
Comprehensive serial biobanking in advanced NSCLC: feasibility, challenges and perspectives
Introduction
With about 1.8 million new cases worldwide each year, non-small cell lung cancer (NSCLC) accounts for over 10% of newly diagnosed malignancies and is the leading cause of cancer-related deaths (1). Most NSCLC patients present with metastases, at first diagnosis. The median life expectancy is below 1.5 years even with palliative chemotherapy, since therapy responses are observed in less than one third of the cases and usually last only for a few months (2). A major improvement has been the application of tyrosine kinase inhibitors (TKI), which result in response and overall survival (OS) rates at least twice higher than the traditional chemotherapy (3-5). However, not every NSCLC patient is eligible for these targeted therapies. Only tumors driven by the corresponding genetic alterations respond to EGFR, ALK, BRAF or ROS1 inhibitors. These types of tumors account for up to 15–20% of all NSCLC cases. For immuno-oncologic (IO) therapies, patient selection currently requires not only the absence of treatable drivers, but also the determination of PD-L1 expression on tumor cells. The latter guides the decisions of optimal use of immune checkpoint inhibitors alone or in combination with other options (6,7). The impending wide adoption of tumor mutational burden (TMB) as an additional, more accurate IO biomarker in the near future will improve therapeutic decision-making for NSCLC patients, but also increase biomaterial demand (8). Similar holds true for plasma ctDNA assays, which are increasingly used as non-invasive, “liquid biopsies” to monitor tumor load and identify baseline or secondary actionable genetic alterations in TKI-, IO- or chemotherapy-treated NSCLC (9-12). Analyses of the Phase III MYSTIC trial data recently demonstrated an apparent association between high blood TMB and response to IO (13).
Currently, tissue rebiopsies at disease progression remain the gold standard. They are necessary in case of a negative “liquid biopsy”, as encountered for example with ctDNA “non-shedders” (12), and are more informative regarding specific secondary tumor changes, for example transformation to small-cell lung cancer (14) or MET amplification in TKI-resistant EGFR+ tumors (15). At all events, availability of blood samples is important for the accuracy of all next-generation sequencing analyses as well as ctDNA analytics in oncology, which should ideally filter out germline and hematopoietic cell mutations (16,17).
In order to support individualized decision-making in patient care and to develop a platform for current and future research projects, the German Centre for Lung Research (DZL) initiated the assembly of a prospective NSCLC patient cohort in 2012. Here we describe the experience, results, challenges and opportunities with this endeavor after 5 years.
Methods
Patient recruitment
Prospective biobanking of NSCLC patients’ materials was performed after informed consent as approved by the ethics committee of Heidelberg University (S-048/2012). Inclusion criteria were inoperable stage IIIB/IV NSCLC according to TNM7, age >40 years and ECOG performance status 0–2. Screening for candidate patients prior to biopsy confirmation was performed by dedicated study nurses (two fulltime positions), who attended the radiologic conferences, checked the clinic and ward patient lists and consulted responsible physicians daily.
Sample collection
For consenting patients, baseline tissue samples were collected at the time of initial biopsy for establishing the diagnosis of NSCLC, either by bronchoscopy or under CT-guidance, or by surgery. At least two to three tumor cores were formalin-fixed and dedicated to routine diagnostics, while additional cores were immediately snap-frozen and stored at −80 °C in the Lung Biobank Heidelberg [LBBH, member of the accredited Tissue Bank of the National Centre for Tumor Diseases (NCT) Heidelberg]. The collection of snap-frozen tissue was performed only in the context of the scheduled rebiopsy procedure due to disease progression.
Complementary serial blood samples were collected at the time of initial biopsy, during subsequent follow-up visits and with every rebiopsy. For patients receiving chemotherapy, longitudinal blood samples were drawn at each restaging, i.e., every 8–12 weeks, while for IO- and TKI-treated patients the intervals were more frequent, namely 2–3 and 4–8 weeks, respectively. Peripheral blood was collected and processed within 1 hour with serum aliquots, plasma aliquots and the buffy coat stored separately at −80 °C for further analyses.
Comprehensive collection of clinical data
In order to ensure detailed clinical annotation, several questionnaires were surveyed by specially trained study nurses. The ad hoc developed clinical report form (CRF) captured basic demographic characteristics including ethnicity, parameters of lung function testing, patient´s own and family history of genetic and cancerous diseases, detailed smoking history including time frames and devices, comprehensive assessment of toxic exposure in private and professional life, diagnosis-related symptoms, results of histopathological and molecular analysis, results of imaging studies including TNM stage, comorbidities and co-medications. In addition, patients were asked to complete standardized, validated questionnaires concerning depression and anxiety (PHQ_4), stress (distress thermometer), comorbidities (Charlson Comorbidity Index) and nutritional status (Minimal Nutritional Assessment) at baseline, restaging and rebiopsy. Disease remission and survival status were actively monitored during follow-up visits for the patients who continued in-house treatment, and by contacting patients or local physicians for those who were treated externally.
Nucleic acid isolation
Nucleic acid isolation from the collected biopsies followed a standard procedure. First, samples were weighted, and then 10–15 tumor cryosections (10–15 µm each) were prepared from each specimen. One section from the centre of each biopsy was stained with hematoxylin and eosin (H&E) and reviewed by an experienced pulmonary pathologist to determine the content of tumor cells vs. non-neoplastic cells, stroma and necrotic areas in the whole sample (18). Frozen sections of tumor were then homogenized with the TissueLyser mixer-mill disruptor (25 sec, 25 Hz, Qiagen, Hilden, Germany), followed by extraction of total RNA and genomic DNA using an AllPrep DNA/RNA/miRNA Universal Kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. RNA and DNA were quantified with the NanoDrop ND-1000 Spectrophotometer (NanoDrop Technologies, Wilmington, MA, USA), and RNA quality was assessed with the Agilent 2100 Bioanalyzer using the RNA 6000 Nano Kit (Agilent Technologies, Boeblingen, Germany).
Results
Feasibility
In order to estimate the proportion of patients with metastatic NSCLC amenable to enrolment, we systematically documented the matching of inclusion criteria (inoperable stage IIIB/IV NSCLC according to TNM7, age >40 years and ECOG performance status 0–2) in the screened patients between June 2016 and September 2017. Overall, 172 patients with metastatic NSCLC who referred to biopsy for primary diagnosis were screened and 39 cases (23%) were included into this cohort. The reasons for exclusion are shown in Figure 1A. Main inclusion criteria were not met for 36% of the patients, because imaging predicted less advanced tumor stages for instance or clinical assessment diagnosed as small cell lung cancer (SCLC). In addition, tumor biopsies for research purposes in addition to the FFPE samples for routine diagnostics could not be collected in 16% of cases, since there was no prospect for tissue acquisition according to the responsible physicians. Purely logistic issues prevented recruitment for another 13% of screened patients, for example insufficient time for patient contact before biopsy or patient’s inclusion in competing biomarker study. Finally, 12% of patients refused to provide consent. Overall, recruitment was possible for 23% of screened patients.
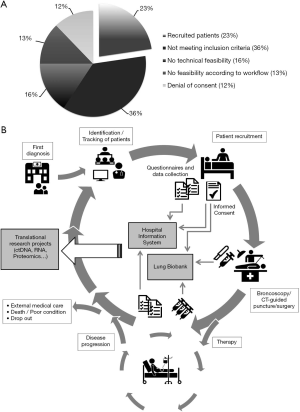
Patient enrolment and clinical characteristics
The recruitment for the cohort started at the time of initial diagnosis when the patient appeared first in the clinical system (Figure 1B). Eligible patients were identified by screening of a variety of clinical documents such as clinic and ward patient lists, attending conferences and contacting responsible physicians. After receiving a signed informed consent, questionnaires and clinical data as well as biomaterial were collected within the frame of the routine biopsy procedure. Data were documented in the hospital information system and the LBBH. During therapy, further questionnaires and blood samples were collected. At progression of disease, additional data and biomaterial were assessed. Biomaterial as well as deep phenotyped data can be provided for future translational research projects.
Over 5-year period of time, 276 patients consented to the sequential biomaterial acquisition, including 16 patients (6%) who received targeted therapies due to detection of activating EGFR (4%) and EML4ALK (2%) alterations. Collection of cryoconserved biopsies for research purposes in addition to FFPE samples as well as a comprehensive phenotyping of these patients were the main goal of this study and were feasible for 87% of these patients (n=241/276, Table 1). Two hundred and five patients matched the inclusion criteria with NSCLC diagnosed by experienced pulmonary pathologists according to the WHO and IASLC/ATS/ERS criteria and were recruited (Figure 2A) (19). Major screening failures were SCLC diagnosis (n=31), deviant TNM (n=19) and other primarius of the tumor (n=8). Collection of cryoconserved biopsies for research purposes was possible in 89% (n=183/205) of these cases, yielding a total of 387 cryoconserved samples (Ø =2.1 cryoconserved biopsies/patient on average). After exclusion of cases with ongoing responses (n=13, Figure 2A) at the time of data analysis or lost-to-follow-up (n=23) with further treatment in other centers closer to their place of residence), 169 patients experienced a disease progression and 43% (n=72/169) underwent a rebiopsy as part of routine management. The rebiopsy rates observed in this study reflect the routine procedures in the participating institutions at the time of the study, i.e., the collection of rebiopsies was not actively controlled and enforced within this project, but left to the discretion of the treating physicians. Main reason for lack of rebiopsy was the clinical deterioration or death (56%, n=95/169), whilst technical non-feasibility was rarely an issue (1%, n=2/169).
Full table
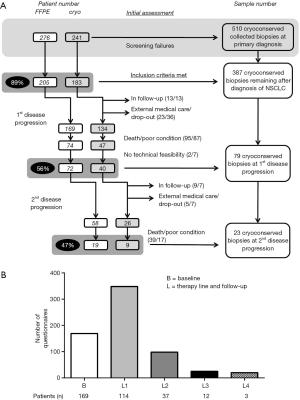
Additional collection of cryoconserved biopsies besides FFPE samples was a main objective and pursued whenever a rebiopsy was requested clinically. This was achievable for 56% of these patients (n=40). Main obstacle was an increased procedural risk due to patient deterioration with disease progression. Comprehensive clinical annotation as described above could be performed for 169 patients at baseline and for 114, 37, 12 and 3 patients during following systemic treatment lines (Figure 2B).
Out of the 72 patients who underwent a routine rebiopsy, 58 patients experienced a second disease progression, 9 patients were still in the second-line therapy at the due date of these analyses, and another 5 patients received further treatment in centers closer to their residence. FFPE biopsies were possible for 33% (n=19/58, Figure 2A) of the patients who had a second disease progression, 47% (n=9/19) of which had additional cryoconserved biopsies.
Main reason for lack of rebiopsy at the second disease progression was the clinical deterioration or death (67%, n=39/58); technical non-feasibility was not an issue.
Cryoconserved biopsy: source and type of acquisition
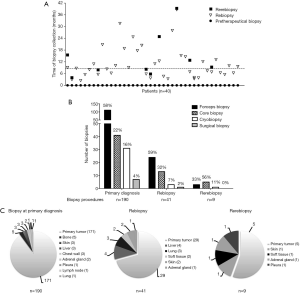
Median time of rebiopsy sampling was 8.9 months after initial biopsy (dotted line in Figure 3A). Of the 183 patients included into the cohort, 190 biopsy procedures were performed to assemble cryoconserved biopsies. Over the course of the disease, the type of biopsy techniques applied changed significantly (Figure 3B). While at baseline the proportion of the forceps biopsies amounted to 58%, it decreased to 33% at re-rebiopsy time. In contrast, the proportion of core biopsies increased from 22% to 56% for the re-rebiopsy collection. Thereby, 171 (90%) biopsies were derived from the primary tumor site (Figure 3C). Other sources of tissue were bone (n=5), skin (3), liver (3), chest wall (3), adrenal gland (2), pleura (1) and lymph node (1). The percentage of biopsies originating from primary tumor decreased from 90% (n=171/190) at baseline to 71% (n=29/41, Figure 3B) and 56% (n=5/9, Figure 3C) at the time of a rebiopsy at first or second disease progression, respectively.
Tissue assessment
Up to the time of this writing, 116 biopsies and rebiopsies have been processed for DNA and RNA isolation for various research projects. The samples were first weighed, followed by preparation of cryosections for the isolation of nucleic acids. A section from the middle of the sample was used for pathological evaluation including determination of the tumor cellularity. The largest tissue samples were obtained with cryobiopsies and had a median weight of 23.8 mg (Figure 4A), which was considerably more than core- and forceps biopsies.
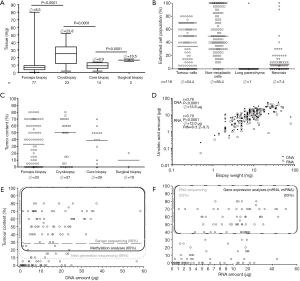
The mean tumor cellularity of the 151 processed biopsies was 34.4% (Figure 4B). Samples predominantly consisted of non-neoplastic cells (56.4%), such as fibroblasts, bronchial cells or endothelial cells of blood vessels, while necrotic or normal lung parenchymal cells were scarce. The different biopsy techniques had no significant influence on the tumor cellularity, which was between 30% and 40% for forceps, cryo- and core-biopsy procedures, respectively (Figure 4C).
Since surgical biopsies are taken only in exceptional cases from stage IIIB/IV patients, for example skin metastases, there are not enough representative data available for the analyzed samples. The amounts of DNA and RNA that could be extracted from the biopsies correlated closely with the weight of the samples (r =0.78 and r =0.70, respectively, Figure 4D). Median yield was 1.17 µg DNA and 0.88 µg RNA per mg tissue, which corresponds to a median amount of 13.6 µg DNA and 12 µg RNA per biopsy and is per se sufficient for most downstream analyses. However, the tumor cell content of the biopsy was a limiting factor in a substantial proportion of cases. While next-generation sequencing approaches may work with a tumor cellularity at around 10% and DNA amounts as low as 10 ng depending on methodology that is being used (feasible in 68% of our processed samples), methylation analyses and Sanger sequencing usually require a higher tumor cellularity of at least 20% to 30%, respectively (feasible in 60% and 56% of our samples, Figure 4E). For gene expression studies and RNA sequencing, a higher tumor cell content of at least 40% is desirable to distinguish between tumor and non-malignant cells, and this was achieved in the half of the processed samples (Figure 4F). Quality controls of the isolated RNA showed a median RNA integrity number (RIN) of 8.0 (Figure 4D). This value was suitable for further downstream analyses. Only 2% (n=3/151) had a RIN lower than 5.
Blood sampling
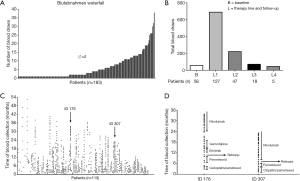
In general, collection of blood samples started concomitantly with the initial biopsy for every patient and was continued during the time of treatment and repeat biopsies. 2.8 mL serum and 2.8 mL plasma were obtained from each blood draw as well as a buffy coat. The number of blood samples per patient varied with a median of 2 (range, 1–38), and in 24% of the patients more than 10 blood samples were taken (Figure 5A). For 127 out of the enrolled 183 patients (69%), follow-up blood samples were collected after primary biopsy (Figure 5B). Follow-up samples during second-line treatment were achieved from 47 patients (26%). The time intervals of blood draws differed from patient to patient depending on their clinical follow-ups (Figure 5C). In contrast to patients receiving chemotherapy, blood samples from patients during immunotherapy and targeted therapy were collected more frequently (Figure 5D). The median total yield of DNA from buffy coat samples was 50.15 µg DNA (125.37 ng/mL) (data not shown).
Discussion
Biomaterial availability is increasingly important for patient management in NSCLC. Selection of patients for targeted and IO therapies depends on the results of biomarker assays, the feasibility and accuracy of which are largely dependent on the quantity and quality of tumor samples (20,21). While FFPE biopsies are in general sufficient for diagnostic purposes (22,23), cryoconserved biopsies increase the diagnostic yield in case of secondary genetic analyses (24), and “liquid”, blood-based biopsies provide an alternative, non-invasive method of molecular profiling (12). With the evolving therapeutic and research landscape of advanced NSCLC in mind, we initiated a pilot project of systematic, prospective sequential biomaterial acquisition in 2012 utilizing broad inclusion criteria and corresponding deep clinical data annotation.
Two hundred and five stage IIIB/IV NSCLC patients were recruited for the described cohort. Extrapolation of data from our feasibility analysis suggests that approximately 890 patients with suspected advanced NSCLC scheduled for a biopsy procedure had to be screened in order to enroll 205 patients. A variety of reasons for exclusion of screened patients was observed. The major component here was the failure to meet inclusion criteria mainly due to the fact that the tumor staging and histologic evaluation was completed only after the bronchoscopy and CT-guided puncture, respectively. Patients diagnosed as stage IV after completion of diagnosis were excluded in this cohort if they did not meet the inclusion criteria at the time of initial biopsy due to a suspected stage I or II. Another issue for exclusion of patients was the inaccessibility of the tumor according to the consulted physician in charge before the bronchoscopy/CT-guided puncture as well as insufficient time for patient contact.
To overcome the obstacles of deviant tumor stage and insufficient time period it might be useful to inform and receive consent from every patient entering the hospital as part of the clinical routine. Additional tissue taken in the biopsy procedures may be collected cryoconserved and initially regarded as diagnostic samples. After completion of diagnosis, these “back-up” samples may be transferred to the biobank and make available for research purposes. The laborious process of tracking and identification of patients could be skipped and man power could be deployed for acquisition and preservation of biomaterials as well as in-depth data collection. The acquisition of additional biopsies entails a slightly increased risk for the patients, therefore the ethical aspects of it should be taken into consideration. By all means, the bright side of it is to avoid repeated biopsy procedures in the case of an impossible histological diagnosis by the pathologist.
The main objective of our study, namely additional collection of cryoconserved biopsies besides the FFPE biopsies for routine diagnostics, was generally feasible for newly diagnosed NSCLC cases with a success rate of 89%. It nevertheless dropped to 56% and 47% at first and second disease progression, respectively. Main obstacle in course of disease was increased procedural risk due to the deterioration of patients´ health conditions. While cryoconserved biopsies were actively pursued in this study, the clinical routine rebiopsies were not enforced. Routine clinical rebiopsies were performed for 43% and 33% of the enrolled patients at first and second disease progression, respectively. A retrospective study investigating rebiopsies for NSCLC patients treated with EGFR inhibitors reported a rebiopsy rate of 63%. In this study specifically pursued collection of rebiopsies was a main endpoint and thoracocentesis for pleural effusions was considered as tissue biopsies (23). Furthermore, patients receiving targeted therapies present a favorable clinical course of disease, and subsequently receive an increased number of therapy lines. This could also contribute to the observed difference, since only 9% of our cryoconserved biopsies derived from patients receiving targeted treatments, and the main obstacle to rebiopsies was patient deterioration. Technically, however, our success rate was higher than 90% which is comparable to those reported by others (25) and with no major notable complications.
The most amount of the tissue acquired was through cryobiopsies compared to core or forceps biopsies. The decreasing rate of cryobiopsy technique applied in the course of disease consequently leads to a reduced amount of tissues and nucleic acids. Nevertheless, DNA and RNA quantity and quality from all processed samples were per se sufficient for downstream assays. Tumor cellularity was the, limiting factor. Only 56–68% of all samples exceeded the threshold value of 40%, 20% or 10%, which are considered adequate for gene expression, methylation or next-generation sequencing assays, respectively. These values were lower compared to a published study with a 70% success rate for microarray analyses (25). However, the authors did not mention the minimum tumor content used for microarray analyses. Due to the preservation of several tissue aliquots in one biopsy procedure, we are able to perform molecular analyses even for most of those patients where the initial aliquot is inadequate. These observations have been made in a study where in 20% of the cases the second aliquot met the QC while the initial tissue specimen did not (25). Liquid biopsies are becoming increasingly part of routine diagnostics. For newly diagnosed NSCLC patients lacking appropriate tumor samples, ctDNA assays provide an alternative method for detection of actionable genetic alterations for targeted therapy stratification (26). Furthermore, for EGFR+ and ALK+ NSCLC patients failing first- and second-generation TKI treatment, liquid biopsies can identify changes in EGFR mutations or ALK resistance mutations and guide subsequent treatment management (11,26). Additional emerging applications of ctDNA testing in NSCLC include early detection of response to targeted therapies (27) and immunotherapy (28), determination of TMB (29) and non-invasive comprehensive molecular profiling (30). Moreover, blood samples can be easily used for detection or validation of prognostic and predictive protein biomarkers in advanced stages (31).
Tumor mutational burden is an emerging predictive biomarker for immunotherapy in multiple tumor entities. The Checkmate 227 trial demonstrated an improved response to the first line immunotherapy combinations in advanced NSCLC patients with high tissue TMB (≥10 mutations per megabase, mut/MB) irrespective of tumor PD-L1 expression level (32). Recent findings from the multicenter phase III MYSTIC trial presented a correlation of blood TMB based on ctDNA at ≥16 mut/MB with tissue TMB (>10 mut/MB), and showed with increasing bTMB (≥20 mut/MB) levels an improved OS and PFS with first-line durvalumab plus tremelimumab versus platinum-based chemotherapy (13). The less invasive bTMB score measured from ctDNA may in addition be more representative of the heterogeneity of advanced NSCLC. The results of the MYSTIC exploratory analysis and the value of bTMB as a predictive biomarker for immunotherapy warrant further investigation.
For 44 out of 183 patients (24%) in our cohort, more than 10 follow-up blood samples were obtained for research purposes with intervals of 2–6 weeks. Besides guidance of therapy, the availability of longitudinal ctDNA samples in association with matched serial tissue biopsies provides excellent opportunities for monitoring of tumor evolution and identification of novel molecular markers. In this respect, the availability of snap-frozen tissue samples has a significant advantage, because cryoconserved material is more suitable for several analyses, such as whole exome sequencing as well as most RNA and protein studies (33). It should be noted that blood samples not only provide ctDNA, but also blood-cell nucleic acids and serum. They can be analyzed in conjunction with the cryoconserved samples of the respective patient for comprehensive genetic, gene expression profiling and proteomic studies (34,35). The possibility to simultaneously characterize local and systemic immunologic parameters by combining routine clinical data with the results of molecular and bioinformatic methods is of particular importance in the era of immunotherapy, the potential of which will require multiparametric approaches for full harnessing (36). Examples include whole exome sequencing for the detection of mutations affecting IO efficacy, determination of TMB and prediction of neoepitopes (37,38), RNA profiling for enumeration of immune cell subsets with CIBERSORT (39) and exploration of immune signatures (40), sequencing of the T-cell receptor repertoire (41) and mass spectrometry for detection of neopeptides and immune effector molecules at the protein level (42,43). Sequencing of RNA isolated from cell nuclei (sNuc-Seq) has become instrumental for interrogating cell types, functional processes and determination of tissue heterogeneity (44). The concomitant availability of high-quality clinical data permits correlation of various parameters with patient outcome for identification of novel prognostic and predictive markers.
Building upon the experience from the first 5 years, we are currently working on better integration of screening procedures into the clinical routine in order to further improve patient recruitment. Sacrifice of research biobanking for the sake of proper diagnostic workup will remain unavoidable in selected cases with limited tissue availability.
Due to the invasiveness of rebiopsies, patient deterioration limits the acquisition of tissue samples during the course of disease. Liquid biopsies are minimal invasive and provide the advantage of capturing spatial and temporal tumor heterogeneity additionally that are associated with the resistance situation. For making the appropriate therapy decision and monitoring of therapy, an overview of the genetic landscape of the tumor is mandatory. Solid biopsies will continue to play a central role in tumor characterization and the complementation of liquid biopsies will extend the therapeutic value and stratification in precision medicine.
In summary, the cohort presented here is the first reported attempt to implement detailed clinical annotation and sequential biomaterial acquisition including cryoconserved biopsies for research purposes in the routine clinical setting. By collecting around 500 tissue and 1,100 blood samples from more than 200 in-depth phenotyped patients, we demonstrate the feasibility and unique value of large-scale systematic biobanking for individualized patient care and translational research. Samples of the cohort have already been used for several high-ranked research projects (27,31,45-47). In particular, routine collection of additional cryoconserved tissue samples is feasible in the large majority (89%) of treatment-naïve NSCLC patients and can significantly extend the possibilities of molecular workup in order to maximize the benefit of targeted and immunooncologic therapies in the future. Nevertheless, the cohort assembly and potential downstream analyses have to be considered in detail before the application of the cryo-conserved biopsy concept because of the time intensive screening/tracking of the patients and costs associated.
Acknowledgments
We thank Magdalena Stephan, Karin Schnorr-Teichert, Saskia Oestringer, Martin Fallenbuechel and Andrea Bopp for patient recruitment, biomaterial collection and processing. We further thank Daniel Kazdal and Anna-Lena Volckmar for technical support. We would like to thank the following people for using their icons from Noun project: Hospital by yanti, People by Shastry, Conference by scott desmond, Screen by Lyhn, Doctor by Zachary Bogard, Caring by Luis Prado, List by hans draiman, Approved list by Anna Sophie, Surgery by Luis Prado, Syringe by Ayub Irawan, Scalpel by arejoenah, Intravenous Therapy by Gan Khoon Lay.
Funding: This study was financially supported in part by the German Centre for Lung Research (DZL, grant number 82DZL00402).
Footnote
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tlcr-20-137
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tlcr-20-137). SW reports grants and personal fees from German Center for Lung Research (DZL), during the conduct of the study. TM reports grants and personal fees from BMBF, German Center for Lung Research (DZL), during the conduct of the study; grants and personal fees from Roche, outside the submitted work. PC reports grants and personal fees from Novartis, grants and personal fees from Roche, grants from AstraZeneca, grants from Takeda, personal fees from Chugai, personal fees from Boehringer, outside the submitted work. MM reports grants from German Center for Lung Research, during the conduct of the study. CPH’s disclosure see the COI statement enclosed. AS reports personal fees from Astra Zeneca, personal fees from Novartis, grants and personal fees from BMS, personal fees from MSD, grants and personal fees from Bayer, personal fees from Seattle Genetics, personal fees from Takeda, personal fees from Eli Lilly, personal fees from Illumina, personal fees from Thermo Fisher, grants from Chugai, personal fees from Roche, personal fees from Pfizer, personal fees from Janssen, during the conduct of the study; serves as an unpaid editorial board member of Translational Lung Cancer Research from Sep 2019 to Sep 2021. RMH reports grants from German Center for Lung Research (DZL), during the conduct of the study. MT reports grants, personal fees and non-financial support from AstraZeneca, grants, personal fees and non- financial support from Bristol-Myers Squibb, personal fees and non-financial support from Boehringer Ingelheim, personal fees and non-financial support from Celgene, personal fees and non-financial support from Chugai, personal fees and non-financial support from Lilly, personal fees and non-financial support from MSD, personal fees and non-financial support from Novartis, personal fees and non-financial support from Pfizer, grants, personal fees and non-financial support from Roche, grants, personal fees and non-financial support from Takeda, outside the submitted work. MAS reports grants and personal fees from German Center for Lung Research (DZL), during the conduct of the study. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Prospective biobanking of NSCLC patients’ materials was performed after informed consent as approved by the ethics committee of Heidelberg University (S-048/2012).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin 2015;65:87-108. [Crossref] [PubMed]
- Pilkington G, Boland A, Brown T, et al. A systematic review of the clinical effectiveness of first-line chemotherapy for adult patients with locally advanced or metastatic non-small cell lung cancer. Thorax 2015;70:359-67. [Crossref] [PubMed]
- Hida T, Nokihara H, Kondo M, et al. Alectinib versus crizotinib in patients with ALK-positive non-small-cell lung cancer (J-ALEX): an open-label, randomised phase 3 trial. Lancet 2017;390:29-39. [Crossref] [PubMed]
- Mok TS, Wu YL, Ahn MJ, et al. Osimertinib or Platinum-Pemetrexed in EGFR T790M-Positive Lung Cancer. N Engl J Med 2017;376:629-40. [Crossref] [PubMed]
- Shaw AT, Kim DW, Mehra R, et al. Ceritinib in ALK-rearranged non-small-cell lung cancer. N Engl J Med 2014;370:1189-97. [Crossref] [PubMed]
- Reck M, Rodriguez-Abreu D, Robinson AG, et al. Pembrolizumab versus Chemotherapy for PD-L1-Positive Non-Small-Cell Lung Cancer. N Engl J Med 2016;375:1823-33. [Crossref] [PubMed]
- Gandhi L, Rodriguez-Abreu D, Gadgeel S, et al. Pembrolizumab plus Chemotherapy in Metastatic Non-Small-Cell Lung Cancer. N Engl J Med 2018;378:2078-92. [Crossref] [PubMed]
- Steuer CE, Ramalingam SS. Tumor Mutation Burden: Leading Immunotherapy to the Era of Precision Medicine? J Clin Oncol 2018;36:631-2. [Crossref] [PubMed]
- Volckmar AL, Sultmann H, Riediger A, et al. A field guide for cancer diagnostics using cell-free DNA: From principles to practice and clinical applications. Genes Chromosomes Cancer 2018;57:123-39. [Crossref] [PubMed]
- Cabel L, Riva F, Servois V, et al. Circulating tumor DNA changes for early monitoring of anti-PD1 immunotherapy: a proof-of-concept study. Ann Oncol 2017;28:1996-2001. [Crossref] [PubMed]
- McCoach CE, Blakely CM, Banks KC, et al. Clinical Utility of Cell-Free DNA for the Detection of ALK Fusions and Genomic Mechanisms of ALK Inhibitor Resistance in Non-Small Cell Lung Cancer. Clin Cancer Res 2018;24:2758-70. [Crossref] [PubMed]
- Merker JD, Oxnard GR, Compton C, et al. Circulating Tumor DNA Analysis in Patients With Cancer: American Society of Clinical Oncology and College of American Pathologists Joint Review. J Clin Oncol 2018;36:1631-41. [Crossref] [PubMed]
- Rizvi NA, Cho BC, Reinmuth N, et al. Durvalumab with or without tremelimumab vs platinum-based chemotherapy as first-line treatment for metastatic non-small cell lung cancer: MYSTIC. Ann Oncol 2018;29:x39-43. [Crossref]
- Oser MG, Niederst MJ, Sequist LV, et al. Transformation from non-small-cell lung cancer to small-cell lung cancer: molecular drivers and cells of origin. Lancet Oncol 2015;16:e165-72. [Crossref] [PubMed]
- Baldacci S, Mazieres J, Tomasini P, et al. Outcome of EGFR-mutated NSCLC patients with MET-driven resistance to EGFR tyrosine kinase inhibitors. Oncotarget 2017;8:105103-14. [Crossref] [PubMed]
- Coombs CC, Zehir A, Devlin SM, et al. Therapy-Related Clonal Hematopoiesis in Patients with Non-hematologic Cancers Is Common and Associated with Adverse Clinical Outcomes. Cell Stem Cell 2017;21:374-82.e4. [Crossref] [PubMed]
- Mandelker D, Zhang L. The emerging significance of secondary germline testing in cancer genomics. J Pathol 2018;244:610-5. [Crossref] [PubMed]
- Muley TR, Herth FJ, Schnabel PA, et al. From tissue to molecular phenotyping: pre-analytical requirements heidelberg experience. Transl Lung Cancer Res 2012;1:111-21. [PubMed]
- Travis WD, Brambilla E, Noguchi M, et al. Diagnosis of lung cancer in small biopsies and cytology: implications of the 2011 International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society classification. Arch Pathol Lab Med 2013;137:668-84. [Crossref] [PubMed]
- Sullivan I, Planchard D. ALK inhibitors in non-small cell lung cancer: the latest evidence and developments. Ther Adv Med Oncol 2016;8:32-47. [Crossref] [PubMed]
- Tomasello C, Baldessari C, Napolitano M, et al. Resistance to EGFR inhibitors in non-small cell lung cancer: Clinical management and future perspectives. Crit Rev Oncol Hematol 2018;123:149-61. [Crossref] [PubMed]
- Hata A, Katakami N, Nanjo S, et al. Rebiopsy of Histological Samples in Pretreated Non-small Cell Lung Cancer: Comparison Among Rebiopsy Procedures. In Vivo 2017;31:475-9. [Crossref] [PubMed]
- Kawamura T, Kenmotsu H, Taira T, et al. Rebiopsy for patients with non-small-cell lung cancer after epidermal growth factor receptor-tyrosine kinase inhibitor failure. Cancer Sci 2016;107:1001-5. [Crossref] [PubMed]
- Hetzel J, Eberhardt R, Herth FJ, et al. Cryobiopsy increases the diagnostic yield of endobronchial biopsy: a multicentre trial. Eur Respir J 2012;39:685-90. [Crossref] [PubMed]
- Stevenson M, Christensen J, Shoemaker D, et al. Tumor acquisition for biomarker research in lung cancer. Cancer Invest 2014;32:291-8. [Crossref] [PubMed]
- Singh AP, Cheng H, Guo X, et al. Circulating Tumor DNA in Non–Small-Cell Lung Cancer: A Primer for the Clinician. JCO Precis Oncol 2017.1-13. [Crossref]
- Riediger AL, Dietz S, Schirmer U, et al. Mutation analysis of circulating plasma DNA to determine response to EGFR tyrosine kinase inhibitor therapy of lung adenocarcinoma patients. Sci Rep 2016;6:33505. [Crossref] [PubMed]
- Goldberg SB, Narayan A, Kole AJ, et al. Early Assessment of Lung Cancer Immunotherapy Response via Circulating Tumor DNA. Clin Cancer Res 2018;24:1872-80. [Crossref] [PubMed]
- Velcheti V, Kim ES, Mekhail T, et al. Prospective clinical evaluation of blood-based tumor mutational burden (bTMB) as a predictive biomarker for atezolizumab (atezo) in 1L non-small cell lung cancer (NSCLC): Interim B-F1RST results. J Clin Oncol 2018;36:12001. [Crossref]
- Dietz S, Schirmer U, Merce C, et al. Low Input Whole-Exome Sequencing to Determine the Representation of the Tumor Exome in Circulating DNA of Non-Small Cell Lung Cancer Patients. PLoS One 2016;11:e0161012. [Crossref] [PubMed]
- Schneider MA, Muley T, Weber R, et al. Glycodelin as a Serum and Tissue Biomarker for Metastatic and Advanced NSCLC. Cancers (Basel) 2018;10:486. [Crossref] [PubMed]
- Hellmann MD, Ciuleanu TE, Pluzanski A, et al. Nivolumab plus Ipilimumab in Lung Cancer with a High Tumor Mutational Burden. N Engl J Med 2018;378:2093-104. [Crossref] [PubMed]
- Oh E, Choi YL, Kwon MJ, et al. Comparison of Accuracy of Whole-Exome Sequencing with Formalin-Fixed Paraffin-Embedded and Fresh Frozen Tissue Samples. PLoS One 2015;10:e0144162. [Crossref] [PubMed]
- Birse CE, Lagier RJ, FitzHugh W, et al. Blood-based lung cancer biomarkers identified through proteomic discovery in cancer tissues, cell lines and conditioned medium. Clin Proteomics 2015;12:18. [Crossref] [PubMed]
- Zander T, Hofmann A, Staratschek-Jox A, et al. Blood-based gene expression signatures in non-small cell lung cancer. Clin Cancer Res 2011;17:3360-7. [Crossref] [PubMed]
- Blank CU, Haanen JB, Ribas A, et al. The “cancer immunogram”. Science 2016;352:658-60. [Crossref] [PubMed]
- Hellmann MD, Nathanson T, Rizvi H, et al. Genomic Features of Response to Combination Immunotherapy in Patients with Advanced Non-Small-Cell Lung Cancer. Cancer Cell 2018;33:843-852.e4. [Crossref] [PubMed]
- Karasaki T, Nagayama K, Kuwano H, et al. Prediction and prioritization of neoantigens: integration of RNA sequencing data with whole-exome sequencing. Cancer Sci 2017;108:170-7. [Crossref] [PubMed]
- Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 2015;12:453-7. [Crossref] [PubMed]
- Faruki H, Mayhew GM, Serody JS, et al. Lung Adenocarcinoma and Squamous Cell Carcinoma Gene Expression Subtypes Demonstrate Significant Differences in Tumor Immune Landscape. J Thorac Oncol 2017;12:943-53. [Crossref] [PubMed]
- Sheng J, Liu X, Jin C, et al. Deep sequencing of the t-cell receptors for monitoring peripheral CD8+T cells in advanced NSCLC Chinese patients treated with anti-PD-L1 antibody. J Clin Oncol 2018;36:e15025. [Crossref]
- Freudenmann LK, Marcu A, Stevanovic S. Mapping the tumour human leukocyte antigen (HLA) ligandome by mass spectrometry. Immunology 2018;154:331-45. [Crossref] [PubMed]
- Marrugal A, Ojeda L, Paz-Ares L, et al. Proteomic-Based Approaches for the Study of Cytokines in Lung Cancer. Dis Markers 2016;2016:2138627.
- Habib N, Avraham-Davidi I, Basu A, et al. Massively parallel single-nucleus RNA-seq with DroNc-seq. Nat Methods 2017;14:955-8. [Crossref] [PubMed]
- Dietz S, Christopoulos P, Gu L, et al. Serial liquid biopsies for detection of treatment failure and profiling of resistance mechanisms in KLC1-ALK-rearranged lung cancer. Cold Spring Harb Mol Case Stud 2019;5:a004630. [Crossref] [PubMed]
- Christopoulos P, Dietz S, Kirchner M, et al. Detection of TP53 Mutations in Tissue or Liquid Rebiopsies at Progression Identifies ALK+ Lung Cancer Patients with Poor Survival. Cancers (Basel) 2019;11:124. [Crossref] [PubMed]
- Schneider MA, Kahn NC, Thomas M, et al. The pregnancy associated protein glycodelin as a follow-up biomarker in a male non-small cell lung cancer patient. Cancer Treat Commun 2015;4:139-42. [Crossref]


