Dysregulation of the Met pathway in non-small cell lung cancer: implications for drug targeting and resistance
Introduction
The last decade or so has seen the management of metastatic non-small cell lung cancer (NSCLC) emerge as a paradigm for personalized medicine (1). This principally follows the development and clinical validation of inhibitors against two key oncogenes; epidermal growth factor receptor (EGFR) and anaplastic lymphoma kinase (ALK). Significantly, the clinical efficacy of inhibiting these two kinases is almost entirely restricted to patients with constitutive activation of the relevant kinase, through mutation (EGFR) or translocation (ALK). This in turn has led to a stratified approach to therapy, with molecular analysis at diagnosis and the use of EGFR or ALK inhibitors if indicated. However, for the majority of patients who do not have mutations in either gene, treatment generally remains chemotherapy, for which survival is relatively unchanged. While there has been recent progress in targeting RET and ROS1 (2,3), mutations in these two genes occur very infrequently, and there is thus a clear need to develop drugs against alternative molecular targets, among which Met is a leading example.
The receptor tyrosine kinase, Met [also known as hepatocyte growth factor receptor (HGFR)], was first discovered 30 years ago as part of a fusion gene, TPR-MET, that was isolated from a osteosarcoma cell line derived through chemical carcinogenesis (4). It has since been shown to have key physiological roles, driving a programme of “invasive growth” that is vital in development and tissue repair (5). Aberrant activation of Met is common in malignancy, most notably hereditary papillary renal carcinoma in which activating mutations in Met are common (6), but also in other malignancies including NSCLC, where, in contrast, signalling is generally driven by increased Met abundance (Table 1).
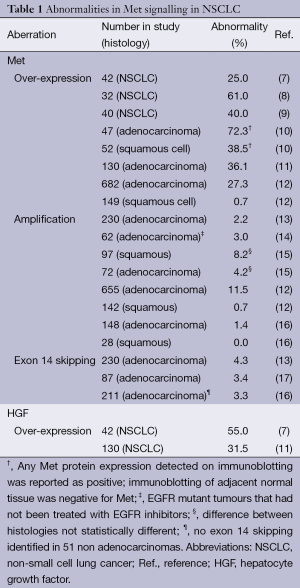
Full table
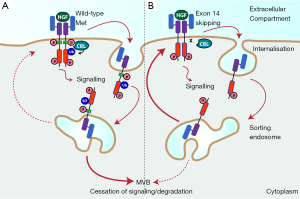
The relative importance of Met in NSCLC has recently been underlined by the findings of a large scale comprehensive molecular profiling study conducted by The Cancer Genome Atlas (TCGA) in lung adenocarcinoma (13). This identified Met as a key targetable driver gene, with approximately 7% of tumours either exhibiting Met amplification or exon 14 skipping. The latter increases Met abundance by decreasing turnover rate (Figure 1). Both these aberrations were mutually exclusive with other known oncogenes, supporting oncogene driver status. Notably only K-RAS, EGFR and BRAF mutations were more frequent. Interest in the clinical potential of targeting Met has also been heightened by studies in which Met amplification was shown to be one of the principle mechanisms by which NSCLC escapes EGFR inhibition (14,18).
In contrast to K-RAS, which is the most commonly mutated driver gene in NSCLC, Met is readily druggable, with a wide range of Met targeted drugs already in clinical development. These fall into several different classes; including small molecule inhibitors, decoy molecules which prevent binding of HGF to Met, and monoclonal antibodies that inhibit either Met or HGF (19,20). Preclinical evidence for these inhibitors is promising, with Met amplified NSCLC cell lines in particular showing exquisite sensitivity (21). However, to date this promise has not been borne out in clinical trials, which have been relatively disappointing. In the last two years, three landmark phase III trials investigating Met targeted agents in combination with erlotinib (an EGFR inhibitor) in pre-treated lung cancer were suspended following interim analyses that indicated no improvement in survival and/or safety concerns (22-24). Further trials are however ongoing with these drugs and it is feasible that subgroup analysis will identify patients who benefit from one or other combination. In addition, several other agents are in development including crizotinib (small molecule inhibitor of Met and ALK kinase) which has shown early evidence of activity in Met amplified NSCLC (25).
Nevertheless, the results of the recent negative trials are sobering and highlight significant gaps in our knowledge, not least in patient selection. It is clear that patient stratification to identify patients most likely to benefit from Met inhibitors will be vital (26), and to this end the ability to detect when Met is acting as a driver oncogene is crucial. In this review we will discuss the various aberrations in Met signalling in NSCLC, and how these may impact on responses to Met inhibitors.
Overview of Met signalling
Met signaling has been extensively reviewed elsewhere (5,20), and we will cover only a few salient points here. Both Met and its ligand HGF are synthesised as single polypeptide chains that are proteolytically cleaved to form the mature protein, each consisting of two polypeptide chains linked by a disulphide bond. Notably, while pro-HGF is capable of high affinity binding to Met, only mature HGF can activate Met signalling (27,28). HGF binding to the extracellular domains of Met (29) leads to its homodimerisation, and transphosphorylation of tyrosine kinase residues Tyr-1234 and Tyr-1235 in the catalytic domain. This is followed by autophosphorylation of residues Tyr-1349 and Tyr-1356 in the c-terminus, which act as a platform for the binding of adaptor proteins. Met has fewer pTyr binding sites than other receptor tyrosine kinases such as EGFR. However, the adapter protein GAB1 expands the palette of sites and is a key co-ordinator of Met signalling, acting as a scaffold for the docking of signalling molecules that include GRB2, PLC, SRC, SHP2 and PI3K (30-33). This leads to activation of a number of downstream pathways that have been shown to be involved in oncogenesis, most notably PI3K, MAPK and STAT3 (34-37).
Physiological Met signalling is tightly regulated, with activation of Met being directly and acutely coupled to its degradation (Figure 1A). Activated Met is rapidly internalised and delivered to the sorting endosome, from which a proportion is recycled back to the membrane, while the rest is directed to the multivesicular body (MVB) and then undergoes degradation in the lysosome (38-42). Ubiquitylation of Met is required for efficient sorting by the endosome, and is dependent on phosphorylation of Tyr-1003 in the Met juxtamembrane domain, which leads to binding of the CBL tyrosine kinase binding (TKB) domain and CBL activation (42). Internalised Met continues to signal from the endosomal platform, although the signalling output is qualitatively different due to the changing palette of substrates present in different subcellular compartments (38,43). Notably, Met receptor in which Tyr-1003 is missing (through exon 14 skipping for example) or mutated, is not directed to the MVB, but is instead trafficked back to the cell surface (Figure 1B) (41,44).
Met signalling in NSCLC
In NSCLC, aberrant activation of the Met pathway in NSCLC may occur through a variety of mechanisms, the most important of which are summarised in Table 1. Over-expression of Met protein is the most commonly reported, with rates of between 25% and 75% in different case series (7-11). Several factors are likely to contribute to this large variability in the literature. Expression of Met is generally assessed through immunohistochemistry and/or immunoblotting, both of which are subject to a large degree of experimental variability, and which are in addition often quantitatively non-linear. The level of Met expression is a continuous variable, and thus reported rates of over-expression also depend partly on the choice of cut-off. The variability in reported rates may additionally reflect true biological differences, for example between histological subtypes, with some studies suggesting over-expression is more frequent in adenocarcinomas than in squamous cell carcinoma (8,10,12). However, even assuming the lower end of range, Met over-expression is a common event in NSCLC. Interestingly, although studies have shown a correlation between Met over-expression and phosphorylation [assessed by Immunohistochemistry (IHC)], not all cases with Met over-expression were positive for phosphorylation or vice versa (11,12). This suggests that Met over-expression may not always be a marker for activated Met signalling.
Met gene amplification is a well-established mechanism by which Met overexpression occurs. Most studies suggest this occurs in about 2-4% of lung adenocarcinomas, and potentially at lower rates in squamous carcinomas (Table 1). In the large-scale, comprehensive TCGA study in adenocarcinoma, 2.2% of cases exhibited amplification (13) compared to only 1% of cases in the comparable study on lung squamous cell carcinomas [curated data from (45) viewed in cBioPortal (46)]. Other smaller studies are generally comparable although there are exceptions (Table 1).
Met amplification has also been shown to be a major mechanism by which cancers develop resistance to EGFR inhibitors. In one study, an EGFR exon 19 mutant NSCLC cell line was exposed to gefitinib at increasing concentrations over 6 months leading to the generation of gefitinib resistance (18). Unlike the parental cell line, the resistant cell line (and six single cell clones) maintained phosphorylation of ERBB3 and Akt in the presence of gefitinib. Copy number analysis revealed a 5-10 fold amplification of Met, and combined inhibition of Met and EGFR restored drug sensitivity. This finding was confirmed in cancer tissue samples, with 4 out of 18 cases of NSCLC that had developed resistance to gefitinib demonstrating Met amplification (18). Further work has shown that treatment with EGFR inhibitors may positively select existing clones with Met amplification, and that EGFR kinase resistance due to either Met amplification or HGF autocrine secretion, can be overcome with the use of Met inhibitors (47). This is supported by a study in which Met amplification was observed in only 3% (2 of 62) of untreated controls, which increased to 21% (9 of 43) in patients with acquired resistance to EGFR inhibitors (14).
In further evidence supporting Met amplification as an important oncogenic event, several studies have shown that Met amplification is associated with phosphorylation and thus activation of Met in cell lines and tumour samples (12,48,49). Met amplification has also been shown to lead to transphosphorylation of other receptor tyrosine kinases such as EGFR and ERBB3 (49). Significantly, studies have shown that Met amplified cell lines are sensitive to Met inhibition (21,50). These included a study which profiled 500 cell lines for sensitivity against a panel of kinase inhibitors in which the 7 which exhibited greatest sensitivity to Met inhibitors were all Met amplified (5 gastric and 2 NSCLC) (21). Interestingly, a few cell lines with Met amplification were not sensitive to the Met inhibitor; these either did not express Met protein or failed to show activation of downstream survival signals (21).
However, Met activation is not observed solely in the presence of amplification, suggesting other mechanisms of activation of signalling. As previously discussed, impaired Met degradation due to exon 14 skipping is a further likely oncogenic driver (Figure 1, Table 1). Splice mutants of Met that lead to skipping of exon 14 and thus lose the juxtamembrane region and Tyr1003 show enhanced stability, prolonged signalling and oncogenic capacity (44). Mutations leading to exon 14 skipping have now been reported in around 3-4% of NSCLC cases (13,16), indicating this to be an important mechanism driving Met overexpression in lung cancer.
Two Met mutations that increase the rate of endocytosis and recycling to the membrane, and reduce rates of degradation leading to Met accumulation have previously been described (51). Both mutations, D1246N and M1268T, involve the catalytic domain, lead to constitutive activation and were identified in papillary renal cell carcinoma (6). A search of the COSMIC and cBioportal databases revealed neither of these mutations in lung cancer, and it is unlikely that either play a significant role in NSCLC. Intriguingly mutations that lead to constitutive activation of Met are almost unheard of in NSCLC. While non-synonymous mutations in other Met domains have been described, these either represent germline polymorphisms or are rare and likely of low oncogenicity (52-54).
The discrepancy between the combined rates of Met amplification and exon 14 skipping (7%), and Met overexpression (at least 25%) is likely due to several other mechanisms, many of which remain to be defined. These include repression of microRNAs leading in turn to increased expression of Met. mir27a is perhaps the best studied in this setting, and has been shown to regulate Met expression in NSCLC both directly, and indirectly through sprouty2 (55). Interestingly miR27a also regulates EGFR and thus may be involved in cross talk between these two receptors (55). In addition, reduced serum expression of mir27a has also been reported in patients with early NSCLC (56). Interestingly, a host of other miRNAs have been implicated in the regulation of Met in NSCLC (miR-449a and miR-7515) or other malignancies (57-74); however their relative importance and interplay remains to be deciphered. Importantly, up regulation of Met through this mechanism is unlikely to be targetable through the use of Met inhibitors, as most miRNAs affect multiple genes. Instead expression of Met could be repressed through the use of miRNA mimetics. The first in class, MRX34, which regulates over 20 genes including Met, is currently undergoing investigation in a phase I trial that is investigating its efficacy in patients with HCC or liver metastases from other malignancies (75). This class of drugs clearly has immense promise, however many challenges remain including drug delivery to target organs as well as potential toxicity issues.
A proportion of Met overexpression and/or activation undoubtedly occurs as a consequence of activation of other oncogenic pathways, which may alter Met transcription, translation or degradation, or indeed directly transactivate Met receptor. EGFR is the best-studied example of the latter, however a host of other molecules can also affect the activation of Met (76). Met activation may also occur through HGF over-expression and autocrine secretion (Table 1), which has also been shown to contribute to gefitinib resistance (77). However, while these processes may facilitate oncogenesis, at present there is limited evidence to suggest that over-expression of Met (apart from that driven by amplification or exon 14 skipping) or HGF acts as a driver.
Targeting Met
Pharmaceutical companies have invested heavily in developing drugs against Met, with most large companies now including one or more such drugs in their pipeline. Several comprehensive reviews of Met inhibitors have been published recently (19,20). There are three main classes of drug, examples of which are summarised in Table 2.
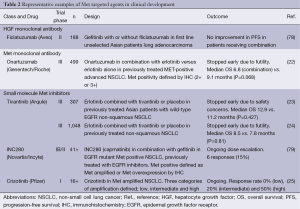
Full table
HGF monoclonal antibodies and decoys
In the first approach, antibodies against HGF or soluble Met fragments act as decoys, binding HGF and thus reducing the concentration of free HGF available to activate Met signalling. Examples of this approach include ficlatuzumab (AV299, Aveo) and rilotumumab (AMG-102, Amgen) which are both monoclonal antibodies, and CGEN241 (Compugen), which is a soluble truncated Met receptor.
The first of these, ficlatuzumab, showed synergism with the EGFR inhibitors erlotinib and cetuximab in a NSCLC xenograft model, prompting a phase I study in which ficlatuzumab was assessed either alone or in combination with erlotinib in solid tumours (78). The combination was well tolerated, however there were no responses, and only 2 out of 8 patients in the combination cohort had stable disease at first assessment. A phase II trial followed in which 188 Asian patients with treatment-naïve NSCLC were recruited and randomised to gefitinib alone or in combination with ficlatuzumab. The results of this trial have been presented and show no benefit for the combination (80).
The second HGF antibody, rilotumumab is currently being investigated in a phase II trial (again in combination with an EGFR inhibitor), while CGEN241 remains in preclinical development.
Met monoclonal antibodies
Binding of HGF to Met involves interactions between multiple surfaces on both proteins, with a recent study identifying four different hotspots that can be inhibited by antibodies (81). There are several antibodies in development, however only onartuzumab (Genentech) is in late clinical development. Met antibodies have been shown to lead to reduced Met signalling, apoptosis and shrinkage of xenografts in a range of tumour types. In addition Met antibodies may drive Met degradation, thus also leading to a reduction in abundance at the membrane (82).
Onartuzumab is a humanised monovalent antibody raised against Met that has recently been assessed in combination with the EGFR inhibitor erlotinib in NSCLC in two trials. The first was a randomised placebo controlled phase II clinical trial, which compared the combination of onartuzumab and erlotinib against erlotinib alone in patients with recurrent NSCLC who had been treated with one or two systemic regimens, and who had not had significant prior exposure to EGFR directed therapy (83). Overall the trial showed no benefit in progression free survival (PFS) or overall survival (OS). However, in patients who were positive for Met by IHC (n=66), both PFS and OS were significantly improved, in contrast to Met negative patients whose outcomes were worse with onartuzumab. These results prompted the institution of a phase III clinical trial with a similar design to the phase II study, with the exception that Met positivity was mandated. Unfortunately the study was suspended following an interim futility analysis, which showed no improvement in either OS or PFS (22).
A related approach that may prove fruitful is the use of conjugated antibodies that are capable of delivering either chemotherapy or radioisotopes to Met expressing cells. An example is anti-Met antibody fragment (FAB) conjugated to doxorubicin which has demonstrated efficacy in preclinical HCC model (84). Proof of principle for this approach is provided from experience with the conjugated Her2 antibody, trastuzumab emtansine, which has demonstrated significant activity in Her2 positive breast cancer (85).
Small molecule inhibitors
Over a dozen different small molecule inhibitors with varying selectivity for Met are in clinical development (19). Two compounds that inhibit Met have been licenced for clinical use, however both of these (crizotinib and cabozantinib) have activity against multiple other kinases. Crizotinib is an inhibitor of ALK kinase and has been licenced for use in NSCLC in which there are ALK translocations, while cabozantinib is a multi targeted kinase inhibitor that has been licenced in prostate cancer.
In a case report, treatment with crizotinib resulted in a rapid and sustained response in a patient with Met amplified NSCLC, with reduction in summated diameters of over 50% (86). The very fact that this case is widely cited is illustrative of the paucity of evidence to date. There is however an ongoing phase I clinical trial of crizotinib in Met amplified NSCLC that was presented at ASCO this year; this has recruited 16 patients, 4 of whom have responded to treatment with a 35 weeks median response. While this trial is still at a very early stage with only a very small number of patients recruited, the degree of Met amplification correlated closely with response rate, with no responses in those classed as low level amplification, and response rates of 20% and 50% in those classed medium or high respectively.
Tivantinib is a specific small molecule inhibitor of Met that is currently in late phase investigation. A phase III trial exploring the efficacy of combining tivantinib and erlotinib in pre-treated metastatic non-squamous lung cancer was halted last year after a preliminary analysis showed it would not achieve its primary objective of increasing overall survival (24). Notably, this trial very closely mirrors the study of onartuzumab, which similarly failed to support activity of the combination. A full subgroup analysis is awaited, and will be extremely valuable in determining whether the combination of a Met targeted drug in combination with erlotinib is a valid strategy.
Future perspectives
Met remains an exciting target for future drug development with significant potential. However, the failure of the three largest trials to date raises significant questions. The results have not been fully published and as such any hypotheses are only tentative. It is feasible that the combination of Met inhibitor and EGFR inhibitor were antagonistic in vivo, or led to increased toxicity (23) and thus reduced dose density. Alternatively, the setting may have contributed; in two out of three trials the patients had been pretreated with chemotherapy and thus are more likely to have developed tumour heterogeneity and/or drug generic resistance mechanisms. However, perhaps the most likely contributory factor is patient selection. Most trials investigating Met have been non-selective or have included all patients with Met (over)expression by IHC. Utilising overexpression as a biomarker is however fraught with difficulties. As described earlier, IHC is subject to considerable variability between users. While it is possible to overcome this with standardisation, expression is a continuous variable, and thus any cutoff level is to some degree arbitrary and not biologically driven. In the onartuzumab study for example, where selection was performed on the basis of Met expression, half of all patients screened were enrolled on the study, which is far higher a proportion than are likely to be Met dependent (87).
Thus far the strongest evidence for a biomarker of response is Met amplification, which predicts for increased sensitivity to Met inhibition in preclinical work (21,88,89). These studies suggest that tumours with Met amplification display oncogene addiction, and this is supported by the trial investigating crizotinib, where there also appears to be a strong correlation between the degree of Met amplification and response, although the results are as yet preliminary (25). Notably, only patients with high-level amplification (defined as a Met to centromere 7 ratio of 5 or more) showed significant response to crizotinib (87). This may explain why Met amplified patients did not show improved outcomes in a subset analysis of the pivotal onartuzumab trial (22).
Rare subgroups that occur at very low frequencies can however be prohibitive for drug development. In the crizotinib study for example, only 0.8% of the population screened had high level amplification (25). While this is clearly a challenge it is not insurmountable with the use of innovative trial designs including adaptive studies such as the BATTLE study (90). In addition exon 14 skipping would intuitively be expected to have a similar effect on sensitivity to Met inhibitors, and combined with Met amplification would allow selection of a significantly higher proportion of patients. While this has not been tested prospectively as yet, evidence to support (or refute) exon 14 skipping as a predictive biomarker may well be obtained from retrospective analyses of the completed phase III trials.
Overall, we remain optimistic that Met inhibition will prove a valuable addition to the therapeutic armamentarium in NSCLC, albeit for a small proportion of patients. Important lessons have been learnt from the recent negative trials; these should strongly influence the design of the next generation of trials, which will need to be rigorously evidence based, and highly selective if they are to unlock the potential of this therapeutic strategy.
Acknowledgements
Disclosure: The authors declare no conflict of interest.
References
- Black A, Morris D. Personalized medicine in metastatic non-small-cell lung cancer: promising targets and current clinical trials. Curr Oncol 2012;19:S73-85. [PubMed]
- Bos M, Gardizi M, Schildhaus HU, et al. Activated RET and ROS: two new driver mutations in lung adenocarcinoma. Transl Lung Cancer Res 2013;2:112-21. [PubMed]
- Shaw AT, Ou SH, Bang YJ, et al. Crizotinib in ROS1-rearranged non-small-cell lung cancer. N Engl J Med 2014;371:1963-71. [PubMed]
- Cooper CS, Park M, Blair DG, et al. Molecular cloning of a new transforming gene from a chemically transformed human cell line. Nature 1984;311:29-33. [PubMed]
- Birchmeier C, Birchmeier W, Gherardi E, et al. Met, metastasis, motility and more. Nat Rev Mol Cell Biol 2003;4:915-25. [PubMed]
- Schmidt L, Duh FM, Chen F, et al. Germline and somatic mutations in the tyrosine kinase domain of the MET proto-oncogene in papillary renal carcinomas. Nat Genet 1997;16:68-73. [PubMed]
- Olivero M, Rizzo M, Madeddu R, et al. Overexpression and activation of hepatocyte growth factor/scatter factor in human non-small-cell lung carcinomas. Br J Cancer 1996;74:1862-8. [PubMed]
- Ma PC, Jagadeeswaran R, Jagadeesh S, et al. Functional expression and mutations of c-Met and its therapeutic inhibition with SU11274 and small interfering RNA in non-small cell lung cancer. Cancer Res 2005;65:1479-88. [PubMed]
- Ma PC, Tretiakova MS, MacKinnon AC, et al. Expression and mutational analysis of MET in human solid cancers. Genes Chromosomes Cancer 2008;47:1025-37. [PubMed]
- Ichimura E, Maeshima A, Nakajima T, et al. Expression of c-met/HGF receptor in human non-small cell lung carcinomas in vitro and in vivo and its prognostic significance. Jpn J Cancer Res 1996;87:1063-9. [PubMed]
- Nakamura Y, Niki T, Goto A, et al. c-Met activation in lung adenocarcinoma tissues: an immunohistochemical analysis. Cancer Sci 2007;98:1006-13. [PubMed]
- Tsuta K, Kozu Y, Mimae T, et al. c-MET/phospho-MET protein expression and MET gene copy number in non-small cell lung carcinomas. J Thorac Oncol 2012;7:331-9. [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014;511:543-50. [PubMed]
- Bean J, Brennan C, Shih JY, et al. MET amplification occurs with or without T790M mutations in EGFR mutant lung tumors with acquired resistance to gefitinib or erlotinib. Proc Natl Acad Sci U S A 2007;104:20932-7. [PubMed]
- Go H, Jeon YK, Park HJ, et al. High MET gene copy number leads to shorter survival in patients with non-small cell lung cancer. J Thorac Oncol 2010;5:305-13. [PubMed]
- Onozato R, Kosaka T, Kuwano H, et al. Activation of MET by gene amplification or by splice mutations deleting the juxtamembrane domain in primary resected lung cancers. J Thorac Oncol 2009;4:5-11. [PubMed]
- Seo JS, Ju YS, Lee WC, et al. The transcriptional landscape and mutational profile of lung adenocarcinoma. Genome Res 2012;22:2109-19. [PubMed]
- Engelman JA, Zejnullahu K, Mitsudomi T, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science 2007;316:1039-43. [PubMed]
- Cui JJ. Targeting receptor tyrosine kinase MET in cancer: small molecule inhibitors and clinical progress. J Med Chem 2014;57:4427-53. [PubMed]
- Gherardi E, Birchmeier W, Birchmeier C, et al. Targeting MET in cancer: rationale and progress. Nat Rev Cancer 2012;12:89-103. [PubMed]
- McDermott U, Sharma SV, Dowell L, et al. Identification of genotype-correlated sensitivity to selective kinase inhibitors by using high-throughput tumor cell line profiling. Proc Natl Acad Sci U S A 2007;104:19936-41. [PubMed]
- Spigel DR, Edelman MJ, O'Byrne K, et al. Onartuzumab plus erlotinib versus erlotinib in previously treated stage IIIb or IV NSCLC: Results from the pivotal phase III randomized, multicenter, placebo-controlled METLung (OAM4971g) global trial. J Clin Oncol 2014;abstr 8000.
- Azuma K, Yoshioka H, Yamamoto N, et al. Tivantinib plus erlotinib versus placebo plus erlotinib in Asian patients with previously treated nonsquamous NSCLC with wild-type EGFR: First report of a phase III ATTENTION trial. J Clin Oncol 2014;abstr 8044.
- ESMO News. Available online: http://www.esmo.org/Conferences/Past-Conferences/European-Cancer-Congress-2013/News/A-Phase-III-Study-of-Tivantinib-Plus-Erlotinib-Did-Not-Meet-a-Primary-Endpoint-in-Patients-with-Locally-advanced-or-Metastatic-Non-squamous-NSCLC, accessed on Jan 22, 2015.
- Camidge DR, Ou SI, Shapiro G, et al. Efficacy and safety of crizotinib in patients with advanced c-MET-amplified non-small cell lung cancer (NSCLC). J Clin Oncol 2014;abstr 8001.
- Koeppen H, Rost S, Yauch RL. Developing biomarkers to predict benefit from HGF/MET pathway inhibitors. J Pathol 2014;232:210-8. [PubMed]
- Naldini L, Tamagnone L, Vigna E, et al. Extracellular proteolytic cleavage by urokinase is required for activation of hepatocyte growth factor/scatter factor. EMBO J 1992;11:4825-33. [PubMed]
- Lokker NA, Mark MR, Luis EA, et al. Structure-function analysis of hepatocyte growth factor: identification of variants that lack mitogenic activity yet retain high affinity receptor binding. EMBO J 1992;11:2503-10. [PubMed]
- Niemann HH. Structural insights into Met receptor activation. Eur J Cell Biol 2011;90:972-81. [PubMed]
- Weidner KM, Di Cesare S, Sachs M, et al. Interaction between Gab1 and the c-Met receptor tyrosine kinase is responsible for epithelial morphogenesis. Nature 1996;384:173-6. [PubMed]
- Schaeper U, Gehring NH, Fuchs KP, et al. Coupling of Gab1 to c-Met, Grb2, and Shp2 mediates biological responses. J Cell Biol 2000;149:1419-32. [PubMed]
- Rodrigues GA, Falasca M, Zhang Z, et al. A novel positive feedback loop mediated by the docking protein Gab1 and phosphatidylinositol 3-kinase in epidermal growth factor receptor signaling. Mol Cell Biol 2000;20:1448-59. [PubMed]
- Gual P, Giordano S, Williams TA, et al. Sustained recruitment of phospholipase C-gamma to Gab1 is required for HGF-induced branching tubulogenesis. Oncogene 2000;19:1509-18. [PubMed]
- Maroun CR, Naujokas MA, Holgado-Madruga M, et al. The tyrosine phosphatase SHP-2 is required for sustained activation of extracellular signal-regulated kinase and epithelial morphogenesis downstream from the met receptor tyrosine kinase. Mol Cell Biol 2000;20:8513-25. [PubMed]
- Rosário M, Birchmeier W. How to make tubes: signaling by the Met receptor tyrosine kinase. Trends Cell Biol 2003;13:328-35. [PubMed]
- Kermorgant S, Parker PJ. Receptor trafficking controls weak signal delivery: a strategy used by c-Met for STAT3 nuclear accumulation. J Cell Biol 2008;182:855-63. [PubMed]
- Lee YY, Kim HP, Kang MJ, et al. Phosphoproteomic analysis identifies activated MET-axis PI3K/AKT and MAPK/ERK in lapatinib-resistant cancer cell line. Exp Mol Med 2013;45:e64. [PubMed]
- Clague MJ. Met receptor: a moving target. Sci Signal 2011;4:pe40. [PubMed]
- Hammond DE, Urbé S, Vande Woude GF, et al. Down-regulation of MET, the receptor for hepatocyte growth factor. Oncogene 2001;20:2761-70. [PubMed]
- Hammond DE, Carter S, McCullough J, et al. Endosomal dynamics of Met determine signaling output. Mol Biol Cell 2003;14:1346-54. [PubMed]
- Abella JV, Peschard P, Naujokas MA, et al. Met/Hepatocyte growth factor receptor ubiquitination suppresses transformation and is required for Hrs phosphorylation. Mol Cell Biol 2005;25:9632-45. [PubMed]
- Peschard P, Fournier TM, Lamorte L, et al. Mutation of the c-Cbl TKB domain binding site on the Met receptor tyrosine kinase converts it into a transforming protein. Mol Cell 2001;8:995-1004. [PubMed]
- Barrow R, Joffre C, Ménard L, et al. Measuring the role for Met endosomal signaling in tumorigenesis. Methods Enzymol 2014;535:121-40. [PubMed]
- Kong-Beltran M, Seshagiri S, Zha J, et al. Somatic mutations lead to an oncogenic deletion of met in lung cancer. Cancer Res 2006;66:283-9. [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature 2012;489:519-25. [PubMed]
- Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. [PubMed]
- Turke AB, Zejnullahu K, Wu YL, et al. Preexistence and clonal selection of MET amplification in EGFR mutant NSCLC. Cancer Cell 2010;17:77-88. [PubMed]
- Kubo T, Yamamoto H, Lockwood WW, et al. MET gene amplification or EGFR mutation activate MET in lung cancers untreated with EGFR tyrosine kinase inhibitors. Int J Cancer 2009;124:1778-84. [PubMed]
- Tanizaki J, Okamoto I, Sakai K, et al. Differential roles of trans-phosphorylated EGFR, HER2, HER3, and RET as heterodimerisation partners of MET in lung cancer with MET amplification. Br J Cancer 2011;105:807-13. [PubMed]
- Tanizaki J, Okamoto I, Okamoto K, et al. MET tyrosine kinase inhibitor crizotinib (PF-02341066) shows differential antitumor effects in non-small cell lung cancer according to MET alterations. J Thorac Oncol 2011;6:1624-31. [PubMed]
- Joffre C, Barrow R, Ménard L, et al. A direct role for Met endocytosis in tumorigenesis. Nat Cell Biol 2011;13:827-37. [PubMed]
- Tengs T, Lee JC, Paez JG, et al. A transforming MET mutation discovered in non-small cell lung cancer using microarray-based resequencing. Cancer Lett 2006;239:227-33. [PubMed]
- Krishnaswamy S, Kanteti R, Duke-Cohan JS, et al. Ethnic differences and functional analysis of MET mutations in lung cancer. Clin Cancer Res 2009;15:5714-23. [PubMed]
- Tyner JW, Fletcher LB, Wang EQ, et al. MET receptor sequence variants R970C and T992I lack transforming capacity. Cancer Res 2010;70:6233-7. [PubMed]
- Acunzo M, Romano G, Palmieri D, et al. Cross-talk between MET and EGFR in non-small cell lung cancer involves miR-27a and Sprouty2. Proc Natl Acad Sci U S A 2013;110:8573-8. [PubMed]
- Heegaard NH, Schetter AJ, Welsh JA, et al. Circulating micro-RNA expression profiles in early stage nonsmall cell lung cancer. Int J Cancer 2012;130:1378-86. [PubMed]
- Luo W, Huang B, Li Z, et al. MicroRNA-449a is downregulated in non-small cell lung cancer and inhibits migration and invasion by targeting c-Met. PLoS One 2013;8:e64759. [PubMed]
- Lee JM, Yoo JK, Yoo H, et al. The novel miR-7515 decreases the proliferation and migration of human lung cancer cells by targeting c-Met. Mol Cancer Res 2013;11:43-53. [PubMed]
- Jiang C, Chen H, Shao L, et al. MicroRNA-1 functions as a potential tumor suppressor in osteosarcoma by targeting Med1 and Med31. Oncol Rep 2014;32:1249-56. [PubMed]
- Huang J, Dong B, Zhang J, et al. miR-199a-3p inhibits hepatocyte growth factor/c-Met signaling in renal cancer carcinoma. Tumour Biol 2014;35:5833-43. [PubMed]
- Takeyama H, Yamamoto H, Yamashita S, et al. Decreased miR-340 expression in bone marrow is associated with liver metastasis of colorectal cancer. Mol Cancer Ther 2014;13:976-85. [PubMed]
- Menges CW, Kadariya Y, Altomare D, et al. Tumor suppressor alterations cooperate to drive aggressive mesotheliomas with enriched cancer stem cells via a p53-miR-34a-c-Met axis. Cancer Res 2014;74:1261-71. [PubMed]
- Yang X, Zhang XF, Lu X, et al. MicroRNA-26a suppresses angiogenesis in human hepatocellular carcinoma by targeting hepatocyte growth factor-cMet pathway. Hepatology 2014;59:1874-85. [PubMed]
- Hagman Z, Haflidadottir BS, Ansari M, et al. The tumour suppressor miR-34c targets MET in prostate cancer cells. Br J Cancer 2013;109:1271-8. [PubMed]
- Xu X, Chen H, Lin Y, et al. MicroRNA-409-3p inhibits migration and invasion of bladder cancer cells via targeting c-Met. Mol Cells 2013;36:62-8. [PubMed]
- Hu Z, Lin Y, Chen H, et al. MicroRNA-101 suppresses motility of bladder cancer cells by targeting c-Met. Biochem Biophys Res Commun 2013;435:82-7. [PubMed]
- Wang LG, Ni Y, Su BH, et al. MicroRNA-34b functions as a tumor suppressor and acts as a nodal point in the feedback loop with Met. Int J Oncol 2013;42:957-62. [PubMed]
- Luo C, Tetteh PW, Merz PR, et al. miR-137 inhibits the invasion of melanoma cells through downregulation of multiple oncogenic target genes. J Invest Dermatol 2013;133:768-75. [PubMed]
- Chen L, Zhang J, Feng Y, et al. MiR-410 regulates MET to influence the proliferation and invasion of glioma. Int J Biochem Cell Biol 2012;44:1711-7. [PubMed]
- Buurman R, Gürlevik E, Schäffer V, et al. Histone deacetylases activate hepatocyte growth factor signaling by repressing microRNA-449 in hepatocellular carcinoma cells. Gastroenterology 2012;143:811-20. [PubMed]
- Yan K, Gao J, Yang T, et al. MicroRNA-34a inhibits the proliferation and metastasis of osteosarcoma cells both in vitro and in vivo. PLoS One 2012;7:e33778. [PubMed]
- Dong F, Lou D. MicroRNA-34b/c suppresses uveal melanoma cell proliferation and migration through multiple targets. Mol Vis 2012;18:537-46. [PubMed]
- Reid JF, Sokolova V, Zoni E, et al. miRNA profiling in colorectal cancer highlights miR-1 involvement in MET-dependent proliferation. Mol Cancer Res 2012;10:504-15. [PubMed]
- Tsuruta T, Kozaki K, Uesugi A, et al. miR-152 is a tumor suppressor microRNA that is silenced by DNA hypermethylation in endometrial cancer. Cancer Res 2011;71:6450-62. [PubMed]
- Bouchie A. First microRNA mimic enters clinic. Nat Biotechnol 2013;31:577. [PubMed]
- Avan A, Quint K, Nicolini F, et al. Enhancement of the antiproliferative activity of gemcitabine by modulation of c-Met pathway in pancreatic cancer. Curr Pharm Des 2013;19:940-50. [PubMed]
- Yano S, Wang W, Li Q, et al. Hepatocyte growth factor induces gefitinib resistance of lung adenocarcinoma with epidermal growth factor receptor-activating mutations. Cancer Res 2008;68:9479-87. [PubMed]
- Patnaik A, Weiss GJ, Papadopoulos KP, et al. Phase I ficlatuzumab monotherapy or with erlotinib for refractory advanced solid tumours and multiple myeloma. Br J Cancer 2014;111:272-80. [PubMed]
- Wu Y, Yang JC, Kim D, et al. Safety and efficacy of INC280 in combination with gefitinib (gef) in patients with EGFR-mutated (mut), MET-positive NSCLC: A single-arm phase lb/ll study. J Clin Oncol 2014; abstr 8017.
- Mok TSK, Park K, Geater SL, et al. A randomized phase (PH) 2 study with exploratory biomarker analysis of ficlatuzumab (F) a humanized hepatocyte growth factor (HGF) inhibitory mAb in combination with gefitinib (G) versus G in Asian patients (pts) with lung adenocarcinoma (LA). ESMO 2012;Abstract 2342.
- Basilico C, Hultberg A, Blanchetot C, et al. Four individually druggable MET hotspots mediate HGF-driven tumor progression. J Clin Invest 2014;124:3172-86. [PubMed]
- Lee JM, Kim B, Lee SB, et al. Cbl-independent degradation of Met: ways to avoid agonism of bivalent Met-targeting antibody. Oncogene 2014;33:34-43. [PubMed]
- Spigel DR, Ervin TJ, Ramlau RA, et al. Randomized phase II trial of Onartuzumab in combination with erlotinib in patients with advanced non-small-cell lung cancer. J Clin Oncol 2013;31:4105-14. [PubMed]
- Chen X, Ding G, Gao Q, et al. A human anti-c-Met Fab fragment conjugated with doxorubicin as targeted chemotherapy for hepatocellular carcinoma. PLoS One 2013;8:e63093. [PubMed]
- Verma S, Miles D, Gianni L, et al. Trastuzumab emtansine for HER2-positive advanced breast cancer. N Engl J Med 2012;367:1783-91. [PubMed]
- Ou SH, Kwak EL, Siwak-Tapp C, et al. Activity of crizotinib (PF02341066), a dual mesenchymal-epithelial transition (MET) and anaplastic lymphoma kinase (ALK) inhibitor, in a non-small cell lung cancer patient with de novo MET amplification. J Thorac Oncol 2011;6:942-6. [PubMed]
- Garber K. MET inhibitors start on road to recovery. Nat Rev Drug Discov 2014;13:563-5. [PubMed]
- Lutterbach B, Zeng Q, Davis LJ, et al. Lung cancer cell lines harboring MET gene amplification are dependent on Met for growth and survival. Cancer Res 2007;67:2081-8. [PubMed]
- Matsubara D, Ishikawa S, Oguni S, et al. Molecular predictors of sensitivity to the MET inhibitor PHA665752 in lung carcinoma cells. J Thorac Oncol 2010;5:1317-24. [PubMed]
- Kim ES, Herbst RS, Wistuba II, et al. The BATTLE trial: personalizing therapy for lung cancer. Cancer Discov 2011;1:44-53. [PubMed]


