diagnosed as having a PD. A plasma sample was obtained at this time and sequenced on an Ion S5™ Sequencer (Thermo Fisher, Palo Alto, CA) using the Oncomine™ Lung cfDNA Assay NGS panel (Thermo Fisher, Palo Alto, CA) to examine circulating tumor DNA (ctDNA). The NGS study revealed the presence of the p.Gly1269Ala (c.3806G>C) resistance mutation in the ALK gene (MAF =0.88%). This mutation was confirmed by dPCR (MAF =0.42%) using a custom TaqMan® assay in conjunction with a QuantStudio® 3D Digital PCR System (Applied Biosystems, South San Francisco, CA, USA). Next, using dPCR, we analyzed the p.Gly1269Ala (c.3806G>C) mutation in a plasma sample collected previously than the former. This technique did not detect the p.Gly1269Ala (c.3806G>C) mutation, correlating with tumor response to crizotinib at that time (November 2015) (Figure 2).
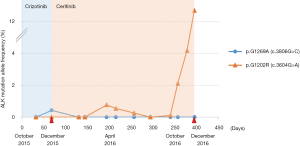
Upon progression to crizotinib treatment, the patient started treatment with ceritinib (600 mg per day). Throughout the course of this treatment there were no adverse events. Furthermore, ten plasma samples were collected and analyzed by dPCR. Partial response was assessed based on a CT scan performed in March 2016, correlating with undetectable plasma levels of p.Gly1269Ala (c.3806G>C) mutation, suggesting that ceritinib was able to eliminate the clone harboring the p.Gly1269Ala (c.3806G>C) mutation (Figure 2).
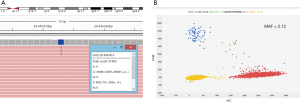
After 9 months of treatment (September 2016), the sequencing of a plasma sample using the Oncomine™ Pan-Cancer Cell-Free Assay NGS panel (Thermo Fisher, Palo Alto, CA) revealed the presence of the p.Gly1202Arg (c.3604G>A) resistance mutation in the ALK locus (MAF =1.28%) (Figure 3A), correlating with the symptomatology of the patient, who had reported a pain in his left hip. This mutation was confirmed by dPCR using another custom TaqMan® assay (MAF =2.12%) (Figure 3B).
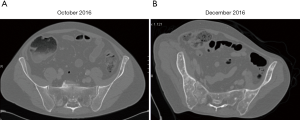
Retrospective analysis of all 12 plasma samples collected by dPCR revealed that the p.Gly1202Arg (c.3604G>A) mutation was not present during the crizotinib treatment, but appeared between the fourth and sixth months (April-June 2016) after the start of the ceritinib treatment, albeit at very low allele frequencies (MAF =0.77% and 0.26%, respectively), coinciding with a hospital admission due to pneumonia. During the last 3 months of ceritinib therapy (October-December 2016), a dPCR study showed a significant increase of p.Gly1202Arg (c.3604G>A) mutation plasma levels (MAF = 2.11% in October 2016, and 6.70% in December 2016) (Figure 2), which was associated with the presence of the PD, as identified by scintigraphy performed in October 2016. Conversely, a CT scan on the same date showed a radiological response of some of the previous bone lesions detected at ceritinib initiation, and there was no evidence of new metastasis. In December 2016 the CT scan revealed PD with new bone blast lesions, especially in the femur and ischium (Figure 4). Finally, the patient died from the PD in December 2016, having attained an overall survival of 30 months.
Discussion
In this case report, we illustrate the usefulness of plasma NGS profiling for detecting drug resistance mechanisms upon disease progression in NSCLC patients with an EML4-ALK rearrangement.
During the course of treatment, two ALK resistance mutations (p.Gly1269Ala (c.3806G>C) and p.Gly1202Arg (c.3604G>A)) were detected and quantified by NGS and dPCR. The p.Gly1269Ala (c.3806G>C) mutation has been described as a frequent event in crizotinib resistance in ALK-positive NSCLC patients, being detected in approximately 4% of cases (8). Next-generation ALK-TKIs such as ceritinib have demonstrated activity against the p.Gly1269Ala (c.3806G>C) mutation (14). In this way, plasma levels of this mutation dropped to undetectable levels soon after ceritinib treatment was initiated. Similarly, the p.Gly1202Arg (c.3604G>A) mutation occurs in 21% of patients following ceritinib treatment (5). Lorlatinib, the next-generation ALK-TKI, has demonstrated its efficacy against cells harboring the p.Gly1202Arg (c.3604G>A) mutation (15,16). Of note, the p.Gly1202Arg (c.3604G>A) resistance mutation was detected before disease progression, as ascertained by CT scan. In the same way, it has been reported that the p.Thr790Met (c.2369C>T) resistance mutation can be detected by ctDNA analysis approximately 50 days before PD is detected by CT scan in EGFR-positive NSCLC patients treated with a TKI (17,18), which suggests that ctDNA can complement imaging techniques to anticipate progression (19,20).
Technologies such as array-based dPCR or NGS allow the dynamic and quantitative analysis of somatic mutations in blood samples with high levels of sensitivity. However, dPCR can detect only a few, already-known genetic mutations in a single sample at a given time, while NGS enables the simultaneous screening of multiple mutations in multiple samples. Moreover, NGS allows to identify novel mutations. In this way the p.Gly1128Ala (c.3383G>C) mutation in ALK locus was recently identified in an ALK-positive NSCLC patient who progressed during treatment with crizotinib (21). As already mentioned, the use of dPCR is limited to monitoring a small number of mutations, and so is an unsuitable approach for screening resistance mutations in the ALK locus. However, as described in this case report, ctDNA profiling by NGS is feasible and can yield clinically useful information. Importantly, the laboratory turnaround time is short which make this a practical method for assisting oncologists in their clinical decision-making.
According to ctDNA analysis, in this case, ceritinib was able to eliminate the tumor cells that carried the resistance mutation p.Gly1269Ala (c.3806G>C), highlighting the high potency of the drug. Likewise, osimertinib, a third-generation EGFR-TKI, seems to be more potent than first- and second-generation TKIs such as erlotinib, gefitinib and afatinib, since it is more effective, in terms of survival, than standard EGFR-TKIs in first-line treatment (22). At the molecular level, unlike the first and second generations of EGFR inhibitors, in which the original EGFR-sensitizing mutation is always detected when disease progression occurs (23), osimertinib removes the tumor cells that carry both the original EGFR sensitizing mutation and the p.Thr790Met (c.2369C>T) resistance mutation in about half of the cases (24).
On the other hand, the two clones p.Gly1269Ala (c.3806G>C) and p.Gly1202Arg (c.3604G>A) were detected at different times during the course of disease, and could be imputed to different cancer lesions upon disease progression. In this way, the bone metastasis detected in the T4 and T10 vertebral lesions could be imputed to the clone harboring the p.Gly1269Ala (c.3806G>C) mutation, since these lesions were diagnosed at the same time as the mutation was detected in the blood. Moreover, both lesions showed partial remission with ceritinib treatment, in association with a decrease in the plasma levels of the p.Gly1269Ala (c.3806G>C) mutation (Figures 1,2). The ischium metastasis could be imputed to the clone harboring the p.Gly1202Arg (c.3604G>A) mutation (Figure 4). The CT scan clearly showed its occurrence to be associated with a significant increase in the plasma levels of this mutation, as assessed by dPCR (Figure 2).
In conclusion, molecular profiling of liquid biopsies using NGS is feasible and can be useful for monitoring tumor heterogeneity and clonal evolution during ALK-TKI treatment of NSCLC patients harboring EML4-ALK translocation, helping clinicians prescribe the most appropriate subsequent treatment lines, and improve the quality of life and outcome of their patients.
Acknowledgments
We thank the patient in participating in our study.
Funding: This study was supported by the Carlos III Institute of Health, the Spanish Ministry of Science and Innovation and the European Regional Development Fund (grant number: PI17/01977 and PI16/01818). ES was funded by the Consejería de Educación, Juventud y Deporte of the Comunidad de Madrid and by the Fondo Social Europeo (Programa Operativo de Empleo Juvenil, and Iniciativa de Empleo Juvenil, PEJ-2017-AI/SAL-6478).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tlcr.2020.02.07). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The patient participated in a research study that aimed to evaluate the clinical utility of liquid biopsies in NSCLC patients. The study protocol was approved by the Hospital Puerta de Hierro Ethics Committee (internal code PIE14/00064). Written informed consent was obtained from the patient for publication of this case report as well as the accompanying images.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sasaki T, Rodig SJ, Chirieac LR, et al. The biology and treatment of EML4-ALK non-small cell lung cancer. Eur J Cancer 2010;46:1773-80. [Crossref] [PubMed]
- Yang L, Ling Y, Guo L, et al. Detection of ALK translocation in non-small cell lung carcinoma (NSCLC) and its clinicopathological significance using the Ventana immunohistochemical staining method: a single-center large-scale investigation of 1,504 Chinese Han patient. Chin J Cancer Res 2016;28:495-502. [Crossref] [PubMed]
- Barrows SM, Wright K, Copley-Merriman C, et al. Systematic review of sequencing of ALK inhibitors in ALK-positive non-small-cell lung cancer. Lung Cancer (Auckl) 2019;10:11-20. [Crossref] [PubMed]
- Singhi EK, Horn L, Sequist LV, et al. Advanced Non-Small Cell Lung Cancer: Sequencing Agents in the EGFR-Mutated/ALK-Rearranged Populations. Am Soc Clin Oncol Educ Book 2019;39:e187-97. [Crossref] [PubMed]
- Gainor JF, Dardaei L, Yoda S, et al. Molecular Mechanisms of Resistance to First- and Second-Generation ALK Inhibitors in ALK-Rearranged Lung Cancer. Cancer Discov 2016;6:1118-33. [Crossref] [PubMed]
- Provencio M, Pérez-Barrios C, Barquin M, et al. Next-generation sequencing for tumor mutation quantification using liquid biopsies. Clin Chem Lab Med 2020;58:306-13. [Crossref] [PubMed]
- Doebele RC, Pilling AB, Aisner DL, et al. Mechanisms of resistance to crizotinib in patients with ALK gene rearranged non-small cell lung cancer. Clin Cancer Res 2012;18:1472-82. [Crossref] [PubMed]
- Katayama R, Shaw AT, Khan TM, et al. Mechanisms of acquired crizotinib resistance in ALK-rearranged lung Cancers. Sci Transl Med 2012;4:120ra17. [Crossref] [PubMed]
- Okada K, Araki M, Sakashita T, et al. Prediction of ALK mutations mediating ALK-TKIs resistance and drug re-purposing to overcome the resistance. EBioMedicine 2019;41:105-19. [Crossref] [PubMed]
- Li J, Huang Y, Wu M, et al. Structure and energy based quantitative missense variant effect analysis provides insights into drug resistance mechanisms of anaplastic lymphoma kinase mutations. Sci Rep 2018;8:10664. [Crossref] [PubMed]
- García-Saenz JA, Ayllón P, Laig M, et al. Tumor burden monitoring using cell-free tumor DNA could be limited by tumor heterogeneity in advanced breast cancer and should be evaluated together with radiographic imaging. BMC Cancer 2017;17:210. [Crossref] [PubMed]
- Kivioja T, Vähärautio A, Karlsson K, et al. Counting absolute numbers of molecules using unique molecular identifiers. Nat Methods 2011;9:72-4. [Crossref] [PubMed]
- Riley DS, Barber MS, Kienle GS, et al. CARE guidelines for case reports: explanation and elaboration document. J Clin Epidemiol 2017;89:218-35. [Crossref] [PubMed]
- Sakamoto H, Tsukaguchi T, Hiroshima S, et al. CH5424802, a Selective ALK Inhibitor Capable of Blocking the Resistant Gatekeeper Mutant. Cancer Cell 2011;19:679-90. [Crossref] [PubMed]
- Zou HY, Friboulet L, Kodack DP, et al. PF-06463922, an ALK/ROS1 Inhibitor, Overcomes Resistance to First and Second Generation ALK Inhibitors in Preclinical Models. Cancer Cell 2015;28:70-81. [Crossref] [PubMed]
- Fontana D, Ceccon M, Gambacorti-Passerini C, et al. Activity of second-generation ALK inhibitors against crizotinib-resistant mutants in an NPM-ALK model compared to EML4-ALK. Cancer Med 2015;4:953-65. [Crossref] [PubMed]
- Provencio M, Torrente M, Calvo V, et al. Prognostic value of quantitative ctDNA levels in non small cell lung cancer patients. Oncotarget 2017;9:488-94. [PubMed]
- Sorensen BS, Wu L, Wei W, et al. Monitoring of epidermal growth factor receptor tyrosine kinase inhibitor-sensitizing and resistance mutations in the plasma DNA of patients with advanced non-small cell lung cancer during treatment with erlotinib. Cancer 2014;120:3896-901. [Crossref] [PubMed]
- Zheng D, Ye X, Zhang MZ, et al. Plasma EGFR T790M ctDNA status is associated with clinical outcome in advanced NSCLC patients with acquired EGFR-TKI resistance. Sci Rep 2016;6:20913. [Crossref] [PubMed]
- De Carlo E, Schiappacassi M, Urbani M, et al. Therapeutic decision based on molecular detection of resistance mechanism in an ALK-rearranged lung cancer patient: a case report. Onco Targets Ther 2018;11:8945-50. [Crossref] [PubMed]
- Ai X, Niu X, Chang L, et al. Next generation sequencing reveals a novel ALK G1128A mutation resistant to crizotinib in an ALK-Rearranged NSCLC patient. Lung Cancer 2018;123:83-6. [Crossref] [PubMed]
- Soria JC, Ohe Y, Vansteenkiste J, et al. Osimertinib in untreated EGFR-Mutated advanced non-small-cell lung cancer. N Engl J Med 2018;378:113-25. [Crossref] [PubMed]
- Gelatti ACZ, Drilon A, Santini FC. Optimizing the sequencing of tyrosine kinase inhibitors (TKIs) in epidermal growth factor receptor (EGFR) mutation-positive non-small cell lung cancer (NSCLC). Lung Cancer 2019;137:113-22. [Crossref] [PubMed]
- Remon J, Caramella C, Jovelet C, et al. Osimertinib benefit in EGFR -mutant NSCLC patients with T790M -mutation detected by circulating tumour DNA. Ann Oncol 2017;28:784-90. [Crossref] [PubMed]


