
Resistance to anaplastic lymphoma kinase inhibitors: knowing the enemy is half the battle won
Introduction
Efficacy outcomes of ALK-translocated NSCLC patients have been strikingly improved with the advent of new generation drugs which inhibit more efficiently aberrant activity of chimeric ALK proteins (1,2). Multiple ALKi are currently available in the clinical setting, nevertheless several open questions arise concerning the correct administration sequence (i.e., stepwise or more potent inhibitor upfront), since TKI may deeply influence the natural history of the tumor and, consequently, patients’ long-term outcomes (3,4).
Similarly to other oncogene-addicted NSCLC, ALK+ patients become sooner or later resistant to targeted therapy, showing clinically progressive disease with a wide range of aggressiveness (5-7). Multiple resistance mechanisms to ALK inhibition have been reported in the past years, even though they can be categorized in “ALK-dependent” or “on-target”, mainly due to intra-kinase domain mutations or ALK gene copy gain, and “ALK-independent” or “off-target”, such as by-pass signaling pathways activation, drug efflux mechanisms or histological transition (8-10). To recognize the specific molecular mechanism underpinning cancer progression is of paramount relevance, since it allows physicians to properly change the treatment, targeting that resistance mechanism at the best (11).
New high-throughput technologies [i.e., next-generation sequencing (NGS)], capable to interrogate tissue and plasma samples thoroughly and rapidly, allow the investigation of changes in the molecular landscape at the time of disease progression (12,13). Even sometimes hard to be accomplished due to the limited accessibility to site of disease progression for re-biopsy (e.g., central nervous system) or technological limitations (poor access to interventional radiology or sequencing platforms), ALK-translocated patients should be re-characterized along their disease story. The elucidation of biological mechanisms of resistance, not exclusively relevant for clinical decisions, will improve our knowledge on this patients’ subset providing physicians with tools to forecast clonal evolution and rapidly adjust therapeutic strategies.
On-target mechanisms of resistance
ALK mutations
A broad spectrum of mutations within the ALK enzymatic domain has been documented, similarly to what previously observed in patients with Philadelphia+ Chronic Myelogenous Leukemia receiving target therapies (i.e., imatinib, dasatinib, etc.) (14). The explanation why mutational landscape of ALK+ NSCLC is notably different from epidermal growth factor receptor (EGFR)+ patients, who develop T790M mutations as main mechanism of resistance to first- and second-generation TKI, may rely into different tumor biology (i.e., genomic instability in fusion-positive tumors and different oncogenic dependency) and distinct TKI properties (i.e., binding modalities, inhibiting potency) (15,16). Thus, ALKi of different generation, with different profiles of activity, generate different mutation profiles within ALK tyrosine-kinase domain (8) (Table 1).

Full table
First generation ALKi (crizotinib)
The reported frequency of secondary mutations acquired following treatment with crizotinib is around 20–30%, a lower proportion of cases compared to second-generation TKIs (8). Data from a large dataset of tumor samples assessed at the time of progression identified different mutations, even if two of them appear in the majority of the cases: L1196M (7%) and G1269A (4%) (8); both of them, the first as a classical gatekeeper mutation and the second one lying in the ATP-binding pocket, alter 3D conformation and hinder TKI binding (17-19). Other mutations (C1156Y/T, L1152P/R, I1151Tins, F1174C/L/V) are supposed to enhance the kinase activity, being localized at the C-terminus or N-terminus of the αC-helix domain (17,20-22). I1171T/N/S, G1202R, S1206Y, E1210K, G1269A mutations also interfere with the binding of TKI determining a conformational alteration of the αC-helix or a steric hindrance (8,23-25). Other rare variants have been identified in individual cases, such as G1128A located in the P-loop and conferring an increased enzymatic activity (26).
Second generation ALKi (ceritinib, alectinib, brigatinib, ensartinib)
The occurrence of acquired mutations following second-generation ALKi increases till 50–70% of patients with G1202R as the most frequent event (35–60% of all mutated samples) (8). In post-ceritinib tissue biopsies secondary mutations were detected in the 56% of the cases—with 17% of double mutations—mainly represented by G1202R (21%), F1174 C/L (17%) and, to a less extent, C1156Y (8%). Moreover, G1202del has been identified (8%) and functional studies demonstrated its partial sensitivity to crizotinib and moderate resistance to second-generation agents, differently from G1202R that confers high level of resistance to first and second generation TKIs (8). At the post-alectinib relapse, acquired mutations have been identified in 53% of the patients, again with G1202R as the most frequent event (29%); other mutations identified are: I1171T/S (12%), V11180L (6%), L1196M (6%) (8,22,24,25). Even though a limited number of samples from patients treated with brigatinib have been analyzed at relapse, the majority reported G1202R (43%) and then E1210K (29%), D1203N (14%), S1206Y/C (14%) (8). Of note, most patients had previously received crizotinib as first ALKi. Compared to other ALKi, ensartinib seems to be the most active inhibitor against G1123S and L1198F mutations, but less potent against G1202R and G1269A. At disease progression two mutations emerged: E1210K and, less relevant, S1206F. Longitudinal changes in ALK mutations were identified during the treatment with ensartinib, showing the relevant role of plasma analyses to track disease evolution (13).
Around 13% of patients who received a first- and second-generation ALKi developed two or more mutations, supporting the concept of progressive multistep genetic complexity. Sequential analyses of mutational profiles help to clarify that resistance is “private” for each ALKi, like F1174V that confers sensitivity and I1171 resistance to alectinib, differently from what has been observed for ceritinib (23,27). In the work by Yoda et al., a single patient, thoroughly analyzed along clonal evolution, developed a E1210K mutation post-crizotinib and two additional different post-brigatinib mutations; this denotes the emergency of two different clones (E1210K + S1206C and E1210K + D1203N) suggesting how TKI administration influences tumor evolution and response to more potent inhibitors (8,9).
Third generation ALKi (lorlatinib) and emergence of compound mutations
The mutational landscape of acquired resistance to lorlatinib has been extensively described, analyzing patients’ samples after different ALKi sequences. Initially, experimental models of acquired resistance have been generated through ENU mutagenesis assays of Ba/F3 cell lines harboring EML4-ALK wt or mut (G1202R) and subsequently these data were validated in a cohort of 20 tissue biopsies of patients progressing on lorlatinib (9).
Almost no patients with primary intrinsic resistance to lorlatinib carried any ALK mutations, suggesting the presence of ALK-independent mechanisms of resistance. On the other hand, 55% of patients with acquired (after initial response) resistance developed compound mutations (9). Consistently with pre-clinical results, patients’ samples showed double or triple mutations due to stepwise accumulation of resistance mechanisms. Inversely, two post-lorlatinib samples lose initially detected mutations. Studying the clonal evolution of three different patients, investigators identified specific pattern of compound mutations, such as: ALK-G1202R/L1196M, ALK-E1210K/D1203N/G1269A, ALK-I1171N/L1198F (9). Another recent study identified acquired compound mutations with different degrees of resistance to lorlatinib: F1174L/G1202R, L1196M/D1203N and C1156Y/G1269A. To what extent such patterns of resistance are sensitive to different ALKi is currently a field of research. It has been showed that, among others, the C1156Y/G1269A mutation maintains responsiveness to lorlatinib, thus indicating the presence of a concomitant off-target mechanism of resistance. On the other hand, the L1196M/D1203N secondary mutation confers a potent lorlatinib resistance, comparable to the L1196M/G1202R mutation that confers primary resistance to third generation ALKi. Conversely, the F1174L/G1202R mutation induces a slight increase in resistance to lorlatinib compared to the single G1202R mutation, which can be overcome by elevated compound concentration in vitro (28).
Taking into account the prevalence of G1202R mutation as mechanism of resistance after second-generation TKI, it could be speculated that the double compound mutation ALK-G1202R/L1196M (identified both through mutagenesis assays and in patient samples) could be the most frequent acquired mutation in post-lorlatinib patients in the clinical setting. Frighteningly, this genotype (comparable to other G1202-coupled mutations) is resistant to all commercially available ALKi, strongly narrowing treatment options of these patients. However, changeable scenarios may emerge such as the case of a patient treated with the sequence of crizotinib-ceritinib-lorlatinib, who developed a post-lorlatinib L1198F mutation, along with the post-crizotinib C1156Y, and surprisingly responded to crizotinib; this double compound mutation C1156Y/L1198F re-sensitizes patient’s tumor to first-generation ALKi (29). Also the I1171S/G1269A mutation, identified in a liver lesion at progression to lorlatinib, turned out to be sensitive to ceritinib and brigatinib and the patient responded to treatment with ceritinib (30).
ALK amplification
ALK gene copy number gain has been observed after exposure to crizotinib with a frequency of 7–18%, occasionally coupled with an ALK kinase mutation (19,21). In a cell line model, the association of amplification and mutation conferred high degree of resistance, while the copy number gain alone was not sufficient to generate a resistant phenotype to intermediate doses of crizotinib (18). However, this biological event was not reported as resistance mechanism after more potent ALKi, thus it might represent a non-clinically relevant phenomenon.
Off-target mechanisms of resistance
If intra-kinase domain mutations justify ~30% and ~50% of resistances following first and second/third generation TKI respectively, other resistance mechanisms, which are relevant in remaining cases, need to be investigated. If patients with ALK-independent mechanisms of resistance post-crizotinib still remain sensitive anyhow to ALK inhibition, due to a lower inhibiting potency of first-generation TKI, ALK-Independence post-second/third generation ALKi seem to no longer respond to ALK inhibition, suggesting how the presence of ALK secondary mutations predict a certain degree of ALK-dependency and, therefore, response to more potent ALKi (23,31,32).
Even if through different mechanisms (genetic mutations, by-pass kinase signaling, gene amplification, etc.), the result is a constitutive activation of intra-cellular downstream signaling pathways, such as RAS-MAPK axis, whose re-activation represents a crucial event driving resistance in ALK positive tumors (33) (Figure 1).
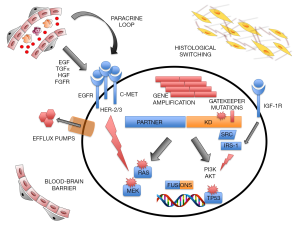
Co-occurring mutations
NGS approaches let the systematic screening of co-occurring genetic mutations as putative mechanisms of resistance to targeted therapy. Non-ALK mutations, in at least one different gene, have been identified in more than half of the cases (56%) of a post-second generation TKI cohort and in a large part of post-lorlatinib samples (8,9). The most frequently disrupted gene was TP53, even though, due to the low amount of pre-therapy samples to be matched, it is not clear if these mutations may pre-exist the development of resistance. Among other genes, rare private alterations at low frequency were identified in BRAF, FGFR2, MET, NRAS and PIK3CA genes (8,34). Among the others, mutations of POLE gene that encodes the catalytic subunit of DNA polymerase epsilon, permissive of the accumulation of a high number of somatic mutations, have been detected in post-crizotinib specimens (34). In a post-ceritinib patient-derived cell line a MAP2K1-K57N mutation was identified and it conferred sensitivity to the dual blockade of ALKi plus selumetinib (MEKi) (35). In the post-lorlatinib samples analyzed by Soda et al., several mutated genes identified may be causative of resistance, such as MAP3K1 disruption or NRAS-G12D activation (9).
Overall, these point mutations occur in a patient-private fashion, since appear as isolated event. Only TP53 mutations have been identified as relevant co-occurring mutations and, recently, their role as prognostic/predictive biomarker has been validated when tested baseline, rather than as a resistance-inducing event (8,34,36). Indeed, Christopoulos et al. recently highlighted the relevant clinical meaning of baseline TP53 mutational status in ALK+ NSCLC, showing that it significantly correlates with worse progression-free survival (PFS) and a shorter overall survival (OS), especially when associated with ELM4-ALK V3 variant. Moreover, the identification of TP53 mutations by liquid biopsies at progression resulted to be associated with a more aggressive disease (37,38).
By-pass kinases and downstream signaling pathways activation
Concomitant activation of other kinases, both on the cell surface and in the cytoplasmic compartment, is frequently the crucial by-pass signaling track that confers resistance to conventional ALK inhibition. Human epidermal growth factor receptor family (EGFR, HER2/HER3, HER4) activation is one of the first and more relevant mechanisms identified (20,21,39). EGFR up-regulation—in absence of mutations—has been identified both in post-crizotinib cell lines and in resistant patients’ samples, sometimes induced by paracrine stimuli, like NRG-1 upregulation (40-42). This event might pre-exist in therapy-naive tumor cells or appear during ALK targeting, rising and falling along treatments in a reversible manner (43,44). A recent study demonstrated that moderate resistance to alectinib in patients with leptomeningeal carcinomatosis (LMC) could be due to amphiregulin (AREG)-triggered EGFR activation, based on the important AREG overexpression found in resistant LMC cells in vitro. Moreover, in this model, the combined use of alectinib and EGFR-TKI, including the third-generation inhibitor osimertinib, significantly led to the control of disease progression at the central nervous system (45). Similarly, Redaelli et al. have identified EGFR phosphorylation as an escape mechanism in lorlatinib-resistant EML4-ALK cell lines, which can be controlled by combination treatment (e.g., lorlatinib plus erlotinib); thus highlighting the role of this adaptive mechanism also during third generation ALK inhibition (46).
Co-stimulation and transactivation of EGFR/MAPK pathway may be sustained also by different players, such as P2Y purinergic receptors that, when upregulated, boost signaling by PKC activation, a mechanism itself capable to induce resistance to ALK inhibition (42). Moreover, Lovly et al. identified a synergistic effect when co-targeting ALK and insulin-like growth factor 1 (IGF-1). IRS-1 was identified as a central adaptor protein for IGF1-R and ALK signaling and inhibition of the IGF1R/IRS1 pathway sensitizes tumor cells to ALK targeting (47). A drug-screening assay, through patient-derived and cell line models, highlighted a relevant synergistic effect of saracatinib (a dual Src and Bcr-Abl inhibitor) and ALKi, pinpointing the cause of resistance in SRC kinase upregulation. In fact, the phosphorylation of SRC substrates was increased after both first and second generation TKI (35,48).
More recently, other molecular changes have been identified. Recondo et al. reported a case of NF2 splicing site mutation that was detected at the progression to crizotinib, subsequently responsive to lorlatinib. At the time of progression to this latter agent, more disrupting events in NF2—a well-known tumor suppressor gene—appeared, causing a stronger bypass activation of the PI3K-AKT-mTOR pathway. Interestingly, cell-lines were sensitive to mTOR inhibition in vitro and in vivo with potential clinical implications (28). Activation of YAP (Yes-associated protein), a major downstream effector of the Hippo pathway, has been recognized as involved in resistance to ALK. YAP overexpression, indeed, hinders the response to alectinib in pre-clinical models and in tumor biopsies of crizotinib-resistant patients. These results, even if still premature, can provide the basis for YAP pathway targeting to overcome ALK-TKI resistance (49,50).
The RAS-MAPK dependency of ALK-rearranged cancers has been clearly elucidated and, being a crucial node where multiple molecular pathways converge, represent a solid pre-clinical evidence for upfront multi agent therapy approach, as emerging also in other types of oncogene addiction (i.e., TRK+ tumors) (33,51).
Gene amplification and translocations
Rather than ALK, different studies reported the occurrence of other genes’ amplification as causative of resistance phenotype. KIT amplification has been identified in one patient progressed on crizotinib; it has been shown that the interaction with stromal paracrine SCF ligand is crucial to confer this phenotype and dual ALK/KIT inhibition might overcome this constraint (21).
Whether alectinib is superior to crizotinib as first line therapy has been demonstrated in the clinical setting (1). However, some investigators claim for the dual ALK/MET inhibitory activity of crizotinib that is not observed with alectinib (52). Initially, anecdotic case reports showed MET amplification as potential cause of massive progression after alectinib administration (53). Nevertheless, HGF/MET amplifications have been found also in crizotinib-progressing samples as proper on-target escape mechanism (34). In sequential analyses of circulating tumor cells derived from a patient who progressed on crizotinib and become primary resistant to ceritinib and alectinib high level of MET amplification was detected; gene amplification developed after crizotinib administration and was confirmed in the liver biopsy at the time of clinical progression and claimed as the causative mechanism of resistance (54). More recently MET gene amplification has been confirmed as relevant mechanism after second and third generation ALKi. Dagogo-Jack et al. identified this event in the 12% and 22% of patients’ samples at progression after second generation or lorlatinib, respectively, therefore highlighting how the probability increases after more potent TKI treatment upfront; interestingly, proper dual inhibition of ALK plus MET, co-administering lorlatinib and crizotinib at low doses, seems a potential strategy to overcome resistance status (55). Similarly, other genes amplification, such as MYC gene copy number gain, may cause resistance to classical ALK inhibition, though these events seldom occur (56).
Genomic instability is the bedrock of acquired resistance mechanisms. An emerging molecular event, observed also in post-osimertinib EGFR+ patients, is the detection of fusion-driven clones (57). Although rarely reported, new gene translocations may restrain ALK+ NSCLC response to TKI. In the above reported paper, MET rearrangements were documented in two cases (one paired to MET amplification) (55). Moreover, a single case report shed light on the potential role of co-occurring NRG1 fusions as potential mechanism of resistance (58).
Phenotypic changes and stem-cell like properties
Among mechanisms of acquired resistance to ALK inhibitors, histological transformation such as epithelial-mesenchymal transition (EMT), switch to small cell (SCLC) or squamous cell lung cancer should be included. Notably, approximately 3‒10% of EGFR+ lung adenocarcinoma, under the selective pressure of EGFR TKIs, undergoes lineage change into SCLC or other neuroendocrine tumors, acquiring TP53 and RB1 mutations (59,60). Multiple clinical reports showed transformation to SCLC even in ALK+ NSCLC at progression on different generations of ALKi. In all the cases, ALK rearrangement was maintained on re-biopsy, suggesting a real histological switch, and not the outgrowth of a baseline pretreatment SCLC clone. Of note, histological switching may appear associated at the same time with acquisition of resistant mutations (e.g., L1196M or G1202R) (61-63). Similarly, transformation to squamous-cell histology has been claimed as causative of ALKi resistance; even though a rare event, morphological and immunohistochemical transformation (i.e., loss of TTF1 and gain of p40 positivity) were reported in the same primary site of initial adenocarcinoma diagnosis (64-66).
In post-ceritinib samples, analyzed by Gainor et al., 42% of the cases expressed immunohistochemical markers of mesenchymal differentiation (i.e., vimentin) and loose E-cadherin, acquiring spindle cell features; of note, 3 of the 5 cases reported also an ALK intra-kinase mutation, meaning that EMT might be one of the mechanisms hindering ALKi sensitivity in these patients (8). Another study suggested the co-existence of L1196M mutation and EMT in a crizotinib-resistant tissue sample, and the administration of HDAC inhibitors seemed to revert the transition both in vitro and in vivo models (67). Further on, Recondo et al. expanded two cell lines derived from patients with EMT during lorlatinib treatment: one associated with C1156Y/G1269A compound mutation, the other apparently without any intra-ALK mutations. Both models, expressing EMT features (high levels of vimentin, ZEB1, FGFR1), showed increased SRC pathway activation, demonstrating that its inhibition could in part re-establish E-cadherin expression. In this context, it is of interest that in vitro fibroblast growth factor receptor (FGFR) inhibitors sensitized ALK-rearranged EMT+ cell lines to lorlatinib, spurring the investigation of combination treatments for patients with phenotypic changes (28).
Molecular signaling underling these phenotypic transformations have not been extensively understood in ALK+ disease. Preclinical studies investigated cell line models matching the resistant EMT phenotype with the emergence of a stemness trait of ALK-translocated cells (68). Nakamichi et al. generated H2228 resistant cell lines to first- and second-generation ALKi and identified the acquisition of EMT and stem-cell like (CSC) features in a drug-tolerant cell subpopulation that had an augmented signaling of ALK and HSP90 pathways. Authors stated that co-targeting these molecules, rather than solely ALK inhibition, might prevent or delay the phenomenon of drug tolerance, due to the inhibitory effect on the associated proteins, clients of the chaperone machine (69,70).
Intrinsic primary resistance
Intrinsic resistance to ALK inhibition has been reported after first- and second/third-generation TKI, when the best-obtained clinical response is disease progression. Around 5–7% of post-crizotinib, 9% of post-ceritinib and 25% of post-lorlatinib cases demonstrated unresponsiveness to treatments and no specific ALK mutations have been identified (8). Multiple factors may underline primary resistance to ALK targeting. First of all, anyone of the above-described mechanisms existing de novo in tumor cells might impinge ALK dependency. If ALK mutations have not been claimed to be causative of this phenomenon, other co-existing by-pass signaling tracks may be taken into consideration. Receptor tyrosine kinases-dependent pathways (e.g., HER members), among others, may represent the co-stimulatory signaling able to re-shape the ALK addiction and may constitute the cause of primary resistance. This concept is indirectly supported by the evidence that, targeting SHP2, a non-receptor protein tyrosine phosphatase on which multiple signals converge, may revert resistance in absence of ALK mutations (71).
Multiple studies investigated whether ALK fusion partner and kinase variant play a role in determining a different degree of response to TKI. Partner genes fused to tyrosine kinase domain play critical roles in the oncogenic transformation, providing oligo-dimerization domain, recruiting scaffold proteins and dictating subcellular localization of the chimera; since a wide number of different partners has been described, a certain degree of intrinsic resistance to therapies may be properly justified (72). Moreover, specific variant (V3 > V1) of the classical EML4-ALK fusion protein seems to confer a more aggressive tumor behavior, with shorter TKI response, high-metastatic spread and reduced PFS/OS. Nevertheless, these retrospective evidences are not supported by data coming from large phase III trials (e.g., ALEX trial) (37,73).
A potential mechanism of intrinsic resistance relies on the augmented capability of the neoplastic cells to eliminate the drug accumulated in the cytoplasm, thus not reaching a proper concentration. This is why crizotinib, being substrate of ATP binding cassette drug efflux transporters, has a low penetration in the cerebrospinal fluid (CSF) not controlling, therefore, disease localization in the central nervous system (CNS) (74). More potent, next-generation TKI have been also developed to overcome this limitation and CNS disease control by these agents is definitively better, considering their capacity of delay brain metastases occurrence and produce more profound intra-cranial objective responses (75).
Lastly, the genomic complexity of cancers bearing translocations should always be taken into account. A major issue is represented by the correct detection of the genomic aberrations and false-positive cases may be a possible cause of the absence of response to TKI. Rosenbaum et al. have recently characterized the presence of “non-productive” rearrangements that are detected by FISH (gold-standard technique) but totally lacking the expression of chimeric protein (76). The harmonization of different diagnostic assays, including NGS, is therefore mandatory and will help refining the definition of ALK+ patients prone to receive TKI.
Conclusions
The Darwinian evolution of cancer cells under drug pressure is an inexhaustible source of resistance mechanisms underpinning disease progression; identification and characterization of these molecular transformations has to be at the crossroads of both preclinical and clinical research. Tumor heterogeneity preexists drug exposure and, multiplying during treatment, may facilitate the selection of cancer clones capable to evade TKI blocking (77). The pre-treatment landscape cooperates in generating the genomic complexity: the more instable is the cancer genome, the higher are the probabilities of acquire additional driver events. Within the molecular portrait of oncogene-positive lung adenocarcinoma, such as ALK-driven tumors, the inactivation of SETD2, a gene involved in the recruitment of DNA repair machinery, frequently occurs (78).
Resistance and disease progression may be due also to incapacity of drug delivery in sanctuary site, such as CNS. This had led to the development of more potent ALK inhibitors, giving the opportunity of sequential TKI administration. However, this strategy may contribute to the emergence of highly resistant mutations (i.e., compound resistance mutations after lorlatinib). The systematic analysis of molecular changes observed at the time of disease progression following upfront next-generation ALKi is matter of research and will be fully clarified in the next years. Other strategies have been investigating how delay or circumvent the emergence of resistance. Intermitting dose or alternate dose schedules in order to relieve drug pressure, administration upfront of dirty pan-inhibitory TKI capable of multi-targeting or early drug combination to maintain and restore drug sensitivity within drug-tolerant cells (70,79,80), are all strategies under investigation. However clinical applicability is not always an easy process and the accessibility into clinical trials for progressing patients remains a stronghold.
The knowledge of the molecular aspects surrounding ALK+ tumors is complex and, often, only marginal information is gained, not representative of the overall adaptive tumor plasticity. Nevertheless, the introduction of novel technologies and the possibility to perform extensive genomic analyses even on liquid specimens (e.g., blood, CSF) is a unique opportunity to speed up the research in this exciting field. The development and application of liquid biopsies on blood specimens—whose discussion is out of the scope of this review—has become a real milestone in the clinical management of oncogene-addicted patients. However, as previously discussed, the emergence of non-oncogene-driven resistance, such as phenotypic changes, underscores the indubitable role of tissue biopsy, when feasible. Differently from other scenarios (e.g., EGFR+ lung cancer, c-KIT+ gastrointestinal stromal tumors), ALK-rearranged NSCLC harbor specific features. The deep understanding of this disease both at baseline and after progression on different drugs is of paramount relevance to increase patients’ survival. At the same time, such wealth of knowledge should be translated to other rare rearrangement-driven tumors.
Acknowledgments
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editors (Silvia Novello, Francesco Passiglia) for the series “Looking for Chimeras in NSCLC: Widen Therapeutic Options Targeting Oncogenic Fusions” published in Translational Lung Cancer Research. The article was sent for external peer review organized by the Guest Editors and the editorial office.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tlcr-20-372). The series “Looking for Chimeras in NSCLC: Widen Therapeutic Options Targeting Oncogenic Fusions” was commissioned by the editorial office without any funding or sponsorship. GVS serves as an unpaid editorial board member of Translational Lung Cancer Research from Feb 2016 to Dec 2020. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Peters S, Camidge DR, Shaw AT, et al. Alectinib versus Crizotinib in Untreated ALK-Positive Non-Small-Cell Lung Cancer. N Engl J Med 2017;377:829-38. [Crossref] [PubMed]
- Camidge DR, Kim HR, Ahn MJ, et al. Brigatinib versus Crizotinib in ALK-Positive Non-Small-Cell Lung Cancer. N Engl J Med 2018;379:2027-39. [Crossref] [PubMed]
- Recondo G, Facchinetti F, Olaussen KA, et al. Making the first move in EGFR-driven or ALK-driven NSCLC: first-generation or next-generation TKI? Nat Rev Clin Oncol 2018;15:694-708. [Crossref] [PubMed]
- Remon J, Tabbò F, Jimenez B, et al. Sequential blinded treatment decisions in ALK-positive non-small cell lung cancers in the era of precision medicine. Clin Transl Oncol 2020;22:1425-9. [Crossref] [PubMed]
- Dagogo-Jack I, Shaw AT. Tumour heterogeneity and resistance to cancer therapies. Nat Rev Clin Oncol 2018;15:81-94. [Crossref] [PubMed]
- Schram AM, Chang MT, Jonsson P, et al. Fusions in solid tumours: diagnostic strategies, targeted therapy, and acquired resistance. Nat Rev Clin Oncol 2017;14:735-48. [Crossref] [PubMed]
- Lin JJ, Riely GJ, Shaw AT. Targeting ALK: Precision Medicine Takes on Drug Resistance. Cancer Discov 2017;7:137-55. b. [Crossref] [PubMed]
- Gainor JF, Dardaei L, Yoda S, et al. Molecular Mechanisms of Resistance to First- and Second-Generation ALK Inhibitors in ALK-Rearranged Lung Cancer. Cancer Discov 2016;6:1118-33. [Crossref] [PubMed]
- Yoda S, Lin JJ, Lawrence MS, Burke BJ, et al. Sequential ALK Inhibitors Can Select for Lorlatinib-Resistant Compound ALK Mutations in ALK-Positive Lung Cancer. Cancer Discov 2018;8:714-29. [Crossref] [PubMed]
- McCoach CE, Le AT, Gowan K, et al. Resistance Mechanisms to Targeted Therapies in ROS1+ and ALK+ Non-small Cell Lung Cancer. Clin Cancer Res 2018;24:3334-47. [Crossref] [PubMed]
- Delmonte A, Burgio MA, Verlicchi A, et al. New generation anaplastic lymphoma kinase inhibitors. Transl Lung Cancer Res 2019;8:S280-9. [Crossref] [PubMed]
- Dagogo-Jack I, Brannon AR, Ferris LA, et al. Tracking the Evolution of Resistance to ALK Tyrosine Kinase Inhibitors through Longitudinal Analysis of Circulating Tumor DNA. JCO Precis Oncol 2018. [Crossref] [PubMed]
- Horn L, Whisenant JG, Wakelee H, et al. Monitoring Therapeutic Response and Resistance: Analysis of Circulating Tumor DNA in Patients With ALK+ Lung Cancer. J Thorac Oncol 2019;14:1901-11. [Crossref] [PubMed]
- Patel AB, O'Hare T, Deininger MW. Mechanisms of Resistance to ABL Kinase Inhibition in Chronic Myeloid Leukemia and the Development of Next Generation ABL Kinase Inhibitors. Hematol Oncol Clin North Am 2017;31:589-612. [Crossref] [PubMed]
- Lobello C, Bikos V, Janikova A, et al. The Role of Oncogenic Tyrosine Kinase NPM-ALK in Genomic Instability. Cancers (Basel) 2018;10:64. [Crossref] [PubMed]
- Cui JJ, Tran-Dubé M, Shen H, et al. Structure based drug design of crizotinib (PF-02341066), a potent and selective dual inhibitor of mesenchymal-epithelial transition factor (c-MET) kinase and anaplastic lymphoma kinase (ALK). J Med Chem 2011;54:6342-63. [Crossref] [PubMed]
- Choi YL, Soda M, Yamashita Y, et al. EML4-ALK mutations in lung cancer that confer resistance to ALK inhibitors. N Engl J Med 2010;363:1734-9. [Crossref] [PubMed]
- Katayama R, Khan TM, Benes C, et al. Therapeutic strategies to overcome crizotinib resistance in non-small cell lung cancers harboring the fusion oncogene EML4-ALK. Proc Natl Acad Sci U S A 2011;108:7535-40. [Crossref] [PubMed]
- Doebele RC, Pilling AB, Aisner DL, et al. Mechanisms of resistance to crizotinib in patients with ALK gene rearranged non-small cell lung cancer. Clin Cancer Res 2012;18:1472-82. [Crossref] [PubMed]
- Sasaki T, Koivunen J, Ogino A, et al. A novel ALK secondary mutation and EGFR signaling cause resistance to ALK kinase inhibitors. Cancer Res 2011;71:6051-60. [Crossref] [PubMed]
- Katayama R, Shaw AT, Khan TM, et al. Mechanisms of acquired crizotinib resistance in ALK-rearranged lung Cancers. Sci Transl Med 2012;4:120ra17. [Crossref] [PubMed]
- Ignatius Ou SH, Azada M, Hsiang DJ, et al. Next-generation sequencing reveals a Novel NSCLC ALK F1174V mutation and confirms ALK G1202R mutation confers high-level resistance to alectinib (CH5424802/RO5424802) in ALK-rearranged NSCLC patients who progressed on crizotinib. J Thorac Oncol 2014;9:549-53. [Crossref] [PubMed]
- Friboulet L, Li N, Katayama R, et al. The ALK inhibitor ceritinib overcomes crizotinib resistance in non-small cell lung cancer. Cancer Discov 2014;4:662-73. [Crossref] [PubMed]
- Katayama R, Friboulet L, Koike S, et al. Two novel ALK mutations mediate acquired resistance to the next-generation ALK inhibitor alectinib. Clin Cancer Res 2014;20:5686-96. [Crossref] [PubMed]
- Toyokawa G, Hirai F, Inamasu E, et al. Secondary mutations at I1171 in the ALK gene confer resistance to both crizotinib and alectinib. J Thorac Oncol 2014;9:e86-7. [Crossref] [PubMed]
- Ai X, Niu X, Chang L, Chen R, et al. Next generation sequencing reveals a novel ALK G1128A mutation resistant to crizotinib in an ALK-Rearranged NSCLC patient. Lung Cancer 2018;123:83-6. [Crossref] [PubMed]
- Ou SH, Milliken JC, Azada MC, et al. ALK F1174V mutation confers sensitivity while ALK I1171 mutation confers resistance to alectinib. The importance of serial biopsy post progression. Lung Cancer 2016;91:70-2. [Crossref] [PubMed]
- Recondo G, Mezquita L, Facchinetti F, et al. Diverse Resistance Mechanisms to the Third-Generation ALK Inhibitor Lorlatinib in ALK-Rearranged Lung Cancer. Clin Cancer Res 2020;26:242-55. [Crossref] [PubMed]
- Shaw AT, Friboulet L, Leshchiner I, et al. Resensitization to Crizotinib by the Lorlatinib ALK Resistance Mutation L1198F. N Engl J Med 2016;374:54-61. [Crossref] [PubMed]
- Takahashi K, Seto Y, Okada K, et al. Overcoming resistance by ALK compound mutation (I1171S + G1269A) after sequential treatment of multiple ALK inhibitors in non-small cell lung cancer. Thorac Cancer 2020;11:581-7. [Crossref] [PubMed]
- Zou HY, Friboulet L, Kodack DP, et al. PF-06463922, an ALK/ROS1 inhibitor, overcomes resistance to first and second generation ALK inhibitors in preclinical models. Cancer Cell 2015;28:70-81. [Crossref] [PubMed]
- Shaw AT, Solomon BJ, Besse B, et al. ALK Resistance Mutations and Efficacy of Lorlatinib in Advanced Anaplastic Lymphoma Kinase-Positive Non-Small-Cell Lung Cancer. J Clin Oncol 2019;37:1370-9. [Crossref] [PubMed]
- Hrustanovic G, Olivas V, Pazarentzos E, et al. RAS-MAPK dependence underlies a rational polytherapy strategy in EML4-ALK-positive lung cancer. Nat Med 2015;21:1038-47. [Crossref] [PubMed]
- Kang J, Chen HJ, Zhang XC, et al. Heterogeneous responses and resistant mechanisms to crizotinib in ALK-positive advanced non-small cell lung cancer. Thorac Cancer 2018;9:1093-103. [Crossref] [PubMed]
- Crystal AS, Shaw AT, Sequist LV, et al. Patient-derived models of acquired resistance can identify effective drug combinations for cancer. Science 2014;346:1480-6. [Crossref] [PubMed]
- Kron A, Alidousty C, Scheffler M, et al. Impact of TP53 mutation status on systemic treatment outcome in ALK-rearranged non-small-cell lung cancer. Ann Oncol 2018;29:2068-75. [Crossref] [PubMed]
- Christopoulos P, Kirchner M, Bozorgmehr F, et al. Identification of a highly lethal V3+ TP53+ subset in ALK+ lung adenocarcinoma. Int J Cancer 2019;144:190-9. [Crossref] [PubMed]
- Christopoulos P, Dietz S, Kirchner M, et al. Detection of TP53 Mutations in Tissue or Liquid Rebiopsies at Progression Identifies ALK+ Lung Cancer Patients with Poor Survival. Cancers (Basel) 2019;11:124. [Crossref] [PubMed]
- Miyawaki M, Yasuda H, Tani T, et al. Overcoming EGFR Bypass Signal-Induced Acquired Resistance to ALK Tyrosine Kinase Inhibitors in ALK-Translocated Lung Cancer. Mol Cancer Res 2017;15:106-14. [Crossref] [PubMed]
- Tanizaki J, Okamoto I, Okabe T, et al. Activation of HER family signaling as a mechanism of acquired resistance to ALK inhibitors in EML4-ALK-positive non-small cell lung cancer. Clin Cancer Res 2012;18:6219-26. [Crossref] [PubMed]
- Yamada T, Takeuchi S, Nakade J, et al. Paracrine receptor activation by microenvironment triggers bypass survival signals and ALK inhibitor resistance in EML4-ALK lung cancer cells. Clin Cancer Res 2012;18:3592-602. [Crossref] [PubMed]
- Wilson FH, Johannessen CM, Piccioni F, et al. A functional landscape of resistance to ALK inhibition in lung cancer. Cancer Cell 2015;27:397-408. [Crossref] [PubMed]
- Voena C, Di Giacomo F, Panizza E. The EGFR family members sustain the neoplastic phenotype of ALK+ lung adenocarcinoma via EGR1. Oncogenesis 2013;2:e43. [Crossref] [PubMed]
- Vaishnavi A, Schubert L, Rix U, et al. EGFR Mediates Responses to Small-Molecule Drugs Targeting Oncogenic Fusion Kinases. Cancer Res 2017;77:3551-63. [Crossref] [PubMed]
- Arai S, Takeuchi S, Fukuda K, et al. Osimertinib overcomes alectinib resistance caused by amphiregulin in a leptomeningeal carcinomatosis model of ALK-rearranged lung cancer. J Thorac Oncol 2020;15:752-65. [Crossref] [PubMed]
- Redaelli S, Ceccon M, Zappa M, et al. Lorlatinib Treatment Elicits Multiple On- and Off-Target Mechanisms of Resistance in ALK-Driven Cancer. Cancer Res 2018;78:6866-80. [Crossref] [PubMed]
- Lovly CM, McDonald NT, Chen H, et al. Rationale for co-targeting IGF-1R and ALK in ALK fusion-positive lung cancer. Nat Med 2014;20:1027-34. [Crossref] [PubMed]
- Tsuji T, Ozasa H, Aoki W, et al. Alectinib Resistance in ALK-Rearranged Lung Cancer by Dual Salvage Signaling in a Clinically Paired Resistance Model. Mol Cancer Res 2019;17:212-24. [Crossref] [PubMed]
- Tsuji T, Ozasa H, Aoki W, et al. YAP1 mediates survival of ALK-rearranged lung cancer cells treated with alectinib via pro-apoptotic protein regulation. Nat Commun 2020;11:74. [Crossref] [PubMed]
- Yun MR, Choi HM, Lee YW, et al. Targeting YAP to overcome acquired resistance to ALK inhibitors in ALK-rearranged lung cancer. EMBO Mol Med 2019;11:e10581. [Crossref] [PubMed]
- Cocco E, Schram AM, Kulick A, et al. Resistance to TRK inhibition mediated by convergent MAPK pathway activation. Nat Med 2019;25:1422-7. [Crossref] [PubMed]
- Huang X. The potential role of HGF-MET signaling and autophagy in the war of Alectinib versus Crizotinib against ALK-positive NSCLC. J Exp Clin Cancer Res 2018;37:33. [Crossref] [PubMed]
- Gouji T, Takashi S, Mitsuhiro T, et al. Crizotinib can overcome acquired resistance to CH5424802: is amplification of the MET gene a key factor? J Thorac Oncol 2014;9:e27-8. [Crossref] [PubMed]
- Berger LA, Janning M, Velthaus JL, et al. Identification of a High-Level MET Amplification in CTCs and cfTNA of an ALK-Positive NSCLC Patient Developing Evasive Resistance to Crizotinib. J Thorac Oncol 2018;13:e243-6. [Crossref] [PubMed]
- Dagogo-Jack I, Yoda S, Lennerz JK, et al. MET Alterations are a Recurring and Actionable Resistance Mechanism in ALK-Positive Lung Cancer. Clin Cancer Res 2020;26:2535-45. [Crossref] [PubMed]
- Rihawi K, Alfieri R, Fiorentino M, et al. MYC Amplification as a Potential Mechanism of Primary Resistance to Crizotinib in ALK-Rearranged Non-Small Cell Lung Cancer: A Brief Report. Transl Oncol 2019;12:116-21. [Crossref] [PubMed]
- Piotrowska Z, Isozaki H, Lennerz JK, et al. Landscape of Acquired Resistance to Osimertinib in EGFR-Mutant NSCLC and Clinical Validation of Combined EGFR and RET Inhibition with Osimertinib and BLU-667 for Acquired RET Fusion. Cancer Discov 2018;8:1529-39. [Crossref] [PubMed]
- Muscarella LA, Trombetta D, Fabrizio FP, et al. ALK and NRG1 Fusions Coexist in a Patient with Signet Ring Cell Lung Adenocarcinoma. J Thorac Oncol 2017;12:e161-3. [Crossref] [PubMed]
- Niederst MJ, Sequist LV, Poirier JT, et al. RB loss in resistant EGFR mutant lung adenocarcinomas that transform to small-cell lung cancer. Nat Commun 2015;6:6377. [Crossref] [PubMed]
- Offin M, Chan JM, Tenet M, et al. Concurrent RB1 and TP53 Alterations Define a Subset of EGFR-Mutant Lung Cancers at risk for Histologic Transformation and Inferior Clinical Outcomes. J Thorac Oncol 2019;14:1784-93. [Crossref] [PubMed]
- Fujita S, Masago K, Katakami N, et al. Transformation to SCLC after Treatment with the ALK Inhibitor Alectinib. J Thorac Oncol 2016;11:e67-72. [Crossref] [PubMed]
- Balla A, Khan F, Hampel KJ, et al. Small-cell transformation of ALK-rearranged non-small-cell adenocarcinoma of the lung. Cold Spring Harb Mol Case Stud 2018;4:a002394. [Crossref] [PubMed]
- Coleman N, Wotherspoon A, Yousaf N, et al. Transformation to neuroendocrine carcinoma as a resistance mechanism to lorlatinib. Lung Cancer 2019;134:117-20. [Crossref] [PubMed]
- Park S, Han J, Sun JM. Histologic transformation of ALK-rearranged adenocarcinoma to squamous cell carcinoma after treatment with ALK inhibitor. Lung Cancer 2019;127:66-8. [Crossref] [PubMed]
- Gong J, Gregg JP, Ma W, et al. Squamous Cell Transformation of Primary Lung Adenocarcinoma in a Patient With EML4-ALK Fusion Variant 5 Refractory to ALK Inhibitors. J Natl Compr Canc Netw 2019;17:297-301. [Crossref] [PubMed]
- Wang F, Qin J, Xie F, et al. Transformation of EML4-ALK fusion-positive adenocarcinoma into squamous cell carcinoma in association with acquired resistance to crizotinib. Lung Cancer 2020;140:118-20. [Crossref] [PubMed]
- Fukuda K, Takeuchi S, Arai S, et al. Epithelial-to-Mesenchymal Transition Is a Mechanism of ALK Inhibitor Resistance in Lung Cancer Independent of ALK Mutation Status. Cancer Res 2019;79:1658-70. [Crossref] [PubMed]
- Guo F, Liu X, Qing Q, et al. EML4-ALK induces epithelial- mesenchymal transition consistent with cancer stem cell properties in H1299 non- small cell lung cancer cells. Biochem Biophys Res Commun 2015;459:398-404. [Crossref] [PubMed]
- Nakamichi S, Seike M, Miyanaga A, et al. Overcoming drug-tolerant cancer cell subpopulations showing AXL activation and epithelial-mesenchymal transition is critical in conquering ALK-positive lung cancer. Oncotarget 2018;9:27242-55. [Crossref] [PubMed]
- Sang J, Acquaviva J, Friedland JC, et al. Targeted inhibition of the molecular chaperone Hsp90 overcomes ALK inhibitor resistance in non-small cell lung cancer. Cancer Discov 2013;3:430-43. [Crossref] [PubMed]
- Dardaei L, Wang HQ, Singh M, et al. SHP2 inhibition restores sensitivity in ALK-rearranged non-small-cell lung cancer resistant to ALK inhibitors. Nat Med 2018;24:512-7. [Crossref] [PubMed]
- Childress MA, Himmelberg SM, Chen H, et al. ALK Fusion Partners Impact Response to ALK Inhibition: Differential Effects on Sensitivity, Cellular Phenotypes, and Biochemical Properties. Mol Cancer Res 2018;16:1724-36. [Crossref] [PubMed]
- Camidge DR, Dziadziuszko R, Peters S, et al. Updated Efficacy and Safety Data and Impact of the EML4-ALK Fusion Variant on the Efficacy of Alectinib in Untreated ALK-Positive Advanced Non-Small Cell Lung Cancer in the Global Phase III ALEX Study. J Thorac Oncol 2019;14:1233-43. [Crossref] [PubMed]
- Costa DB, Kobayashi S, Pandya SS, et al. CSF concentration of the anaplastic lymphoma kinase inhibitor crizotinib. J Clin Oncol 2011;29:e443-5. [Crossref] [PubMed]
- Gadgeel S, Peters S, Mok T, et al. Alectinib versus crizotinib in treatment-naive anaplastic lymphoma kinase-positive (ALK+) non-small-cell lung cancer: CNS efficacy results from the ALEX study. Ann Oncol 2018;29:2214-22. [Crossref] [PubMed]
- Rosenbaum JN, Bloom R, Forys JT, et al. Genomic heterogeneity of ALK fusion breakpoints in non-small-cell lung cancer. Mod Pathol 2018;31:791-808. [Crossref] [PubMed]
- Bhang HE, Ruddy DA, Krishnamurthy Radhakrishna V, et al. Studying clonal dynamics in response to cancer therapy using high-complexity barcoding. Nat Med 2015;21:440-8. [Crossref] [PubMed]
- Lee JJ, Park S, Park H, et al. Tracing Oncogene Rearrangements in the Mutational History of Lung Adenocarcinoma. Cell 2019;177:1842-57.e21. [Crossref] [PubMed]
- Foo J, Chmielecki J, Pao W, Michor F. Effects of pharmacokinetic processes and varied dosing schedules on the dynamics of acquired resistance to erlotinib in EGFR-mutant lung cancer. J Thorac Oncol 2012;7:1583-93. [Crossref] [PubMed]
- Shrestha N, Nimick M, Dass P, et al. Mechanisms of suppression of cell growth by dual inhibition of ALK and MEK in ALK-positive non-small cell lung cancer. Sci Rep 2019;9:18842. [Crossref] [PubMed]


