
CD5 and CD6 as immunoregulatory biomarkers in non-small cell lung cancer
Introduction
The immune system’s natural ability to detect and eliminate malignant cells is currently considered the best weapon in the battle against cancer. The study of immune biomarkers is critical to diagnose, prevent and choose the appropriate immunotherapy strategy in cancer treatment. The presence, localization, and proportion of helper and especially cytotoxic T cells (CTLs) in tumour-infiltrating lymphocytes (TILs) from aggressive cancers such as non-small cell lung cancer (NSCLC) and melanoma has been associated with a favorable prognosis (1-3). In recent years, gene expression patterns and single nucleotide polymorphism studies have contributed to cancer prognosis (4). Particularly, inducible immunoregulatory genes like immune checkpoints blockers (CTLA-4 or PD-1) expressed in TILs, are among the most predictive biomarkers for cancer clinical outcome and targets for immunotherapy (5,6). Identification of new immune biomarkers is still needed to fully understand the mechanisms of immune evasion and facilitate subsequent development of novel immunotherapies.
The lymphocyte surface co-receptors CD5 and CD6 are immunomodulators involved in the development, activation, differentiation and survival of lymphocytes (7,8). They are encoded by homologous genes derived from duplication of a common ancestral gene (9), and both are constitutively expressed by all T cells and the small B1a cell subset. CD5 and CD6 are signal-transducing receptors that physically associate with the T and B cell antigen-specific clonotypic receptor (TCR and BCR, respectively) at the centre of the immune synapse (10,11). The endogenous CD6 ligands involve CD166/ALCAM (12), Galectins 1 and 3 (13) and CD318 (14), all broadly distributed on immune, epithelial, mesenchymal and/or cancer cells. In contrast, the nature of the CD5 ligand/s is ill-defined because no reported candidate (CD72, IgVH framework region, gp200, gp150, gp40-80, CD5 itself and IL-6) has been firmly validated by independent groups (7,15). Based on monoclonal antibody in vitro data, CD5 and CD6 were initially considered co-stimulatory molecules (7,8). However, analysis of CD5 and CD6 knockout mice unveiled their negative modulatory role for thymocyte (and B1a) activation and differentiation signals upon clonotypic receptor cross-linking (16-19). Interestingly, this immunomodulatory role occurs even in the absence of ligand interaction (20,21). This implies that lymphocyte function can be up- or down-regulated by CD5 and CD6 expression. Accordingly, anergic T and B cells show upregulated surface CD5 expression (22,23). CD5 and CD6 expression also parallels TCR/CD3 levels and is predictive of TCR avidity and survival of T cells (24-27). Moreover, regulation of CD5 expression by TCR engagement has been reported in peripheral T cells (28). Regarding antitumour responses, in situ regulation of CD5 expression by CTLs is thought to adapt their sensitivity to intra-tumour peptide-major histocompatibility complex (p-MHC) levels (29). Indeed, CTL clones from lung cancer patients show that CD5 expression is inversely proportional to their anti-tumour cytolytic activity, preventing activation-induced cell death (AICD) during T cell overactivation (29).
CD5 and CD6’s modulation of lymphocyte activation and survival supports the hypothesis that their overall intra-tumour expression levels alter the anti-tumour immune response and can be therefore used as prognosis biomarkers in cancer. Accordingly, this study investigated the prognostic value of CD5 and CD6 gene expression in a cohort of 186 patients with resectable NSCLC. The results show that high intra-tumour levels of both CD5 and CD6 associate to better prognosis as measured by overall survival and relapse-free survival. This was validated in silico using NSCLC biopsy information from The Cancer Genome Atlas (TCGA) database.
Methods
NSCLC patient cohort and sample collection
The training cohort included 186 patients with resected and non-pretreated stage I to IIIA NSCLC from Consorcio Hospital General Universitario de Valencia. Between 2004 and 2017, 186 fresh-frozen tumour and normal tissue samples were obtained from surgical resection and preserved in RNAlater® (Applied Biosystems, USA). Patients who had received neoadjuvant treatment and those with a follow-up shorter than 1 month were excluded. This study abides by the Declaration of Helsinki (as revised in 2013) and was approved by the institutional review board. All patients had signed the informed consent prior to sample collection.
Real-time PCR analysis of NSCLC patients
RNA was isolated using TRI Reagent® (Sigma, USA) and retrotranscribed using High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, USA) in a MasterCycler® thermocycler (Eppendorf, Germany) following the manufacturer’s instructions. CD3D, CD3E, CD4, CD5, CD6 and CD8 gene expression was analysed by RTqPCR using Taqman® hydrolysis probes and Taqman® Gene Expression Master Mix (Applied Biosystems, USA) in a LightCycler 480 thermocycler (Roche, Switzerland).
Relative gene expression was calculated by Pfaffl formula, taking into consideration expression differences between normal and tumour tissue, as well as RTqPCR efficiencies of each TaqMan® assay. Reference gene expression corresponds to the geometric mean of ACTB, CDKN1B and GUSB, endogenous controls used after evaluation with GeNorm software (30,31).
Immunohistochemistry (IHC)
CD5 and CD6 relative gene expression data were normalized against CD4+ and CD8+ lymphocyte infiltration in NSCLC samples (n=60). To do so, CD4 and CD8 expression was evaluated in 60 Formalin-Fixed Paraffin Embedded (FFPE) samples using a Dako Autostainer Link 48 and the Dako EnVisionTM FLEX detection system (Dako, Canada). After section drying and antigen dewaxing in a PT Link instrument, the endogenous peroxidase activity was quenched with peroxidase blocking reagent in the Autostainer Link 48 instrument. Immunostaining was carried out with Dako FLEX Ready to-Use format for CD4 (Clone 4B12, Dako) and CD8 (Clone C8/144B, Dako). Briefly, a detection system chromogen (3,3'-diaminobenzidine, DAB) was used after primary antibody incubation, followed by washing and counterstaining of sections with hematoxylin, dehydration and mounting. CD4+ and CD8+ lymphocytes were counted in 10 high power fields (HPF) (magnification ×400) for tumour areas. Negative controls and normal human tonsil positive control tissue were included. IHC staining quantification was performed by two independent evaluators.
TCGA database search for CD5 and CD6 mRNA expression levels in NSCLC biopsies
Online information available at TCGA database (https://cancergenome.nih.gov/) for NSCLC patients was downloaded and used as independent validation cohorts. Patients with resected NSCLC and available gene expression data for CD5 and CD6 in normal and tumour tissue samples were selected (n=97). Patients who had received neoadjuvant treatment or with follow-up shorter than 1 month were excluded.
Statistical analyses
Relative gene expression was dichotomized using median as a cut-off value. Non-parametric tests were used for correlations between clinico-pathological and analytical variables. Survival analyses were performed considering relapse-free survival (RFS) and overall survival (OS). RFS spans from surgery to relapse or exitus dates, and OS from surgery to exitus dates, following the Response Evaluation Criteria in Solid Tumours (RECIST). For patients who neither relapsed nor died, the last recorded follow-up was considered. Gene prognostic value was assessed using Kaplan-Meier curves (Log-rank test) and univariate Cox regression analyses, followed by a multivariate Cox regression analysis, using all significant variables to establish independent prognostic biomarkers. Statistical analyses were performed using the Statistical Package for Social Sciences (SPSS) 15.0 software (Chicago, USA), considering significant P<0.05.
Results
Analysis of CD5 and CD6 expression in NSCLC samples from training cohort
The NSCLC training cohort was composed of 186 patients (see Table S1 for demographic and clinicopathological data), mainly men (85.0%) and current or former smokers (88.7%). Histology was squamous cell carcinoma in 47.9% and adenocarcinoma in 41.4% of all cases. During the follow-up (median 34.2 months), 85 patients relapsed (45.7%) and 91 died (48.9%). Non-parametric tests were conducted to determine an association of relative gene expression with clinicopathological variables.
Full table
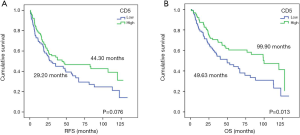
The survival analyses revealed that high CD5 expression had significant biomarker prognostic value for OS (OS, 49.63 vs. 99.90 months, P=0.013). Furthermore, a statistical trend toward significant RFS was detected (RFS, 29.20 vs. 44.30 months, P=0.076) (Table 1; Figure 1). In contrast, similar analyses did not provide significant results for CD6 expression (Table 1). Multivariate Cox regression analysis revealed high CD5 expression as a potential prognostic biomarker for OS in resected NSCLC patients (HR=0.554; 95% CI, 0.360–0.853; P=0.007) (Table 2).

Full table

Full table
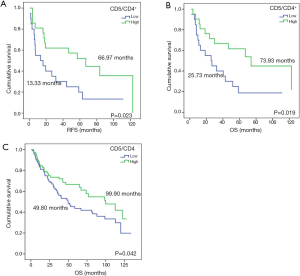
CD5 and CD6 are constitutively expressed lymphocytic receptors, whose expression can be regulated during lymphocyte development and activation events (32,33). Thus, mRNA level correlation to lymphocyte infiltration or to up-regulation in infiltrating lymphocytes was assessed. To this end, CD5 and CD6 gene expression was normalized to CD4+ and CD8+ lymphocyte infiltration by IHC, and the CD5/CD4+, CD5/CD8+, CD6/CD4+ and CD6/CD8+ ratios were calculated in 60 samples. Survival analysis indicated that patients with higher CD5/CD4+ ratios had significantly improved prognosis (RFS, 13.33 vs. 66.97 months, P=0.023; OS, 25.73 vs. 73.93 months, P=0.019) (Figure S1). Furthermore, a tendency for improved RFS and OS was observed for CD5/CD8+, CD6/CD4+ and CD6/CD8+ ratios (Table S2).

Full table
In order to confirm these results, relative CD5 and CD6 expression using RTqPCR was normalized against CD4 and CD8 expression in 186 NSCLC patients from our training cohort. Survival analysis confirmed that higher CD5/CD4 expression was associated to improved OS (OS, 49.80 vs. 99.90 months, P=0.042) (Figure S1), but no significant results were obtained neither for the rest of the ratios nor for RFS (Table S3). Additionally, and taking into account that the Spearman’s test correlated CD5 and CD6 to CD3D and CD3E, the CD5/CD3D, CD5/CD3E, CD6/CD3D and CD6/CD3E ratios were evaluated but showed no significant association to prognosis (Table S3).
Full table
In silico analysis of CD5 and CD6 expression from NSCLC TCGA database
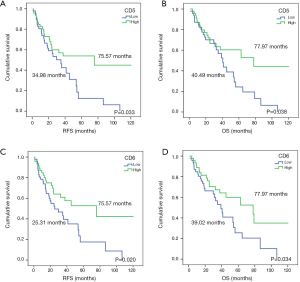
Online information of 97 NSCLC patients from the TCGA database was used as a validation cohort. Their median follow-up was 27.61 months, time in which 27 (32.1%) patients had relapsed and 46 (47.4%) were exitus. The statistical analysis confirmed that higher expression of both CD5 and CD6 improved prognosis in NSCLC patients, as it was associated with increased RFS (34.98 vs. 75.57 months, P=0.033; 25.31 vs. 75.57 months, P=0.020, respectively) and OS (40.49 vs. 77.97 months, P=0.038; 39.02 vs. 77.97 months, P=0.034, respectively) (Table 3; Figure 2). Multivariate analysis did not provide significant results on account of the small sample size (n=97). In all, our data supports that higher CD5 expression in early-stage NSCLC patients associates to increased OS. Regarding CD6, the validation cohort indicates a NSCLC biomarker potential that should be explored further.

Full table
Discussion
The presence of immune cells in the tumour microenvironment plays a key role in NSCLC and melanoma prognosis (34,35). Our work provides a prognostic value for intratumour CD5 and CD6 mRNA expression, two lymphocyte co-receptors involved in modulation of T (and B1a) cell development, activation, differentiation and survival (7,8). High CD5 expression associates to favourable prognosis in 186 fresh-frozen samples of a NSCLC training cohort, validated further with online data from the NSCLC TCGA database. The analysis of CD6 expression in NSCLC tumour samples reveals similar associations. Furthermore, CD5 and CD6 expression correlate to the number of CD4+ and CD8+ lymphocytes and these ratios indicate better prognosis. Importantly, better prognosis also correlates with CD5 overexpression by TILs.
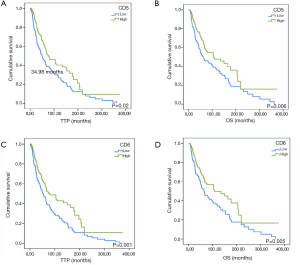
In order to explore the prognostic potential of CD5 and CD6 expression in other cancer types, melanoma information from the TCGA database was analyzed. As illustrated in Figure S2, better survival rate (“alive” group) presented significantly higher CD5 and CD6 expression, together with other T cell (CD3, CD4, CD8) and B cell (CD19) markers. There was also a significant overall difference rate between the “alive” and “dead” groups regarding time to progression (TTP) and OS considering high vs. low CD5 (Figure S3A,B) or CD6 expression (Figure S3C,D).
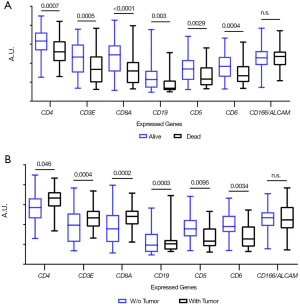
In addition, patients with better survival and tumour regression (no tumour in last follow-up) showed higher CD5 and CD6 expression, but lower CD3, CD4, CD8, CD19 levels (Figure S2B), in support of infiltrating lymphocytes over-expressing CD5 and CD6 with better prognosis in melanoma.
Tumour microenvironment genetics has identified inhibitory or stimulatory lymphocyte accessory molecules (e.g., PD-1/CD279, CTLA-4, LAG-3, TIM-3 and 4-1BB/CD137) that can be regulated to modulate T-cell activation and survival. Interestingly, these biomarkers distinguish tumour-specific T cells from unrelated T cells in the tumour infiltrate (36,37). This is illustrated by PD-1—an inhibitory receptor inducibly expressed on activated T cells—as a marker of the tumour-reactive CD8+ T cell fraction in melanoma tumours (37) and of high avidity CD8+ T cells specific for Melan-A (38) or neoantigens (39). PD-1 expression is related to TCR signal strength, and thus to the functional avidity of specific T cells, underlining the complex significance of PD-1 expression on tumour-specific T cells (38).
Similar to PD-1 and CTLA-4, the CD5 and CD6 co-receptors act as negative regulators of T cell activation (8,40). CD5- and CD6-deficient thymocytes are hyper-reactive to TCR/CD3 cross-linking (16,18). Surface CD5 and CD6 expression is set in the thymus, and parallels TCR/CD3, becoming predictive of TCR avidity (16,18,19,24,25). Thus, thymocytes binding self-peptide-MHC (self-pMHC) with high affinity consequently deliver strong TCR-mediated activation signals, and express higher CD5 and CD6 surface levels to overcome negative selection. Surface CD5 and CD6 promote thymocyte survival by different mechanisms (25,41). Post-positively selected (peripheral) CD4+ and CD8+ T cells with high CD5/CD6 expression (CD5/6hi) respond to foreign peptides with increased activation and survival (42-44). In other words, T cells with TCRs of stronger avidity for self-pMHC dominate in responses to foreign antigens and accumulate in aging individuals, revealing that positive selection contributes to effective immunity (42). This would be in line with our finding that high intratumour CD5 (and CD6) expression correlates with better NSCLC and Melanoma prognosis. Accordingly, higher intratumour CD5 (and CD6) levels reflect infiltrating T cells with higher avidity for tumour antigens and more resistant to activation-induced cell death (29).
Self-pMHC contact modulates CD5 expression, survival and homeostatic proliferation of naïve T cells in the periphery (28,45) suggesting modulation of CD5 on TILs at the tumour site. Indeed, in situ CD5 expression adaptation to pMHC levels of TILs from a lung carcinoma patient has been reported (46). The decreased MHC class I expression, observed in tumour escape from CD8+ T-cell killing, would induce TILs down-modulation of CD5 to prevent tumour evasion. In line with this, CD5lo CTL clones from lung carcinoma patients displayed higher tumour-specific cytotoxicity than CD5hi clones and increased susceptibility to tumour-induced AICD (29). This is in agreement with CD5’s prevention of AICD by negatively regulating T-cell activation (47). Higher AICD susceptibility of CD5lo CTLs would explain the transient control of tumour growth observed in CD5-deficient mice challenged with melanoma (48). Our work demonstrates that protection of CD5-deficient T cells from AICD by adenoviral-mediated expression of soluble Fas-Fc results in reduced melanoma growth (48). The latter may reflect a therapeutic strategy for patients showing low intratumour CD5 mRNA levels. In contrast, potentiation of TILs efficiency in tumours with high CD5 expression should include CD5 and Fas-FasL blocking strategies. A necessary CD5 blockade is supported by improved anti-tumour responses in mice expressing the soluble human CD5 (shCD5) transgene or injected with recombinant shCD5 (49,50).
Recent IHC analyses in small series (n=30) of untreated advanced-stage NSCLC patients concluded that intra-tumour high CD3+ and low CD5+ infiltrates associate to poor prognosis (51). Melanoma patient genetics has revealed that the hypofunctional CD5 haplotype (isoform Pro224-Ala471, a poor down-regulator of TCR/CD3-mediated T-cell activation) associates to better survival (29).
This is the first study of CD6 expression in tumour-resected samples from early-stage NSCLC patients. No significance has been obtained in our NSCLC training cohort, but higher CD6 expression correlates with improved RFS and OS in our analyses of NSCLC and melanoma TCGA data. There is little information on the function of CD6. However, the homology between CD5 and CD6, may enable some degree of functional redundancy. Knockout mice show that both receptors share modulatory roles in thymocyte activation (negative) and survival (positive) (16,18,19). Moreover, Nur-77 levels -indicative of TCR signaling strength- are elevated in CD6hi compared to CD6lo peripheral T cells (18). CD6lo/neg peripheral T cell populations are less responsive to T-cell activators, more susceptible to apoptosis and enriched in regulatory T cells (Treg) (27,52). High TCR avidity and survival of CD6hi T cells would be compatible with high intratumour CD6 expression and favourable cancer prognosis, suggesting its biomarker potential only awaits confirmation.
In order to determine if CD5 expression association to favourable prognosis is due to higher lymphocyte infiltration, CD5 and CD6 ratios to CD4 and CD8 were calculated. Our results confirm their prognostic value and support that higher CD5 expression and lymphocyte infiltration associate to increased antitumour immune responses and improved patient prognosis in early-stage NSCLC.
Conclusions
This study points to a positive prognostic role for two lymphocyte inhibitory co-receptors, CD5 and CD6, in early-stage NSCLC (and in Melanoma). This conclusion is compatible with high surface levels of both CD5 and CD6 associated to TCR avidity and resistance to AICD. This evidence suggests that CD5 and CD6, along with other checkpoint inhibitors (e.g., PD-1 and CTLA-4), may be additional markers of tumour-specific T cells. Further studies deciphering the exact role of CD5, CD6 and their ligands in cancer would benefit patient stratification for personalized immunotherapies and development of new and more efficient strategies.
Acknowledgments
The authors thank Marcos Isamat for critical edition of the manuscript.
Funding: This work was supported by Spanish Health Institute Carlos III (ISCII, Fondo de Investigación Sanitaria; PI15-00753 to RS and P118/00226 to EJ and CC), Spanish Ministry of Economy, Industry and Competitiveness (MINECO, Plan Nacional de I+D+i; SAF2016-80535-R to FL) -co-financed by European Development Regional Fund “A way to achieve Europe” ERDF; Worldwide Cancer Research (14-1275 to FL), Fundació La Marató TV3 (201319-30-31-32 to FL and YS), and Agència de Gestió d’Ajuts Universitaris i de Recerca (AGAUR) from Generalitat de Catalunya (2017/SGR/1582 to FL). AM is recipient of a PhD scholarship from Asociación Española Contra el Cáncer (AECC) Scientific Foundation and Junta Provincial Asociada de Valencia AECC; IS, FA and EC are recipients of fellowships from Fundação para a Ciência e a Tecnologia (SFRH/ BD/75738/2011), Sara Borrell Program from ISCIII (CD15/00016), and European Community Seventh Framework Program (BIOTRACK, FP7/2007/2013; 229673), respectively.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tlcr-19-445). RR serves as an unpaid editorial board member of Translational Lung Cancer Research from Jun 2019 to May 2022. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was approved by institutional ethics board of Hospital General Universitario de Valencia (Approval date: 28th May 2015). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013) and all patients had signed the informed consent prior to the collection of their biological samples.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Fortes C, Mastroeni S, Mannooranparampil TJ, et al. Tumor-infiltrating lymphocytes predict cutaneous melanoma survival. Melanoma Res 2015;25:306-11. [Crossref] [PubMed]
- Reynders K, De Ruysscher D. Tumor infiltrating lymphocytes in lung cancer: a new prognostic parameter. J Thorac Dis 2016;8:E833-5. [Crossref] [PubMed]
- Lee N, Zakka LR, Mihm MC, et al. Tumour-infiltrating lymphocytes in melanoma prognosis and cancer immunotherapy. Pathology 2016;48:177-87. [Crossref] [PubMed]
- Baxevanis CN, Anastasopoulou EA, Voutsas IF, et al. Immune biomarkers: how well do they serve prognosis in human cancers? Expert Rev Mol Diagn 2015;15:49-59. [Crossref] [PubMed]
- Kerr KM, Tsao MS, Nicholson AG, et al. Programmed Death-Ligand 1 Immunohistochemistry in Lung Cancer: In what state is this art? J Thorac Oncol 2015;10:985-9. [Crossref] [PubMed]
- Usó M, Jantus-Lewintre E, Bremnes RM, et al. Analysis of the immune microenvironment in resected non-small cell lung cancer: the prognostic value of different T lymphocyte markers. Oncotarget 2016;7:52849-61. [Crossref] [PubMed]
- Consuegra-Fernández M, Aranda F, Simões I, et al. CD5 as a Target for Immune-Based Therapies. Crit Rev Immunol 2015;35:85-115. [Crossref] [PubMed]
- Santos RF, Oliveira L, Carmo AM, Tuning T. Cell Activation: The Function of CD6 At the Immunological Synapse and in T Cell Responses. Curr Drug Targets 2016;17:630-9. [Crossref] [PubMed]
- Lecomte O, Bock JB, Birren BW, et al. Molecular linkage of the mouse CD5 and CD6 genes. Immunogenetics 1996;44:385-90. [Crossref] [PubMed]
- Brossard C, Semichon M, Trautmann A, et al. CD5 inhibits signaling at the immunological synapse without impairing its formation. J Immunol 2003;170:4623-9. [Crossref] [PubMed]
- Gimferrer I, Calvo M, Mittelbrunn M, et al. Relevance of CD6-mediated interactions in T cell activation and proliferation. J Immunol 2004;173:2262-70. [Crossref] [PubMed]
- Bowen MA, Patel DD, Li X, et al. Cloning, mapping, and characterization of activated leukocyte-cell adhesion molecule (ALCAM), a CD6 ligand. J Exp Med 1995;181:2213-20. [Crossref] [PubMed]
- Escoda-Ferran C, Carrasco E, Caballero-Baños M, et al. Modulation of CD6 function through interaction with Galectin-1 and -3. FEBS Lett 2014;588:2805-13. [Crossref] [PubMed]
- Enyindah-Asonye G, Li Y, Ruth JH, et al. CD318 is a ligand for CD6. Proc Natl Acad Sci 2017;114:E6912-21. [Crossref] [PubMed]
- Masuda K, Ripley B, Nyati KK, et al. Arid5a regulates naive CD4 + T cell fate through selective stabilization of Stat3 mRNA. J Exp Med 2016;213:605-19. [Crossref] [PubMed]
- Tarakhovsky A, Kanner SB, Hombach J, et al. A role for CD5 in TCR-mediated signal transduction and thymocyte selection. Science 1995;269:535-7. [Crossref] [PubMed]
- Bikah G, Carey J, Ciallella JR, et al. CD5-mediated negative regulation of antigen receptor-induced growth signals in B-1 B cells. Science 1996;274:1906-9. [Crossref] [PubMed]
- Orta-Mascaró M, Consuegra-Fernández M, Carreras E, et al. CD6 modulates thymocyte selection and peripheral T cell homeostasis. J Exp Med 2016;213:1387-97. [Crossref] [PubMed]
- Li Y, Singer NG, Whitbred J, et al. CD6 as a potential target for treating multiple sclerosis. Proc Natl Acad Sci U S A 2017;114:2687-92. [Crossref] [PubMed]
- Bhandoola A, Bosselut R, Yu Q, et al. CD5-mediated inhibition of TCR signaling during intrathymic selection and development does not require the CD5 extracellular domain. Eur J Immunol 2002;32:1811-7. [Crossref] [PubMed]
- Oliveira MI, Gonçalves CM, Pinto M, et al. CD6 attenuates early and late signaling events, setting thresholds for T-cell activation. Eur J Immunol 2012;42:195-205. [Crossref] [PubMed]
- Stamou P, de Jersey J, Carmignac D, et al. Chronic exposure to low levels of antigen in the periphery causes reversible functional impairment correlating with changes in CD5 levels in monoclonal CD8 T cells. J Immunol 2003;171:1278-84. [Crossref] [PubMed]
- Hippen KL, Tze LE, Behrens TW. CD5 maintains tolerance in anergic B cells. J Exp Med 2000;191:883-90. [Crossref] [PubMed]
- Azzam HS, Grinberg A, Lui K, et al. CD5 expression is developmentally regulated by T cell receptor (TCR) signals and TCR avidity. J Exp Med 1998;188:2301-11. [Crossref] [PubMed]
- Singer NG, Fox DA, Haqqi TM, et al. CD6: expression during development, apoptosis and selection of human and mouse thymocytes. Int Immunol 2002;14:585-97. [Crossref] [PubMed]
- Kassiotis G, Zamoyska R, Stockinger B. Involvement of Avidity for Major Histocompatibility Complex in Homeostasis of Naive and Memory T Cells. J Exp Med 2003;197:1007-16. [Crossref] [PubMed]
- Carrasco E, Escoda-Ferran C, Climent N, et al. Human CD6 Down-Modulation following T-Cell Activation Compromises Lymphocyte Survival and Proliferative Responses. Front Immunol 2017;8:769. [Crossref] [PubMed]
- Smith K, Seddon B, Purbhoo MA, et al. Sensory adaptation in naive peripheral CD4 T cells. J Exp Med 2001;194:1253-61. [Crossref] [PubMed]
- Tabbekh M, Mokrani-Hammani M, Bismuth G, et al. T-cell modulatory properties of CD5 and its role in antitumor immune responses. Oncoimmunology 2013;2:e22841. [Crossref] [PubMed]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 2001;29:e45. [Crossref] [PubMed]
- Vandesompele J, De Preter K, Pattyn F, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 2002;3:research0034.1-research0034.11.
- Arman M, Aguilera-Montilla N, Mas V, et al. The human CD6 gene is transcriptionally regulated by RUNX and Ets transcription factors in T cells. Mol Immunol 2009;46:2226-35. [Crossref] [PubMed]
- Arman M, Calvo J, Trojanowska ME, et al. Transcriptional Regulation of Human CD5: Important Role of Ets Transcription Factors in CD5 Expression in T Cells. J Immunol 2004;172:7519-29. [Crossref] [PubMed]
- Thomas NE, Busam KJ, From L, et al. Tumor-Infiltrating Lymphocyte Grade in Primary Melanomas Is Independently Associated With Melanoma-Specific Survival in the Population-Based Genes, Environment and Melanoma Study. J Clin Oncol 2013;31:4252-9. [Crossref] [PubMed]
- Usó M, Jantus-Lewintre E, Calabuig-Fariñas S, et al. Analysis of the prognostic role of an immune checkpoint score in resected non-small cell lung cancer patients. Oncoimmunology 2016;6:e1260214. [Crossref] [PubMed]
- Pu X, Hildebrandt MAT, Lu C, et al. Inflammation-Related Genetic Variations and Survival in Patients With Advanced Non-Small Cell Lung Cancer Receiving First-Line Chemotherapy. Clin Pharmacol Ther 2014;96:360-9. [Crossref] [PubMed]
- Gros A, Robbins PF, Yao X, et al. PD-1 identifies the patient-specific CD8+ tumor-reactive repertoire infiltrating human tumors. J Clin Invest 2014;124:2246-59. [Crossref] [PubMed]
- Simon S, Vignard V, Florenceau L, et al. PD-1 expression conditions T cell avidity within an antigen-specific repertoire. Oncoimmunology 2015;5:e1104448. [Crossref] [PubMed]
- Gros A, Parkhurst MR, Tran E, et al. Prospective identification of neoantigen-specific lymphocytes in the peripheral blood of melanoma patients. Nat Med 2016;22:433-8. [Crossref] [PubMed]
- Freitas CM, Johnson D, Weber K. T Cell Calcium Signaling Regulation by the Co-Receptor CD5. Int J Mol Sci 2018;19:1295. [Crossref] [PubMed]
- Mier-Aguilar CA, Cashman KS, Raman C, et al. CD5-CK2 Signaling Modulates Erk Activation and Thymocyte Survival. Baldwin TA, editor. PLoS One 2016;11:e0168155.
- Mandl JN, Monteiro JP, Vrisekoop N, et al. T cell-positive selection uses self-ligand binding strength to optimize repertoire recognition of foreign antigens. Immunity 2013;38:263-74. [Crossref] [PubMed]
- Persaud SP, Parker CR, Lo WL, et al. Intrinsic CD4+ T cell sensitivity and response to a pathogen are set and sustained by avidity for thymic and peripheral complexes of self peptide and MHC. Nat Immunol 2014;15:266-74. [Crossref] [PubMed]
- Fulton RB, Hamilton SE, Xing Y, et al. The TCR’s sensitivity to self peptide-MHC dictates the ability of naive CD8+ T cells to respond to foreign antigens. Nat Immunol 2015;16:107-17. [Crossref] [PubMed]
- Ernst B, Lee DS, Chang JM, et al. The peptide ligands mediating positive selection in the thymus control T cell survival and homeostatic proliferation in the periphery. Immunity 1999;11:173-81. [Crossref] [PubMed]
- Dorothée G, Vergnon I, El Hage F, et al. In situ sensory adaptation of tumor-infiltrating T lymphocytes to peptide-MHC levels elicits strong antitumor reactivity. J Immunol 2005;174:6888-97. [Crossref] [PubMed]
- Axtell RC, Xu L, Barnum SR, et al. CD5-CK2 binding/activation-deficient mice are resistant to experimental autoimmune encephalomyelitis: protection is associated with diminished populations of IL-17-expressing T cells in the central nervous system. J Immunol 2006;177:8542-9. [Crossref] [PubMed]
- Tabbekh M, Franciszkiewicz K, Haouas H, et al. Rescue of Tumor-Infiltrating Lymphocytes from Activation-Induced Cell Death Enhances the Antitumor CTL Response in CD5-Deficient Mice. J Immunol 2011;187:102-9. [Crossref] [PubMed]
- Fenutría R, Martinez VG, Simões I, et al. Transgenic expression of soluble human CD5 enhances experimentally-induced autoimmune and anti-tumoral immune responses. PLoS One 2014;9:e84895. [Crossref] [PubMed]
- Simões IT, Aranda F, Carreras E, et al. Immunomodulatory effects of soluble CD5 on experimental tumor models. Oncotarget 2017;8:108156-69. [Crossref] [PubMed]
- Dirican N, Karakaya YA, Günes S, et al. Association of Intratumoral Tumor Infiltrating Lymphocytes and Neutrophil-to- Lymphocyte Ratio Are an Independent Prognostic Factor in Non-Small Cell Lung Cancer. Clin Respir J 2017;11:789-96. [Crossref] [PubMed]
- Garcia Santana CA, Tung JW, Gulnik S. Human treg cells are characterized by low/negative CD6 expression. Cytometry A 2014;85:901-8. [Crossref] [PubMed]


