Genetic and microenvironmental differences in non-smoking lung adenocarcinoma patients compared with smoking patients
Introduction
Lung cancer is the most common cancer worldwide. Non-small cell lung cancer (NSCLC) (1), the main subtypes of which include adenocarcinoma and squamous cell carcinoma, accounts for approximately 85% of the cases of lung cancer (2). Lung cancer is the most common malignant tumor and has the highest morbidity and mortality (1). According to the World Health Organization Report on the Global Tobacco Epidemic (3), there are 1.4 billion tobacco users aged 15 years and older worldwide. It is well accepted that tobacco smoking is the primary cause of lung cancer, especially lung squamous cell carcinoma (3-5) since Richard Doll and Austin Bradford Hill first proved the link between smoking and lung cancer in the 1950s (6).
However, approximately 25% of lung cancer cases worldwide, mainly adenocarcinoma, cannot be attributed to tobacco smoking; lung cancer in never smokers is the seventh leading cause of cancer deaths worldwide (7). According to clinical experience, different epidemiology and natural history are observed between lung cancers in never smokers and those in smokers (8), suggesting that lung cancer in never smokers is a ‘different’ disease, with specific etiology and molecular differences (9). With the advent of RNA sequencing technology, more studies have focused on specific genetic or molecular mechanisms (10); however, these have remained unclear until now in non-smoker-related lung adenocarcinoma (LUAD).
To study the differences in non-smoking compared with smoking LUAD patients, clinical data, gene expression, somatic mutations, immune infiltration, and stemness indices were analyzed to determine the role of smoking in LUAD. The effects were also confirmed in LUAD cells. This study will improve our understanding of the causes of oncogenesis of non-smoking LUAD and provide a reference for therapeutic decisions in clinical treatments.
We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/tlcr-20-276).
Methods
The Cancer Genome Atlas (TCGA) data collection
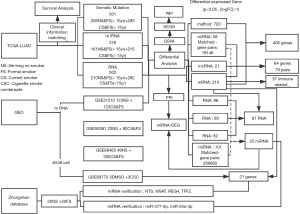
Detailed information for patients with LUAD was downloaded from TCGA. Clinical data, RNA expression data, microRNA (miRNA) expression and somatic mutation (VarScan MAF files) data were downloaded from NCI’s Genomic Data Commons (https://gdc.cancer.gov/) (TCGA-LUAD). After matching mRNA expression, miRNA expression, and somatic mutation data with clinical data, 502 cases of RNAs, 376 cases of miRNAs, and 501 cases of somatic mutations were obtained to explore the differences in the genomes of smoking and non-smoking LUAD patients at different levels (see Figure 1 for details). According to the smoking data described in TCGA, patients who had never smoked or who had quit smoking for more than 15 years were classified as non-smokers while those who were current smokers, who quit smoking less than 15 years ago, or quit smoking but the years were unknown were classified as smokers (Figure 1).
GEO data collection
The mRNA expression data of LUAD from current, former, and never smokers were download from Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/). GSE31210, GSE50081, and GSE68465 datasets (11-13), published on February 20, 2008, were collected to determine the coding RNA expression signature of cigarette smoking.
The GSE69770 dataset (14), published on February 7, 2018, performed RNA sequencing of dimethyl sulfoxide (DMSO)- or cigarette smoke condensate (CSC)-treated A549 human LUAD cells. Cells were treated for 48 h or 2 weeks.
Immune infiltration data
Immune infiltration data was collected from a previous study (15) and matched with the clinical data downloaded from the TCGA database. A total of 503 samples were finally enrolled; 210 were classified as non-smokers and 293 were classified as smokers.
Stemness indices
Stemness indices were collected from a previous study (16) and matched with the clinical data downloaded from the TCGA database. A total of 496 samples were finally enrolled; 207 were classified as non-smokers and 289 were classified as smokers.
Tissue samples
All samples were obtained from patients at the Department of Thoracic Surgery, Zhongshan Hospital, Fudan University. Tumor samples were derived from surgical resection of LUAD between January 2019 and April 2019. All samples were quickly frozen in liquid nitrogen after removal and then stored at −80 °C before used. Finally, 20 pairs of non-smoking LUAD tissue and smoking adenocarcinoma (smoking index ≥20 pack-years) tissues from 40 patients were obtained for qRT-PCR validation.
RNA, miRNA preparation and qRT-PCR analysis
To detect the expression of NTS, NNAT, TFF2, REG4, miR-512, miR-372 in 20 pairs of samples, RT-qPCR was carried out on an ABI Prism 7500 real-time PCR system (Applied Biosystems) with proper PCR parameters.
Total RNAs were extracted by TRIzol (Beyotime, China). The first-strand cDNA was synthesized using PrimeScriptTM RT reagent Kit with gDNA Eraser (Real Time Perfect) (TaKaRa, Tokyo, Japan) according to the manufacturer’s instructions. Then SYBR Premix Ex TaqTM II (Tli RNaseH Plus) (TaKaRa) was used with the following PCR parameters, 1 cycle of 30 s at 95 °C, 40 cycles of 5 s at 95 °C and 34 s at 60 °C. β-actin was used as the reference. Primers used in this study are listed in Table S1.

Full table
Total miRNAs were extracted with miRcute miRNA Isolation Kit (TianGen Biotech, Beijing, China) and then reversed by miRcute Plus miRNA First-Strand cDNA Kit (TianGen). miRcute Plus miRNA qPCR Kit (SYBR Green) (TianGen) was used in the PCR system with the following PCR parameters, 1 cycle of 2 min at 94 °C, 40 cycles of 20 s at 94 °C and 34 s at 60 °C. U6 was used as the reference. All miRNA primers were obtained from TianGen.
All the samples were repeated three times.
Statistical analysis
The distribution of patients’ characteristics (e.g., sex, race, age group, primary site, differentiation grade, and chemotherapy) was summarized using counts and percentages. Statistical analysis was performed with SPSS 23.0 software (IBM). Differential analysis of RNA, miRNA, and immune infiltration data was performed with R (version 3.6.1). All data were normalized and standardized by constructing relevant expression matrices using edgfR. Differential genes [P<0.05, false discovery rate (FDR) <0.05 for somatic mutations, miRNAs, and others] were sorted according to logFoldChange values (|logFC| >1) to identify significantly different expression between smoking and non-smoking patients. A merged protein-protein interaction (PPI) network was constructed and analyzed with Cytoscape software (version 3.7.2). Differences in stemness indices and verification were determined by Student’s t-test with Graphpad Prism 8. The test level was α=0.05, and differences were considered statistically significant at P<0.05. Survival analysis was performed with the Kaplan-Meier method with R (version 3.6.1).
All procedures performed in this study were in accordance with the Declaration of Helsinki (as revised in 2013) and was approved by the Ethics Committee of Zhongshan Hospital, Fudan University, China (B2019 – 377R). Because of the retrospective nature of the re-search, the requirement for informed consent was waived.
Results
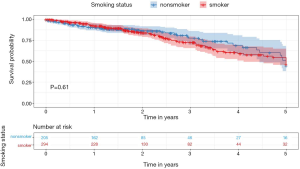
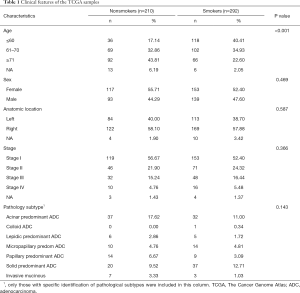
Table 1 shows the basic clinical characteristics of the patients (e.g., sex, age group, location, and stage) summarized using counts and percentages. The smokers were younger than the non-smokers. Most of the cases of LUAD in non-smokers were acinar, papillary predominant, and invasive mucinous pathology subtype. Smokers had lower 3-year survival rates [smokers (S), 72.11% vs. non-smokers (NS), 77.56%], although this difference was not statistically significant (Figure 2).
Full table
Differences in somatic mutations
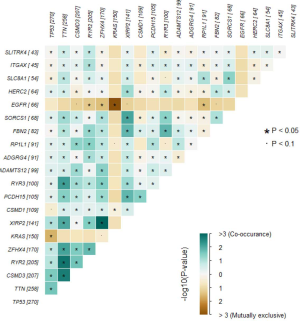
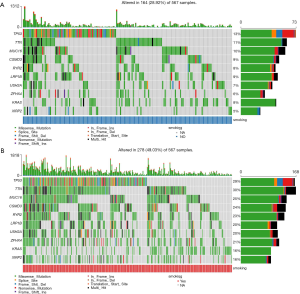
The distribution patterns of somatic mutations (the overall pattern is shown in Figure S1) were investigated between the non-smoker and smoker groups. After performing enrichment analysis, we identified 174 enriched mutations with FDR <0.05 between the non-smoker and smoker groups (Figure 3). The somatic mutation rates of tumor protein p53 (TP53; S, 56.31% vs. NS, 35.10%); titin (TTN; S, 57.34% vs. NS, 29.33%); KRAS proto-oncogene (KRAS; S, 31.40% vs. NS, 22.60%), and ryanodine receptor 2 (RYR2; S, 45.39% vs. NS, 22.12%), were higher in smokers, while epidermal growth factor receptor (EGFR; S, 6.83% vs. NS, 19.71%) were higher in smokers. The mutation rate of human epidermal growth factor receptor-2 (HER2; S, 17.41% vs. NS, 8.17%) and phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha (PIK3CA; S, 8.60% vs. NS, 0) showed no difference between smokers and nonsmokers.
Using the somatic interactions function, which performed a pair-wise Fisher’s exact test to detect significant pairs of genes, we detected the relationships among the top 50 genes with different somatic mutations (Figure S2). Almost all of the mutated genes were tightly linked to each other, except EGFR, which was strongly excluded. Thus, these mutations might be linked to the development of LUAD.
Differentially expressed coding genes and functional annotation in TCGA-LUAD
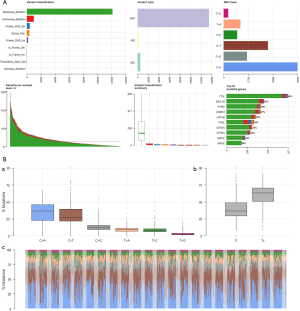
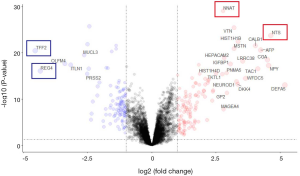
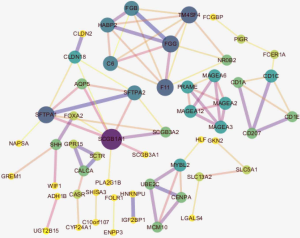
To determine whether smoking can change biological characteristics, expression profiles were examined for differentially expressed genes (DEGs). Of the 60,483 ensemble numbers, 13,747 were successfully converted to gene names by org.Hs.eg.db, and the edgeR algorithm identified 231 DEGs [210 mRNAs and 21 long non-coding RNAS (lncRNAs)] (P<0.05) with an obvious fold change (|logFC| >1), of which 124 were upregulated and 107 were downregulated (Figure 4). A PPI network (Figure S3) was also constructed to reveal the association between proteins encoded by DEGs.
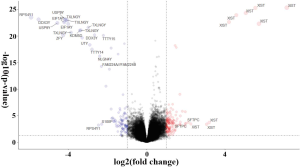
We also applied the same differential analysis method for the data from the GEO databases (GSE31210, GSE50081, and GSE68465) to identify DEGs between smokers and non-smokers (Figure S4). A total of 81 RNAs were differentially expressed in both the TCGA-LUAD and GEO datasets.
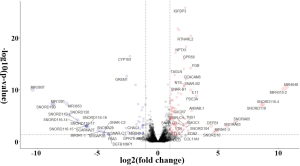
The expression of genes in LUAD cells has been linked to smoking-related transversion mutations in lung tumors. To identify tobacco smoke-inducible enhancers at the cellular level, we used the same method to analyze the expression profile of A549 human LUAD cells treated with DMSO or CSC for 2 weeks (GSE69770; Figure S5). A total of 21 genes (upregulated: MALAT1, RSPO3, CPLX2, BAAT, CLDN2, TM4SF4, NTS, and VTN; downregulated: IGF2, HIST1H3F, HIST1H4C, HIST1H4D, HIST1H4E, HIST1H3B, HIST1H2AI, HIST1H1E, HIST1H2AM, HIST1H2BE, HIST1H3C, HIST1H1D, and HIST1H1B) were both differentially expressed between smoking and non-smoking LUAD patients and between CSC- and DMSO-treated A549 cells, which more reliably revealed that changes in these genes were caused by smoking and their upregulation or downregulation might play a more important role.
Gene functional analysis
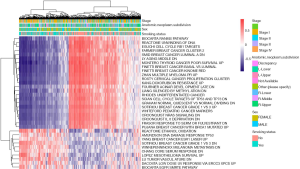
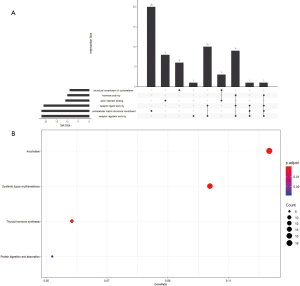
Gene set variation analysis (GSVA) of RNA sequencing data converts biological function annotations (GO and KEGG pathway analyses) into new expression matrices and treats them as general matrix files for differential expression analysis. This analysis revealed 42 significant pathways between smokers and non-smokers (Figure 5); e.g., a Reactome for ethanol oxidation and the Amundson DNA damage response TP53 were downregulated and Ly aging middle DN, and Montero thyroid cancer poor survival up were upregulated in nonsmokers.
We also performed Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses of the 231 genes that had differential somatic mutations or were differentially expressed based on the TCGA data. Gene functional enrichment analysis showed that six GO functional groups exhibited significant differences between smokers and non-smokers: extracellular matrix structural constituent, receptor ligand activity, structural constituent of cytoskeleton, actin filament binding, hormone activity, and receptor regulator activity (Figure S6A). For KEGG pathway analysis, four pathways differed between smokers and non-smokers: systemic lupus erythematosus, alcoholism, thyroid hormone synthesis, and protein digestion and absorption (Figure S6B).
Differentially expressed miRNAs and functional annotation in TCGA-LUAD
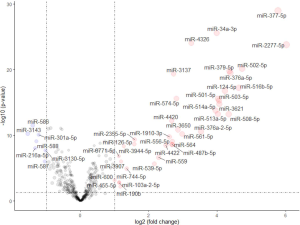
Of the 1,881 miRNAs expressed in the TCGA-LUAD with clinical information, 38 miRNAs were significantly upregulated (P<0.05, |logFC| >1) and 7 were downregulated between smokers and non-smokers using the edgeR algorithm (Figure S7).
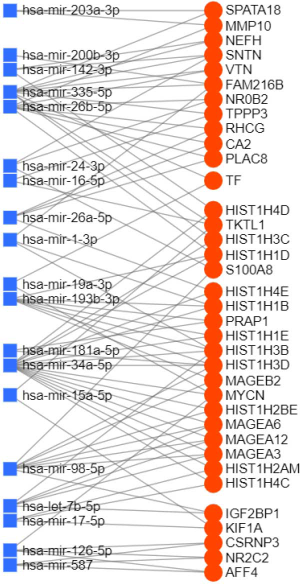
To determine the regulatory relationship between DEGs and miRNAs, we looked for links between the genes and miRNAs using the miRNet platform and obtained 7,945 pairs of miRNA-DEGs,while in consideration of the appearance of the figure, only the closest 85 pairs were shown in Figure 6 (differential genes were defined as P<0.001 and |logFC| >1 and differential miRNAs were defined as P<0.05). miR-335-5p and miR-34a-5p are located at the core of the network, suggesting these miRNAs and their binding genes may play an important role in the differences between smoking and non-smoking lung cancer mechanisms.
Validation of mRNA and miRNA expression by qRT-PCR
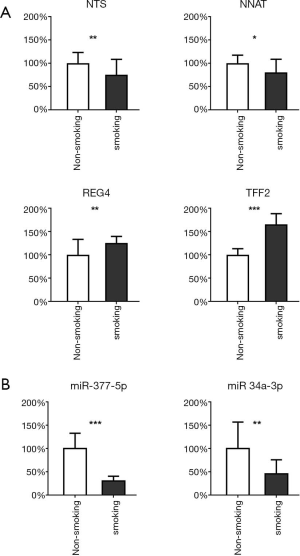
After comprehensively considering the significance of differences and the FC value, we selected 4 highly significantly differential genes and 2 miRNAs. NTS, NNAT and miR-377-5p, miR-34a-3p are the most up-regulated factors in non-smoking adenocarcinoma and the TFF2 and REG4 are the most down-regulated in TCGA data as shown in Figure 4 and Figure S7. Therefore, we used qRT-PCR to measure the expression levels of them in 20 pairs of non-smoking LUAD and smoking adenocarcinoma samples. As shown in Figure 7, the alteration of the factors’ expression in the results of qRT-PCR was generally consistent with that shown in the bioinformatics analyses though the FCs were not as large as those in the bioinformatics results.
Microenvironmental differences
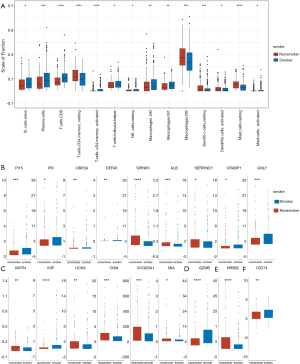
Tumor immune cell infiltration refers to the migration of immune cells from the blood to the tumor tissue, where they exert their function. Infiltrating immune cells can be isolated from the tumor tissue. Resting mast cells, M2 macrophages, memory resting CD4+ T cells, and resting dendritic cells were upregulated in non-smokers, while CD8+ T cells, plasma cells, activated CD4+ T cells, M1 macrophages, and follicular helper T cells were downregulated (Figure 8A). Furthermore, we looked for differences in immune-related gene expression. Nine genes related to antimicrobials (Figure 8B), six genes related to cytokines (Figure 8C), granzyme B (GZMB) for natural killer (NK) cell cytotoxicity (Figure 8D), and nuclear receptor subfamily 0 group B member 2 (NR0B2) for cytokine receptors (Figure 8E) were found to be significantly differentially expressed, indicating that cytotoxic and antimicrobial function may be one reason for the differences in the immune microenvironment between smokers and non-smokers. For programmed death-ligand 1 (PD-L1) (CD274), a significant difference in expression was also observed between smokers and non-smokers (Figure 8F), but with a low fold change value (logFC =0.09).
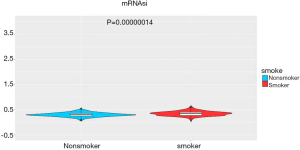
The stemness indices (mRNAsi) were calculated for the TCGA data. mRNAsi is an index calculated based on expression data, which ranges from 0 to 1; a value close to 1 indicates a lower degree of cell differentiation and stronger stem cell characteristics (16). We found a significant difference in stemness indices between the groups (496 samples; 207 non-smokers and 289 smokers, P<0.05, Figure 9), which confirmed the original results that non-smokers or long-term reformed smokers exhibit lower stemness than current and recently reformed smokers, as previously reported (16).
Discussion
The study of cancer predisposition and cancer-specific genes is an important research direction. In this study, we observed quantitative differences between the LUAD genome of never smokers compared with smokers. Many studies before concluded and verified that smoke would change human bronchial and pulmonary alveolar epithelial cells via affecting mitochondrial function and regulating the expression of several genes, especially PP2A, which might be connected with oncogenesis (17-20). However, these studies still focused more on how smoking affects lung acinar epithelial cells and causes chronic lung disease rather than tumors.
We proved that these differences existed at different levels of the genome, including somatic mutations, RNAs, and miRNAs, and some, mainly RNAs, showed significant increases or decreases at the cellular level.
Boeckx et al. (21) found that the frequency of EGFR mutations is higher, and the frequency of TP53 mutations, is reduced in never smokers compared with smokers. However, in our study, these genes only showed differences at the somatic level, and not the RNA level, which is in contrast to previous studies of smoking-related genomic and/or transcriptional alterations. These differences may be due to the selection of different patient populations, tumor characteristics, and cohort sizes (22). In our previous study (23), we found that over half of our patients carried EGFR mutations, followed by KRAS and HER2 mutations, the first two of which also show different rate of mutation in this study, while HER2 rem ALK, RET, and ROS1 translocations, PIK3CA, BRAF, and NRAS mutations were rare and all were identified in less than 2% of patients. PD-1 positive tumors were identified in 14 (1.9%) patients, and PD-L1 positive tumors in 95 (13.0%) patients.
In the present study, NTS and NNAT were upregulated in non-smoking LUAD patients, DMSO-treated A549 LUAD cells and our own samples. NTS, along with its receptor NTSR, has been shown to be important in lung cancer outcomes (24,25) and therapies against NTS can decrease tumor growth and metastasis (26,27). These data suggested that the overexpression of NTS in non-smokers may be a trigger for the development of LUAD. NNAT mRNA is mainly expressed in endocrine and adipose tissues in a hormone- and nutrient-sensitive manner (28-30) and has the potential to be used as a marker differentiating between Large-Cell Neuroendocrine Carcinoma (LCNEC) and small cell lung cancer (SCLC) as it more highly expressed in LCNEC, making it different from smoker NSCLC LUAD (31,32).
TFF2 and REG4 were both downregulated in non-smoking LUAD in TCGA and our own samples. Lung macrophages rely upon TFF2 to promote epithelial proliferation, while in the absence of TFF2, lung epithelia were unable to proliferate and expressed reduced lung mRNA transcript levels (33,34). In non-smoking LUAD patients, down-regulation of TFF2 might indicate weaker lung repair ability, which promotes tumor development. The overexpression of REG4, which was shown in the smoking group previously, was proven to be closely correlated to the carcinogenesis in some types of cancer (35). Also, REG4 plays an important role in KRAS-driven lung cancer pathogenesis and is a novel biomarker of LUAD subtype (36). However, whether its expression can be used to distinguish non-smoking from smoking LUAD needs further experiments to verify, as it showed unstable CT values when verifying.
A group of histones including HIST1H3F, HIST1H4C, HIST1H4D, HIST1H4E, HIST1H3B, HIST1H2AI, HIST1H1E, HIST1H2AM, HIST1H2BE, HIST1H3C, HIST1H1D, and HIST1H1B differentially expressed between smokers and nonsmokers in our study. Histone variants act as transcriptional activators or repressors of cancer-related genes. Several researches about HIST1H2, HIST1H3, HIST1H4 found that they were related to the progression and metastasis of tumor cell (37-39). However, there reports on how smoking regulates the expression of these histones so far.
Overexpression of miR-377-5p, which was more often in the non-smokers in our study, was reported remarkably downregulated in NSCLC tissues, and inhibited cell proliferation and development as a tumor‐suppressor gene (40,41). miR-34a overexpressed among the nonsmokers. Previous study found that p53 regulates PDL1 via miR-34 and SART3 overexpression increased miR-34a levels, which may affect cell cycle progression in NSCLC cells, while miR-34a inhibits NSCLC tumor growth and metastasis through targeting EGFR (42-44). Furthermore, it was also considered to be a target in the therapy of lung cancer (45,46).
We also focused on gene function. The leukocyte fraction varied substantially across the immune subtypes (15). We found that CD8+ T cells, plasma cells, activated CD4+ T cells, M1 macrophages, and follicular helper T cells were comparatively downregulated in non-smokers. Previously Kinoshita et al. found a high FOXP3/CD4 ratio in smokers with adenocarcinoma, a low number of CD20+ B cells in non-smokers was identified as an independent unfavorable prognostic factor in resected NSCLC, and infiltrating CD8+ T cells may not be activated sufficiently in the immunosuppressive microenvironment in non-smokers with adenocarcinoma (47,48). Li et al. (49) concluded that aberrant activation of mast cells and CD4+ memory T cells plays a crucial role in cigarette smoking-induced immune dysfunction in the lung, leading to tumor development and progression. Generally, tobacco smoking patients with NSCLC showed a higher PD-L1 tumor proportion score (50-52) and experience a higher response rate with immunotherapy, mainly pembrolizumab and nivolumab, than non-smokers (50,53-57). In the present study, we also found a significant difference in expression of the PD-L1 gene (CD274) between the two groups but only with a small fold-change value. All of this evidence suggested that smoking created a different immune environment with abnormal immune cells. Smokers appeared to develop LUAD more easily because their immune cells are not as limited as those of non-smokers.
A strong association between mRNAsi and tobacco smoking status in LUAD was shown in this study. This confirmed previous research, which suggested that the stemness of LUAD tumors might be activated in response to environmental stimuli such as smoking, and might influence tumor aggressiveness (16).
There are some limitations to this study. Smoking status data in the TCGA database did not include information regarding whether the patients were frequently exposed to second-hand smoke or kitchen fumes, which are also important causes of lung cancer. Also, our research results were mainly based on the data of TCGA and verified with GEO datasets. We were unable to verify our results with other databases because of a lack of high-quality data on smoking. Lung cancer caused by smoking is a long-term process, so it is difficult to determine the role of these factors. Additional studies will be necessary to further explore and verify these differences.
Conclusions
In summary, our study characterized thorough patterns of genome alterations and the microenvironment in non-smoking LUAD. Compared with smokers, non-smoking LUAD patients had a lower mutation rate in somatic cells, RNAs, and miRNAs with a different microenvironment, including immune cell infiltration and stemness, which revealed the complex association between molecular mechanisms and clinical outcomes. We believe this study will improve our understanding of the mechanism underlying non-smoking LUAD, and effectively prevent, diagnose, and treat LUAD.
Acknowledgments
The authors thank International Science Editing (http://www.internationalscienceediting.com) for editing this manuscript.
Funding: This work was supported by FDUROP (Fudan’s Undergraduate Research Opportunities Program), FDUROP (19028), National University Student Innovation Program Zhengyi Scholar Foundation of School of Basic Medical Sciences, Fudan University (S22-11) and the Training Program for the Talents of Zhongshan Hospital, Fudan University (2019ZSYXGG06).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/tlcr-20-276
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tlcr-20-276). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in this study were in accordance with the Declaration of Helsinki (as revised in 2013) and was approved by the Ethics Committee of Zhongshan Hospital, Fudan University, China (B2019 – 377R). Because of the retrospective nature of the research, the requirement for informed consent was waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Yoon JY, Sigel K, Martin J, et al. Evaluation of the Prognostic Significance of TNM Staging Guidelines in Lung Carcinoid Tumors. J Thorac Oncol 2019;14:184-92. [Crossref] [PubMed]
- WHO report on the global tobacco epidemic 2019. Available online: https://apps.who.int/iris/bitstream/handle/10665/326043/9789241516204-eng.pdf?ua=1, accessed 12 December 2019: Geneva: World Health Organization 2019.
- WHO highlights huge scale of tobacco-related lung disease deaths. In: World No Tobacco Day 2019: Don’t let tobacco take your breath away. Geneva: World Health Organization. Available online: https://www.who.int/news-room/detail/29-05-2019-who-highlights-huge-scale-of-tobacco-related-lung-disease-deaths. Accessed 12/12 2019.
- Glantz S, Gonzalez M. Effective tobacco control is key to rapid progress in reduction of non-communicable diseases. Lancet 2012;379:1269-71. [Crossref] [PubMed]
- Doll R, Hill AB. The mortality of doctors in relation to their smoking habits; a preliminary report. Br Med J 1954;1:1451-5. [Crossref] [PubMed]
- Parkin DM, Bray F, Ferlay J, et al. Global Cancer Statistics, 2002. CA Cancer J Clin 2005;55:74-108. [Crossref] [PubMed]
- Sun S, Schiller JH, Gazdar AF. Lung cancer in never smokers — a different disease. Nat Rev Cancer 2007;7:778-90. [Crossref] [PubMed]
- Toh CK, Gao F, Lim WT, et al. Never-Smokers With Lung Cancer: Epidemiologic Evidence of a Distinct Disease Entity. J Clin Oncol 2006;24:2245-51. [Crossref] [PubMed]
- Akhtar N, Bansal JG. Risk factors of Lung Cancer in nonsmoker. Curr Probl Cancer 2017;41:328-39. [Crossref] [PubMed]
- Der SD, Sykes J, Pintilie M, et al. Validation of a histology-independent prognostic gene signature for early-stage, non-small-cell lung cancer including stage IA patients. J Thorac Oncol 2014;9:59-64. [Crossref] [PubMed]
- Shedden K, Taylor JMG, Enkemann SA, et al. Gene expression-based survival prediction in lung adenocarcinoma: a multi-site, blinded validation study. Nat Med 2008;14:822-7. [Crossref] [PubMed]
- Okayama H, Kohno T, Ishii Y, et al. Identification of genes upregulated in ALK-positive and EGFR/KRAS/ALK-negative lung adenocarcinomas. Cancer Res 2012;72:100-11. [Crossref] [PubMed]
- Stueve TR, Li WQ, Shi J, et al. Epigenome-wide analysis of DNA methylation in lung tissue shows concordance with blood studies and identifies tobacco smoke-inducible enhancers. Hum Mol Genet 2017;26:3014-27. [Crossref] [PubMed]
- Thorsson V, Gibbs DL, Brown SD, et al. The Immune Landscape of Cancer. Immunity 2018;48:812-30.e14. [Crossref] [PubMed]
- Malta TM, Sokolov A, Gentles AJ, et al. Machine Learning Identifies Stemness Features Associated with Oncogenic Dedifferentiation. Cell 2018;173:338-54.e15. [Crossref] [PubMed]
- Doherty DF, Nath S, Poon J, et al. Protein Phosphatase 2A Reduces Cigarette Smoke-induced Cathepsin S and Loss of Lung Function. Am J Respir Crit Care Med 2019;200:51-62. [Crossref] [PubMed]
- Liu CY, Huang TT, Chen YT, et al. Targeting SET to restore PP2A activity disrupts an oncogenic CIP2A-feedforward loop and impairs triple negative breast cancer progression. EBioMedicine 2019;40:263-75. [Crossref] [PubMed]
- Ballweg K, Mutze K, Königshoff M, et al. Cigarette smoke extract affects mitochondrial function in alveolar epithelial cells. Am J Physiol Lung Cell Mol Physiol 2014;307:L895-L907. [Crossref] [PubMed]
- Mizumura K, Cloonan SM, Nakahira K, et al. Mitophagy-dependent necroptosis contributes to the pathogenesis of COPD. J Clin Invest 2014;124:3987-4003. [Crossref] [PubMed]
- Boeckx B, Shahi RB, Smeets D, et al. The genomic landscape of nonsmall cell lung carcinoma in never smokers. Int J Cancer 2020;146:3207-18. [Crossref] [PubMed]
- Karlsson A, Ringnér M, Lauss M, et al. Genomic and Transcriptional Alterations in Lung Adenocarcinoma in Relation to Smoking History. Clin Cancer Res 2014;20:4912. [Crossref] [PubMed]
- Zhao M, Zhan C, Li M, et al. Aberrant status and clinicopathologic characteristic associations of 11 target genes in 1,321 Chinese patients with lung adenocarcinoma. J Thorac Dis 2018;10:398-407. [Crossref] [PubMed]
- Reinecke M. Neurotensin. Immunohistochemical localization in central and peripheral nervous system and in endocrine cells and its functional role as neurotransmitter and endocrine hormone. Prog Histochem Cytochem 1985;16:1-172. [PubMed]
- Alifano M, Souazé F, Dupouy S, et al. Neurotensin receptor 1 determines the outcome of non-small cell lung cancer. Clin Cancer Res 2010;16:4401-10. [Crossref] [PubMed]
- Younes M, Wu Z, Dupouy S, et al. Neurotensin (NTS) and its receptor (NTSR1) causes EGFR, HER2 and HER3 over-expression and their autocrine/paracrine activation in lung tumors, confirming responsiveness to erlotinib. Oncotarget 2014;5:8252-69. [Crossref] [PubMed]
- Wu Z, Fournel L, Stadler N, et al. Modulation of lung cancer cell plasticity and heterogeneity with the restoration of cisplatin sensitivity by neurotensin antibody. Cancer Lett 2019;444:147-61. [Crossref] [PubMed]
- Millership SJ, Da Silva Xavier G, Choudhury AI, et al. Neuronatin regulates pancreatic β cell insulin content and secretion. J Clin Invest 2018;128:3369-81. [Crossref] [PubMed]
- Usui H, Morii K, Tanaka R, et al. cDNA cloning and mRNA expression analysis of the human neuronatin. High level expression in human pituitary gland and pituitary adenomas. J Mol Neurosci 1997;9:55-60. [Crossref] [PubMed]
- Joseph R, Dou D, Tsang W. Molecular cloning of a novel mRNA (neuronatin) that is highly expressed in neonatal mammalian brain. Biochem Biophys Res Commun 1994;201:1227-34. [Crossref] [PubMed]
- Nass N, Walter S, Jechorek D, et al. High neuronatin (NNAT) expression is associated with poor outcome in breast cancer. Virchows Arch 2017;471:23-30. [Crossref] [PubMed]
- Okubo C, Minami Y, Tanaka R, et al. Analysis of differentially expressed genes in neuroendocrine carcinomas of the lung. J Thorac Oncol 2006;1:780-6. [Crossref] [PubMed]
- Hung LY, Oniskey TK, Sen D, et al. Trefoil Factor 2 Promotes Type 2 Immunity and Lung Repair through Intrinsic Roles in Hematopoietic and Nonhematopoietic Cells. Am J Pathol 2018;188:1161-70. [Crossref] [PubMed]
- Hung LY, Sen D, Oniskey TK, et al. Macrophages promote epithelial proliferation following infectious and non-infectious lung injury through a Trefoil factor 2-dependent mechanism. Mucosal Immunol 2019;12:64-76. [Crossref] [PubMed]
- Jin J, Lv H, Wu J, et al. Regenerating Family Member 4 (Reg4) Enhances 5-Fluorouracil Resistance of Gastric Cancer Through Activating MAPK/Erk/Bim Signaling Pathway. Med Sci Monit 2017;23:3715-21. [Crossref] [PubMed]
- Sun S, Hu Z, Huang S, et al. REG4 is an indicator for KRAS mutant lung adenocarcinoma with TTF-1 low expression. J Cancer Res Clin Oncol 2019;145:2273-83. [Crossref] [PubMed]
- Park SM, Choi EY, Bae M, et al. Histone variant H3F3A promotes lung cancer cell migration through intronic regulation. Nat Commun 2016;7:12914. [Crossref] [PubMed]
- Ju J, Chen A, Deng Y, et al. NatD promotes lung cancer progression by preventing histone H4 serine phosphorylation to activate Slug expression. Nat Commun 2017;8:928. [Crossref] [PubMed]
- Hsu CC, Zhao D, Shi J, et al. Gas41 links histone acetylation to H2A.Z deposition and maintenance of embryonic stem cell identity. Cell Discov 2018;4:28. [Crossref] [PubMed]
- Wu H, Liu HY, Liu WJ, et al. miR-377-5p inhibits lung cancer cell proliferation, invasion, and cell cycle progression by targeting AKT1 signaling. J Cell Biochem 2018. [Crossref] [PubMed]
- Zhang J, Zhao M, Xue ZQ, et al. miR-377 inhibited tumorous behaviors of non-small cell lung cancer through directly targeting CDK6. Eur Rev Med Pharmacol Sci 2016;20:4494-9. [PubMed]
- Cortez MA, Ivan C, Valdecanas D, et al. PDL1 Regulation by p53 via miR-34. J Natl Cancer Inst 2015;108:djv303. [PubMed]
- Sherman EJ, Mitchell DC, Garner AL. The RNA-binding protein SART3 promotes miR-34a biogenesis and G cell cycle arrest in lung cancer cells. J Biol Chem 2019;294:17188-96. [Crossref] [PubMed]
- Li YL, Liu XM, Zhang CY, et al. MicroRNA-34a/EGFR axis plays pivotal roles in lung tumorigenesis. Oncogenesis 2017;6:e372. [Crossref] [PubMed]
- Xue W, Dahlman JE, Tammela T, et al. Small RNA combination therapy for lung cancer. Proc Natl Acad Sci U S A 2014;111:E3553-61. [Crossref] [PubMed]
- Jiang ZQ, Li MH, Qin YM, et al. Luteolin Inhibits Tumorigenesis and Induces Apoptosis of Non-Small Cell Lung Cancer Cells via Regulation of MicroRNA-34a-5p. Int J Mol Sci 2018;19:447. [Crossref] [PubMed]
- Kinoshita T, Kudo-Saito C, Muramatsu R, et al. Determination of poor prognostic immune features of tumour microenvironment in non-smoking patients with lung adenocarcinoma. Eur J Cancer 2017;86:15-27. [Crossref] [PubMed]
- Kinoshita T, Muramatsu R, Fujita T, et al. Prognostic value of tumor-infiltrating lymphocytes differs depending on histological type and smoking habit in completely resected non-small-cell lung cancer. Ann Oncol 2016;27:2117-23. [Crossref] [PubMed]
- Li X, Li J, Wu P, et al. Smoker and non-smoker lung adenocarcinoma is characterized by distinct tumor immune microenvironments. Oncoimmunology 2018;7:e1494677. [Crossref] [PubMed]
- Gainor JF, Shaw AT, Sequist LV, et al. EGFR Mutations and ALK Rearrangements Are Associated with Low Response Rates to PD-1 Pathway Blockade in Non-Small Cell Lung Cancer: A Retrospective Analysis. Clin Cancer Res 2016;22:4585-93. [Crossref] [PubMed]
- Rangachari D, VanderLaan PA, Shea M, et al. Correlation between Classic Driver Oncogene Mutations in EGFR, ALK, or ROS1 and 22C3-PD-L1 ≥50% Expression in Lung Adenocarcinoma. J Thorac Oncol 2017;12:878-83. [Crossref] [PubMed]
- Pan Y, Zheng D, Li Y, et al. Unique distribution of programmed death ligand 1 (PD-L1) expression in East Asian non-small cell lung cancer. J Thorac Dis 2017;9:2579-86. [Crossref] [PubMed]
- Mok TSK, Wu YL, Kudaba I, et al. Pembrolizumab versus chemotherapy for previously untreated, PD-L1-expressing, locally advanced or metastatic non-small-cell lung cancer (KEYNOTE-042): a randomised, open-label, controlled, phase 3 trial. Lancet 2019;393:1819-30. [Crossref] [PubMed]
- Sundar R, Soong R, Cho BC, et al. Immunotherapy in the treatment of non-small cell lung cancer. Lung Cancer 2014;85:101-9. [Crossref] [PubMed]
- Soria JC, Gettinger S, Gordon M, et al. 1322pbiomarkers associated with clinical activity of pd-l1 blockade in non-small cell lung cancer (NSCLC) patients (pts) in a phase I study of MPDL3280A. Ann Oncol 2014;25:iv465. [Crossref]
- Gandhi L, Rodríguez-Abreu D, Gadgeel S, et al. Pembrolizumab plus Chemotherapy in Metastatic Non-Small-Cell Lung Cancer. N Engl J Med 2018;378:2078-92. [Crossref] [PubMed]
- Borghaei H, Paz-Ares L, Horn L, et al. Nivolumab versus Docetaxel in Advanced Nonsquamous Non-Small-Cell Lung Cancer. N Engl J Med 2015;373:1627-39. [Crossref] [PubMed]