
A narrative review of digital pathology and artificial intelligence: focusing on lung cancer
Advances in digital pathology
Advances in the speed of technology for digitizing glass slides and the drop in storage prices has significantly expanded the adoption of whole slide images (WSIs), also known as virtual slides. WSIs enable users to examine slides digitally on electronic displays under different magnifications as seamlessly as in the Google Maps application.
With numerous feasibility tests (1-4), tools for digital pathology imaging, mostly WSIs, have been approved for clinical use by regulatory agencies in major countries. As a first step, the College of American Pathologists (CAP) has published guidelines for the use of digital pathology (5). Shortly thereafter, the WSI scanner introduced by Philips was the first to acquire CE marking in 2014. This approval, along with others, has increased the interest in digital pathology with many labs shifting to digitization across the world. Such movement is analogous to the transition from films to DICOM in diagnostic radiology, which has taken place over the last 2–3 decades (6).
The transition to digital pathology has been slower owing to many factors, including the difficulty in assessing the return of investment, and it remains challenging even with regulatory approval of multiple commercial systems. Digitizing a pathology practice requires significant changes in workflow and histology lab practices, including slide preparation, quality control, labeling, and custom integration with existing laboratory information systems (LIS). Digital practice can generate petabytes of data that must be stored long term to perceive many of the benefits of digitization. Capital investments in storage systems and staff to maintain them are expensive.
Despite many discussions regarding the benefits of digitization, a vast majority of pathologists have been hesitant to accept the change in workflow and have avoided the implementation of digital practice (6,7). Following a decade with no major progress, WSIs have now been adopted for routine clinical practice only at several major hospitals across Europe, Asia, and the US.
Needless to say, there are many benefits of digitization—the most noteworthy being its use for telepathology and computational image analysis. Advances over the past decade in artificial intelligence (AI) technology point toward a potentially significant impact on the practice of pathology and other diagnostic fields in medicine.
Telepathology has existed in some form using still images or video for more than 30 years (8). Consultation using still images, an easy method that can be performed even with smartphones, is popular among pathologists globally (9,10). SNS is another popular tool in which captured images are uploaded and cases are discussed on a daily basis (11). Simplicity and convenience have been key factors in the dissemination of digital tools, especially among young pathologists.
Full remote diagnosis by scanning slides has become a reality with WSIs that capture an entire slide at high magnification, and the rise of 5G technology is expected to accelerate the use of WSIs in remote diagnosis. As it was pointed out above, currently, there are a few operating remote diagnostic networks in various areas of the world (12-14). This trend will accelerate, allowing institutions to develop remote consultation systems that include outside expert pathologists more easily. Digital remote consultation will significantly reduce turn-around time and replace the current consultation procedure of sending actual glass slides which takes longer and involves the risks of breaking or losing the sent materials (15,16).
Remote diagnosis not only supports hospitals with an insufficient number of pathologists, but it also benefits academic institutes. Its effect is maximized when used in combination with telecommunication services such as Skype, WebEx, Zoom, Google Handouts, Spark, or TeamViewer. Such use is best suited for local university hospitals, for example, the Japanese national universities hosting medium-sized affiliated hospitals. These institutions handle on average 7,000–15,000 histopathological cases annually—a workload relatively lower than that in Western universities. Accordingly, the number of board-certified pathologists per such laboratory is typically limited to a few experts.
Remote diagnosis is particularly beneficial to small-sized academic institutes, wherein the development of training programs covering all pathology subspecialties is challenging. Via a web communication, these institutes can share their expertise in subspecialties, digital infrastructure, and faculty; they can together develop comprehensive educational programs for trainees.
Our team established multi-institutional digital pathology network in 2017. Nagasaki-Kameda digital pathology network connects academic institution (Nagasaki University Hospital), large-scale hospital (Kameda Medical Center), and several independent and affiliated centers (Figure 1). It includes around 40 pathologists, both general and specialized, who are responsible for diagnosing over 40,000 cases annually. Together the hospitals in this network cover a broad range of subspecialty experts, including thoracic, gastrointestinal, pancreatobiliary, genitourinary, soft tissue, head and neck, breast, renal, endocrine medicine, hematopathology, and more.
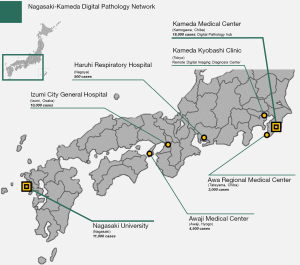
Remote sign-out using a telecommunication system
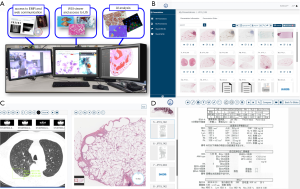
We routinely use a virtual conferencing system to sign out pulmonary consultation cases received from all over Japan to assure the quality of diagnosis. A secure communication channel (Cisco Webex, Milpitas, CA, USA) is open for all participating labs during working hours and allows remote sites to contact the consulting site via a persistent video-conferencing connection. Scanned pathology images are uploaded by remote sites to a HIPAA-compliant secure cloud database (PathPresenter, New York, USA), along with radiology DICOM images and PDF reports including clinical data following complete removal of patient personal identifiers. The data for this single case is then accessible to pathologists at the consulting and providing sites, and annotations and comments can be added dynamically during the session by multiple pathologists. The driver of the session, typically the attending consultant, shares their desktop via network for all members to see as the attending consultant navigates the slide, enabling consensus diagnosis. Histopathological specimens are routinely correlated with findings of digital cytology, which is particularly important in the field of lung pathology. Multidisciplinary team (MDT) discussions with radiologists and pulmonologists from multiple institutes all over Japan are routinely held using the same system.
We have previously reported that the diagnosis of interstitial pneumonia is significantly improved when using a nationwide cloud-based integrated database of clinical data, radiological DICOMs, and pathological WSIs, along with a virtual conferencing system, to create a virtual central MDT diagnostic center (17). The expansion of functions within the cloud-oriented setting will significantly contribute to the improvement in diagnostic accuracy and education system to train pathologists (Figure 2); these functions include seamless viewing, searching, annotating, and commenting.
Basics of artificial intelligence in pathology
Digitization of the pathology practice creates opportunities for the application of various computational approaches, including AI and machine learning techniques. These approaches may improve the accuracy of diagnosis, aid in exploring and defining new diagnostic and prognostic criteria, and play a role in helping pathology labs handle increased workloads and expertise shortages. Herein, we discuss the potential advantages of and challenges encountered in advancing these methods for use in routine clinical practice. We begin by describing fundamental principles of the technology and later discuss their applications in the field of lung cancer. We present the following article/case in accordance with the Narrative Review reporting checklist (available at http://dx.doi.org/10.21037/tlcr-20-591).
History of AI
The concept of AI emerged in the 1950s with Dr. Alan Turing who described the notion as “Computing Machinery and Intelligence” (18). Since the 1950s, AI has witnessed periods of success and decline owing to methodological advances, advances in computing technology, and the accumulation and generation of labeled datasets for developing and validating AI systems.
The specific term artificial intelligence was first used to describe “thinking machines” that could solve problems typically reserved for humans at the Dartmouth summer research project conference organized by Dr. John McCarthy in 1955 (19). Researchers in the field attempted to develop intelligence similar to that of the human brain by modifying factors called “reasoning as search.” However, only limited progress, such as being able to solve puzzles and simple games, were made. This was far from the level required to make it a useful technology. When researchers failed to deliver the desired results, the funding for AI reduced significantly.
The second boom was around the 1980s when computers were more accessible to the public as “personal computers”. The main research of this period was called the expert system in which the accumulated knowledge of experts was used for machine training to be used in problem solving (20). However, its area of application was very limited, and the boom ended without any significant breakthrough.
Since the late 2000s, significant advances have been made in computing technology and data accumulation, which have enabled breakthrough results. The creation of databases such as ImageNet by Fei-Fei Li at Stanford in 2009 created an open benchmark of more than 14 million images that researchers could use to develop methods and compare their relative success. This was formalized by the ImageNet Large Scale Visual Recognition Challenge (ILSVRC), where teams submit algorithms to perform classification and detection in a challenge environment. The establishment of ImageNet and ILSVRC is now recognized as a significant factor in accelerating the development of AI methods (21).
Types and fundamental structure of AI
Machine learning
AI is a broad term used for certain types of computer technology, some of which is called machine learning. Machine learning refers to a system in which a computer repeatedly learns from data, and the computer can derive an answer by the learning effect without a person providing any guidance. There is a technique in machine learning called deep learning, in which artificial neural networks of calculating “cells” are multilayered to resemble the human brain.
Machine learning constructs predictive models from data to identify patterns or to perform tasks like regression or classification. There are two main types of machine learning methods-supervised learning and unsupervised learning. In supervised learning, data are structured as paired features (e.g., images or other measurements) and their labels (ground truth). These data serve as examples for the algorithm to learn the relationships between the features and labels in a process called training. Models that have been trained are validated on independent data to assess their performance, and can be applied to new data prospectively to make predictions. One example of a classification task is predicting whether a WSI contains cancer (here the classes are “cancer” and “non-cancer”).
In pathology the training data is often derived as annotations which can be made at various levels (Figure 3). Annotations range from those that delineate areas of tumor, to labels of square patches as containing cancer cells (or not), to annotation of individual nuclei as neoplastic, stromal, or inflammatory cells. To train an accurate model, a large number of annotations may be required depending on the intrinsic difficulty of the problem, variability in pre-analytic factors like staining, and complexity of the algorithm used. Annotation process presents many challenges, among them being subjectivity of the annotation, and the requirement of an expert pathologist to supervise the process and to produce or approve large numbers of annotations.
In unsupervised learning there are no labels, and the goal is to identify patterns in the features such as how they tend to aggregate. The most common unsupervised learning task is called clustering, in which the AI identifies similar properties in data and sorts them into groups, although there are other tasks that attempt to learn to accurately model visual patterns in image data.
Deep learning
Deep learning most often refers to a neural network composed of many layers (hence the description “deep”). These adaptive algorithms have demonstrated a remarkable ability to learn from complex data like images with unrivaled accuracy. One way to conceptualize these algorithms is that each layer transforms the data to produce a new representation of the problem. As these layers are stacked, the algorithm can learn to represent complex phenomena through successive transformation of the input data via the layers and by repeated exposure to the data during training. This characteristic enables learning directly from “raw” data like images without needing any intermediate representation.
Prior to this development, research in machine learning for pathology focused on developing methods to transform pathology images into intermediate features that capture what humans think is important. For example, for classifying whether an image contains cancer, one might start by using image processing algorithm to delineate individual cell nuclei, and then making morphologic measurements of the shapes and textures of these nuclei that serve as features for training. In contrast to deep learning, this approach has the advantage of being transparent and explainable. The accuracy of these methods is typically inferior since the definition of features is not adaptive but determined a priori. Deep learning avoids this bias, learning features in a way that is entirely driven by labels in an unbiased manner. As a consequence, trained deep learning algorithms cannot be readily explained and are referred to as “black box” algorithms. This lack of explainability presents problems for validation, and black box algorithms may fail in unpredictable ways which is dangerous in clinical applications.
Convolutional neural networks (CNNs)
CNNs are a type of neural network for processing images. These networks explicitly model the spatial structure of images using convolution operations. The introduction of deep CNNs (DCNN) in the 2010s led to significant improvements in many image analysis tasks and so DCNNs have become the predominant approach for image analysis today. To describe how CNNs function we first introduce neural networks and then describe the convolution operation.
The fundamental element in a neural network is a computational model of a neuron (Figure 4A) which is inspired by biological neurons. In this model, input signals (x1, x2, .., xn) are weighted, summed, and transformed to obtain an output (y). Each input has a corresponding weight (w1, w2, .., wn) that is adjusted through training to learn how to optimally combine the inputs to minimize prediction error.
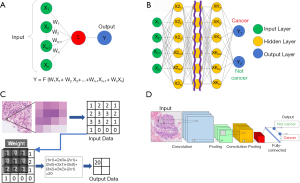
When these artificial neurons are layered in a neural network (NN), the outputs of one neuron become the inputs of another. Figure 4B presents a schematic of the entire neural network. Here, the weight symbols are omitted to simplify the drawing. In this case, m different weighting factors are assigned to the inputs (x1, x2, .., xn) to obtain m outputs (x1, x2, .., xm). By using this as input for another layer of neurons, a cascade of layers can be formed. The first (x1, x2, .., xn) is called the input layer, the final (y1, y2) is called the output layer, and the intermediate layers are called the hidden layers. Each pixel of the pathological image is input as (x1, x2, .., xn). If the input image is a cancer image, the output is y1 > y2, and if the input image is a non-cancer image, the output is y1 < y2. Training involves the adaptation of all weights to explain the relationships between the input features and corresponding labels. The number of weights can be large, easily in the millions. Therefore, a computer with a specialized device called a graphics processing unit (GPU) is required to efficiently carry out the mathematical operations used in training in parallel.
For dealing with images as inputs, such as high-power fields from a pathology image, it is beneficial for the weights to represent the grid-like structure of the inputs. The values of neighboring pixels in an image are correlated, and by organizing the weights similarly, a CNN can learn to represent the patterns observed in images of histology like the shapes or textures of cells at high magnification, or cell orientation and tissue architecture at lower magnifications. An arithmetic method called convolution is used when the weights are structured to represent the spatial relationships of pixels. As illustrated in Figure 4C, the CNN performs an operation and derives its result from 9 or 25 nearby pixels using a filter consisting of grids, such as a 3×3 or a 5×5 grid, and repeats the same operation to make the layer deeper as is done in the case of a neural network. WSIs are extremely large, containing billions of pixels, and typically cannot be analyzed directly by a CNN without subdividing them into smaller parts. This process often involves tiling the WSI with a regular determined to be appropriate for the application, and these patches are then analyzed individually during training and prediction (Figure 4D).
Developing a CNN involves many design decisions. How many layers should be used? How many neurons should each layer have? The answers to these design questions are made based on a combination of experience and evidence from experiments. In general, more complex networks are needed for more difficult tasks, however, these networks also require a larger number of training examples to be able to realize their potential. If data is inadequate then prediction error on new data will be high.
Generative neural networks
Recently, generative AI, including variational autoencoder and generative adversarial network (GAN) techniques, have attracted attention for its various applications. An autoencoder is a neural network that encodes data into lower-dimensional embeddings and decodes embeddings back to the original data. Typically, encoded embeddings have lower dimensions than the original data. Thus, they handle and maintain the original information and can be used in other analyses.
GANs are a recent development that seeks to train networks to synthesize realistic data. Proposed by Goodfellow et al., the GAN applies the autoencoder technology and consists of two neural networks—a generator and a discriminator (22). The generator learns structure of the data and attempts to generate synthetic examples that are similar enough to the training data so as to fool the discriminator. The discriminator, in contrast, attempts to accurately classify images as being either real (originating from the training data) or synthetic (originating from the generator). The GAN is the combination of these two neural networks that improve by competing with each other, so that the generator becomes capable of creating images that are indistinguishable from real image data (Figure 5A).
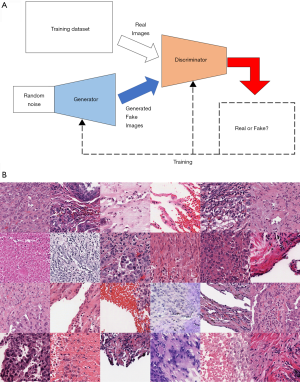
With the introduction of the deep convolutional GAN, which uses the DCNN as a part of the GAN algorithm, GAN has significantly improved, and its application method has been widely expanded. Figure 5B is a series of fake lung-cancer images created by the GAN using lung-cancer pathological images in the TCGA database.
Various fake images of not only cancer cells but also the surrounding stroma and the normal lung around the tumor are created using this method, which also allows users to create fake cancer cases. These fake images created by the GAN can be applied to education as an atlas, to correct out-of-focus images, to build 3D images, to extract features, and to create images based on words expressed by pathologists (23-25).
Application of deep learning in pathology
Potential benefits in clinical applications
The benefits of applying AI in pathology include standardization and reproducibility, improvements in diagnostic accuracy, expansion of the availability of subspecialty expertise, and increased efficiency (26). Pathologic diagnosis is widely acknowledged to feature significant inter-observer variability whether in lung pathology (27,28) or any other subspecialty (29,30). Misdiagnosis clearly leads to medical errors in treatment and can also obscure the findings of clinical research studies and trials. By assisting pathologists in diagnoses, AI can potentially help in reducing errors and identifying cases where consultation is required; this technology can also provide a support in an area beyond the expertise of the attending pathologist. For example, in neuroendocrine tumors of the gastrointestinal tract or breast cancer, the rate of MIB-1 positive cells in 500–2,000 tumor cells is one of the criteria for grading (31,32). Insufficient reproducibility is often observed in judgments directly related to treatment, e.g., companion diagnosis, where high reproducibility is required. This has led to the development of image analysis tools and guidelines for evaluating immunohistochemical markers in tumors, such as breast cancer (33). This is the type of task that AI algorithms excel at, and their applications here can improve reproducibility and efficiency.
At present, a quality of pathological diagnosis is assured through a double check performed by a different pathologist. Applying AI as the initial screening to identify cases for further expert evaluation requires near-perfect sensitivity. The familiar challenge of balancing sensitivity with specificity and the hazards of “alarm fatigue” emerge in this scenario. Another possible application of AI in diagnostics is for an algorithm to provide a computational second read to identify cases where the pathologist and algorithm reach different conclusions. These discordant cases can then be further analyzed, and the visualization of the algorithm’s output can help the pathologists in detecting cancer regions or areas that the algorithm is relying on to make a diagnosis. This may be, for example, a situation in which a pathologist has diagnosed adenocarcinoma, but AI shows a high probability for squamous cell carcinoma. In return, the pathologist can take an action such as performing additional immunostaining to confirm the diagnosis that is correct. In this case, since the initial screening is performed by a human, the threshold for sensitivity is reduced.
State-of-the-art lung cancer applications
The types of AI that can be applied to pathological diagnosis are detailed in Table 1. In addition to the detection or histologic subtyping, many of these platforms utilize computer analysis by including tasks beyond the skills of a pathologist.

Full table
Table 2 summarized the most influential reports on AI applications in lung cancer published in 2017–2019, starting with a report of a model that simply recognizes cancer (34), followed by models that predicts prognosis (41,42,51), a model that predicts gene mutation (44), and a model that determines PD-L1 expression and estimates its expression level from hematoxylin and eosin (H&E) staining (52). All of these studies revealed the high potential of AI, but the clinical application of none of these algorithms is possible immediately. Translational research on how such models can be connected to clinical practice is expected. The following section gives a more detailed description based on some publications and our own data.
Full table
Detecting cancer regions
The most fundamental task in AI for pathology is the development of algorithms to detect and delineate cancer regions in WSIs. This task often referred to as “image segmentation” is essential for many other downstream applications such as histological subtyping, mutation prediction, quantification of biomarkers on immunostaining, and clinical prognosis. However, when the sensitivity for cancer recognition is increased, the specificity typically decreases, and false positive results, recognizing non-cancerous parts as cancer, increase. While it would be best to create a perfect model that features both high sensitivity and specificity, it is extremely difficult in a real-life setting where staining and tissue preparation can vary significantly even within a single lab.
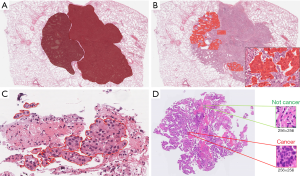
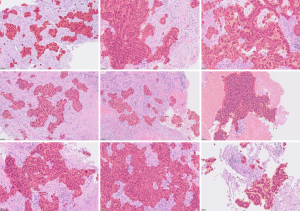
Figure 6 presents the results of a lung cancer segmentation model created by training a VGG implemented in HALO-AI® (Indica Lab, Albuquerque). Although the model is capable of recognizing lung cancer with an accuracy exceeding 90%, close observations reveal false positive and negative regions predicted as cancer (Figure 7). Most errors are morphologically similar to cancer, such as activated non-neoplastic bronchial epithelial cells and airspace macrophages, and understandable as confounders. However, some errors are significantly morphologically distinct from cancer, and the reason for their identification as cancer is not clear. Missed cancer regions occur specifically when the degree of cellular atypia is minimal, or the case exhibits rare histology. It is also reported that the performance degrades when applied to images of tissues from different pathology labs with different staining protocols and/or generated by different scanners (57). This sensitivity to tissue processing and imaging is the most significant barrier to the commercial development of viable tools intended for multi-institutional use. To overcome this problem, a large number of cases representing these variations, along with validated ground-truth annotations, are necessary.
Predicting histologic subtypes of cancer
Recurrence rate and prognosis are largely different by histological subtype and their accurate recognition is clinically important especially for adenocarcinoma (58). Algorithms for recognizing subtypes of lung adenocarcinoma have been studied by several groups (37-39). These algorithms can significantly aid pathologists in determining the ratio and distribution of histologic subtypes/patterns in a specimen, which is currently a tedious process. The low inter-observer agreement in histological subtyping, specifically the diagnostic inconsistency between invasive and non-invasive cancer subtypes, has been a major problem where AI may make significant contributions toward standardization. One challenge in this area is the reliability of the annotations used to train and validate such algorithms as these are based on the H&E judgement of pathologists and can vary from one pathologist to another or from institution to institution. One example is a report by Wei et al. where the agreement between pathologists indicated a Kappa value of 0.485, and the agreement between the trained AI and a pathologist indicated a similar low value of 0.525 (37). This is understandable as AI internalizes any bias or can be influenced by discordant annotation labels during training. Significant attention must be directed towards the generation of ground-truth standards and variability in labels in studies on AI. AI publications must clarify these ground-truth definitions and perform validation in independent cohorts, where possible.
Detection of lymph node metastases
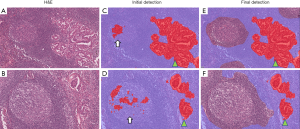
Using AI for the detection of metastases in WSIs of lymph node sections has attracted significant interest, being the subject of the international competitions, so-called AI challenges. This task is ideal for the application of AI as it involves an exhaustive search of large tissue areas, has clear clinical need and impact which determines the disease stage and clinical outcome after the surgery, and is easily framed as an AI/machine learning problem. Manual screening of lymph nodes is a primary duty of pathologists in the staging of cancer, but it is laborious and prone to errors (with significant consequences). Mistakes are frequent because the pathologist has to keep track of the inspected regions in large tissue areas that can be visually very similar. To evaluate the potential of machine learning in this field, open international competitions called CAMELYON were held in 2016 and 2017, where teams competed to identify lymph-node metastases of breast cancers in WSIs (35,59). Teams from all across the world submitted AI algorithms that showed different levels of performances; the algorithm that delivered the best performance outperformed a pathologist operating under a time constraint (35,59). This application presents familiar challenges in balancing the sensitivity of micrometastasis identification with a false positive rate, specifically for the detection of isolated tumor cells. Our team has also conducted a study to identify a lymph node metastasis of lung cancer using a deep learning platform, where false positive segmentation was successfully excluded through the combination of two deep learning platforms, explained in Figure 8 (36).
Measuring tumor cellularity for genomic analysis
With introduction of targeted therapy, lung cancer specimens often require an estimate of the percentage of tumor cells or “purity” prior to molecular testing to confirm sufficient amount of tumor cell DNA (otherwise, genomic assays may return false negative results), and to assist interpreting the allelic fraction of mutations. Visually counting the ratio of tumor cells present is a challenging task and is another field where AI can make a significant contribution.
Noteworthy, the concordance of pathologists regarding assessment of tumor cellularity is low (60). Figure 9A,B presents the result voting at a meeting where 59 pathologists were asked to estimate tumor cellularity in a given specimen of lung adenocarcinoma. The accurate confirmed tumor content was 28%, and only 3 out of 59 pathologists could guess the correct range (26–30%).
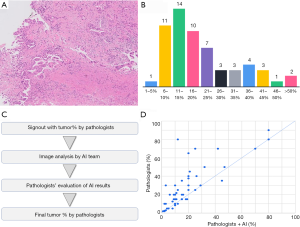
Our team created a platform for measuring percentage of tumor cells in lung adenocarcinoma by developing an AI model for recognizing tumor regions combined with a model for detecting distinct nuclei, which allowed to count the number of cells in each area (49). Once a specimen requiring molecular testing is submitted, the pathological diagnosis is confirmed by a pathologist, and a WSI from this case is passed over to the AI analysis team that uses the model to segment the tumor regions and detect the nuclei. The percent tumor cells calculated from region segmentation and nuclei counts are recorded. The calculated percent tumor cell content is presented to multiple pathologists during sign out. The pathologist observes the original image, estimates the tumor content according to a consensus, evaluates the quality of segmentation delivered by the AI, and decides the numerical value or the multiplicative factor required to calculate the correct answer simultaneously (Figure 9C). After the accuracy of nuclear detection is confirmed, AI analysis results are referenced to incorporate the calculated percent tumor cells into the pathological report.
A prospective study of 53 biopsy cases using this method revealed that the values initially determined by pathologists based on a consensus and the final decisions determined after considering the AI results were the same in only 13 out of the 53 cases. Initial values determined by the pathologists were higher than the final values of the same pathologists after the evaluation of the AI results in 34 cases and lower in 6 cases. The study confirmed that pathologists often correct their decisions by referring to the results of an AI analysis. Our preliminary analysis also revealed that pathologists tended to overestimate the proportion of cancer (Figure 9D).
Prediction of mutations from H&E morphology
One of the exciting possibilities is that AI may reveal subtle and even latent features that have not been appreciated by pathologists. For example, a few studies reported that driver gene mutations or microsatellite instability can be predicted from H&E images of different malignancies (44,61,62). Coudray et al. (44) used a DCNN (Inception-V3) to develop an AI platform that classifies lung cancer into adenocarcinoma, squamous cell carcinoma, and normal lung tissue. In addition, they trained the AI to create a model that predicts the 10 most frequent gene mutations from histopathological images of lung adenocarcinoma; the predicted AUCs of 6 out of 10 genes (STK11, EGFR, FAT1, SETBP1, KRAS, TP53) ranged from 0.733 to 0.856. The reported sensitivity and specificity of the platform do not reach the threshold of being diagnostically meaningful, but in the future, these approaches may improve and find use as a pre-screening tool to select samples for sequencing, assess potential clonality or heterogeneity, or better understand how genetic alterations impact the function and morphology. Additional research to analyze genetic signatures in heterogenous tumor foci such as by using laser capture microdissection can help in verifying some of these findings.
Multiplex image analysis
Fluorescent multiplex immunohistochemistry is an assay to visualize multiple antigens simultaneously on the tissue. Not only identifying multiple markers on the tumor cells, in response to recent advances in research on immune cells in cancer, many research findings based on the multiplex test that analyzes the distribution of several different types of immune cells on the same tissue slide have been reported (63-66).
The clinical application of the multiplex test is not expected at this point due to its limitation of expensive costs. There are many unknowns associated with the precise mechanism of immune microenvironment related to PD-L1, PD-1, and immune checkpoints. It is believed that the multiplex tests will make a significant contribution towards revealing the background biology of how immune cells are involved in antitumor host defense. Conducting such an analysis via human observation has significant limitations. The implementation of an AI-based image analysis will generate new discoveries that will considerably contribute to the development of new therapeutics. Currently, many analyses of multiplex tests are conducted not by deep learning but by simple computer-based image analysis and machine learning such as the random forest method. Because multiplex analysis becomes more complicated, deep learning can be the key tool for analysis, leading to new discoveries and advances in medicine.
Creation of multimodal platforms
As the image analysis technology advances, pathological diagnosis goes beyond the boundaries of histopathological images. Pathological diagnosis, radiological diagnosis, and genomic data produce diagnostic information that guide the treatment of patients. These sets of data are currently presented separately and are often contradictory in what they suggest about the patient. Clinicians must integrate these findings to form a single diagnosis. Presenting these modalities and findings in a single integrated platform may offer many benefits.
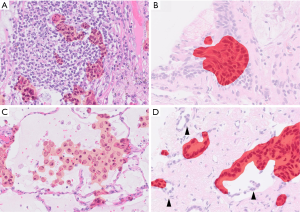
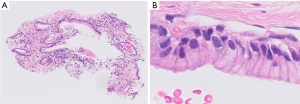
Pathological appearance can often be very similar for different diseases, and supporting data from other diagnostic modalities can facilitate pathologic diagnosis. One of the examples is detection of mucin producing atypical epithelia (Figure 10), particularly in small sized specimens where background morphology is not available for evaluation. This histologic appearance can be observed in several pathologic conditions, such as invasive mucinous adenocarcinoma, metastatic pancreatic cancer, metaplasia around the fibrotic scar, and ciliated muconodular papillary tumor. In such cases, the inputs from radiology and clinical information are indispensable for correct diagnosis (e.g., data such as radiological images, endoscopic images, genetic data, blood test data, age, and gender, in addition to images of pathological specimens). Furthermore, providing additional information such as prognostic indication and prediction of treatment effect on top of diagnosis is more suitable for clinical needs. Such integrated pathologic diagnoses may be practiced in the future. The development of AI methods around such multimodal data has been reported in a few fields, although the evidence at the moment is fairly low (67,68). It is an area where future development is anticipated.
Barriers to clinical translation of AI
As described above, AI has the potential to have a widespread and significant impact on the practice of pathology; while the clinical translation of the AI technology is expected to come to pass, hurdles remain in its introduction to clinical practice. The most significant hurdle is the digitization of pathology workflows and the transition from light microscopy to whole-slide imaging, as mentioned earlier. In addition to the high costs associated with the digital infrastructure, pathologists in general have insufficient education and knowledge of digital pathology.
Reimbursement is an important factor in popularizing the digitization of pathology. Even if hurdles are cleared and WSIs can be used in daily practice, the use of AI platforms will be low if different AI platforms are developed with different functions, requiring users to launch different software for each purpose or to repeatedly download and upload images. In other words, the development of a simplified user interface - an AI that can be embedded into the workflow as part of the WSI and reporting system - is a key factor in the successful implementation of the actual clinical practice. In the future, it is assumed that AI will be integrated into laboratory information system and electronic medical records. Pathologists must investigate and identify the practice that would be the best, considering all variety of vendors.
The value of annotated datasets for development and validation of AI technology
One of the popular motivations for AI in pathology is that there is a current shortage of pathologists, which is anticipated to worsen while caseloads and responsibilities only continue to increase. The essential ingredient for the development of many AI approaches is a well-annotated ground truth. These data must be generated or supervised by pathologists; however, the time available to them to generate the annotation data is limited. Some studies have succeeded in significantly reducing the burden of annotation using a technique called weakly supervised learning that relies on less granular labels that are easier to generate (such as drawing a box around a structure rather than tracing its boundaries), but these methods often require a large number of cases to overcome the lack of annotations (56,69). A large database with good clinical information and annotations are highly difficult to obtain. Currently, TCGA and NLST are the only large public databases for lung cancer cases, but significant image markups for them are not available.
Approval as a medical device vs. laboratory developed test
The clinical use of AI is often discussed in the context of a universally applicable commercial platform that has received approval from a regulatory agency such as the FDA or CE marking. However, it is known that the diagnostic accuracy of AI deteriorates when it employs these approaches outside the institutions where they were developed and where staining and tissue processing can be different (57). The modification or adaptation of these platforms at new deployment sites would not be possible, given the FDA and CE marking regulations; therefore, this approach is currently not widely considered, although it offers several benefits. Continued challenges in deploying universal multi-site AI platforms and decreases in the diagnostic accuracy due to variability may result in this being reconsidered at some point.
In contrast, the pathology lab has a long history of internal development and validation of laboratory tests, and it may be possible to develop an in-house AI platform within this paradigm. Here, the challenges include the limited number of cases, software development resources, and infrastructure required to successfully develop and validate an algorithm. Recruiting data scientists in pathology departments is unrealistic for most institutes, and multiple experts would likely be required. In larger academic institutions, this may be possible. Decisions must be made on application and regulation as professional societies, platforms not approved by the FDA or CE marking as laboratory development test (LDT).
Difficulty in defining the ground truth
The most commonly used approach for AI algorithms is for pathologists to annotate structures and train and validate algorithms to detect and classify these structures; in such case, the trained AI platform will internalize all the biases of the pathologists who created the ground truth.
Results generated by AI would be reviewed by a responsible pathologist who signs out the case and who would assess the accuracy of AI predictions and whether they should be used or not. What is important here is how the AI algorithm defines the correct answer. Ground truth definitions vary among algorithms. For example, the ground truth for an algorithm developed under the supervision of a leading expert pathologist and the ground truth for a platform developed based on strong and convincing correlation to clinical information such as prognosis and therapeutic effects would be different. Hence, explaining how ground truth was defined for each algorithm is an essential part of AI algorithm development.
The standardization of pathology diagnosis is currently not an issue that is widely discussed in the field of AI, but it is, without doubt, a critical issue. For example, in the histopathological classification of prostate cancer using the Gleason scoring system, although the histological image of each pattern is well defined and depicted in the textbooks, the subjective judgment by pathologists often results in the significant interobserver variation (70,71). Similarly, in the case of lung cancer, judgement of small cell carcinoma vs. non-small cell carcinoma is known for its high concordance among pathologists. However, when examining the histological subtypes, the interobserver agreement rate is low even among the authorities (72,73).
The standardization of diagnosis is hence highly important for AI development. If the ground-truth data is prepared in an unstandardized field, the data will be dispersed by various annotators, thereby causing significantly negative effects on the AI analysis accuracy.
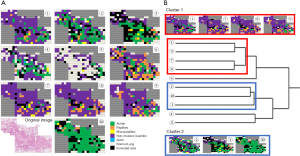
Herein, we present our results of one lung cancer case for which we cropped one representative WSI into 218 high-magnification still images and determined the variation in the judgement of the pathologists regarding histological subtypes. A total of 10 pathologists classified the 218 images into the following histological subtypes—lepidic, papillary, acinar, solid, mucinous, and micropapillary—along with non-cancer. As a result, the overall Kappa value among the invasion, non-invasion, and non-tumor cases was 0.24. Figure 11A presents a reconstructed image of the case with the selected subtypes highlighted by different colors. When participating pathologists were classified via cluster analysis based on the diagnosis, they ended up in two clusters (Figure 11B). The agreements of the clusters yielded Kappa values of 0.45 and 0.23. With further survival analysis using a higher number of cases, a favorable ground truth can be identified.
Challenges in explaining AI inferences
One of the major barriers to the clinical adoption of AI methods is the black box nature of the more successful and accurate methods such CNNs. Although deep learning has the ability to deliver results that are superior to those produced by humans in some applications, the prediction mechanisms of these algorithms often cannot be explained, making it difficult to predict the modes in which they fail, or to formulate protocols to establish their analytic validity. This presents regulatory challenges and can pose problems when explaining a diagnosis to the patient.
Explainability and interpretability are widely studied in the fields of computer science and engineering where AI algorithms are applied to solve general problems (49), but currently, there are no solutions that adequately address pathology applications. Several proponents of AI in pathology have suggested that the power of algorithms such as the DCNN must be directed to accurately detecting and classifying histologic features that are known to human experts, including necrosis, mitoses, immune infiltrates, and tumor and stromal cells. From these detailed maps of the tissue, quantitative features can be defined, describing the abundance and morphology of these components (e.g., pleomorphism), allowing the development of diagnostic and prognostic models that can be linked back to these concepts. Alternatives for understanding DCNNs include visualizing how the units and layers respond to different features, determining the input sites that change the result (74). Unsupervised learning can also play a role in understanding patterns using models such as autoencoders to arrange images based on the calculated features and to group them perceptually.
Conclusion: the role of pathologists in the AI age
AI has the potential to provide gains in quality, accuracy, and efficiency through the automation of tasks such as detecting metastases, identifying tumor cells, and counting mitoses in the not so distant future. The introduction of AI as a device assisting pathological diagnosis is expected to not only reduce the workload of pathologists but also to help standardize the otherwise subjective diagnosis that can lead to suboptimal treatment of patients.
Risks in deploying AI in daily clinical diagnostics include leakage of personal information, high operating costs, misdiagnosis, and assignment of responsibility for such misdiagnosis. In its current state, AI will need to be closely supervised in diagnostic tasks. However, with the use of advanced algorithms outlined above, AI should be able to perform diagnoses in collaboration with human pathologists. Developing integrative platforms that bring together multimodal data to suggest prognosis and/or the choice of therapy will be another substantial benefit of digitization and provide an additional sanity check on AI generated predictions.
When such advanced (“next generation”) pathological diagnosis will enter medical practice, it is likely that demands of clinicians would not be satisfied with the level of current pathological diagnosis offered by pathologists using solely a microscope. Pathologists who reject digital pathology and AI may face a diminished role in the future pathology practice.
“Will AI replace pathologists?” is a question frequently asked by pathologists today. Although the achievements in some areas are impressive, AI remains unsuited for many tasks and will require close supervision in clinical use for the foreseeable future. Most pathologists will agree that help automating tedious tasks and providing second reads can help as they increasingly struggling with growing caseloads. Like any technology AI is a tool that can be a strong ally or a foe depending on how pathologists decide to use it. We are convinced that as AI takes root in clinical practice, pathologists who are skilled in the use of AI and understand its limitations will reap significant benefits.
Acknowledgments
Funding: This work was partly supported by the New Energy and Industrial Technology Development Organization (NEDO), the US National Institutes of Health National Cancer Institute grants U24CA19436201, U01CA220401 and National Institute of Biomedical Imaging and Bioengineering U01CA220401.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editor (Helmut H. Popper) for the series “New Developments in Lung Cancer Diagnosis and Pathological Patient Management Strategies” published in Translational Lung Cancer Research. The article was sent for external peer review organized by the Guest Editor and the editorial office.
Reporting Checklist: The authors have completed the Narrative Review reporting checklist. Available at http://dx.doi.org/10.21037/tlcr-20-591
Peer Review File: Available at http://dx.doi.org/10.21037/tlcr-20-591
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tlcr-20-591). The series “New Developments in Lung Cancer Diagnosis and Pathological Patient Management Strategies” was commissioned by the editorial office without any funding or sponsorship. LADC reports grants from US National Cancer Institute, during the conduct of the study; grants from Roche Tissue Diagnostics, personal fees from Konica Minolta, outside the submitted work. JF reports grants from NEDO (New Energy and Industrial Technology Development Organization), other from PathPresenter, other from ContextVision, other from Sony, other from Future Corp, during the conduct of the study; other from Pathology Institute Corp, other from N Lab Corp, outside the submitted work; in addition, JF has a patent PCT/JP2020/000424 pending. The other authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Tabata K, Mori I, Sasaki T, et al. Whole-slide imaging at primary pathological diagnosis: Validation of whole-slide imaging-based primary pathological diagnosis at twelve Japanese academic institutes. Pathol Int 2017;67:547-54. [Crossref] [PubMed]
- Snead DR, Tsang YW, Meskiri A, et al. Validation of digital pathology imaging for primary histopathological diagnosis. Histopathology 2016;68:1063-72. [Crossref] [PubMed]
- Mukhopadhyay S, Feldman MD, Abels E, et al. Whole Slide Imaging Versus Microscopy for Primary Diagnosis in Surgical Pathology: A Multicenter Blinded Randomized Noninferiority Study of 1992 Cases (Pivotal Study). Am J Surg Pathol 2018;42:39-52. [PubMed]
- Bauer TW, Slaw RJ. Validating whole-slide imaging for consultation diagnoses in surgical pathology. Arch Pathol Lab Med 2014;138:1459-65. [Crossref] [PubMed]
- Pantanowitz L, Sinard JH, Henricks WH, et al. Validating whole slide imaging for diagnostic purposes in pathology: guideline from the College of American Pathologists Pathology and Laboratory Quality Center. Arch Pathol Lab Med 2013;137:1710-22. [Crossref] [PubMed]
- Griffin J, Treanor D. Digital pathology in clinical use: where are we now and what is holding us back? Histopathology 2017;70:134-45. [Crossref] [PubMed]
- Williams BJ, Lee J, Oien KA, et al. Digital pathology access and usage in the UK: results from a national survey on behalf of the National Cancer Research Institute's CM-Path initiative. J Clin Pathol 2018;71:463-6. [Crossref] [PubMed]
- Weinstein RS, Bloom KJ, Rozek LS. Telepathology and the networking of pathology diagnostic services. Arch Pathol Lab Med 1987;111:646-52. [PubMed]
- Hartman DJ, Parwani AV, Cable B, et al. Pocket pathologist: A mobile application for rapid diagnostic surgical pathology consultation. J Pathol Inform 2014;5:10. [Crossref] [PubMed]
- Emanuel P, Patel R, Liu S, et al. Use of dynamic telepathology utilizing a smartphone in margin control cutaneous surgery. ANZ J Surg 2019;89:982-3. [Crossref] [PubMed]
- Gardner JM, McKee PH. Social Media Use for Pathologists of All Ages. Arch Pathol Lab Med 2019;143:282-6. [Crossref] [PubMed]
- Retamero JA, Aneiros-Fernandez J, Del Moral RG. Complete Digital Pathology for Routine Histopathology Diagnosis in a Multicenter Hospital Network. Arch Pathol Lab Med 2020;144:221-8. [Crossref] [PubMed]
- Huang Y, Lei Y, Wang Q, et al. Telepathology consultation for frozen section diagnosis in China. Diagn Pathol 2018;13:29. [Crossref] [PubMed]
- Baidoshvili A, Stathonikos N, Freling G, et al. Validation of a whole-slide image-based teleconsultation network. Histopathology 2018;73:777-83. [Crossref] [PubMed]
- Vergani A, Regis B, Jocolle G, et al. Noninferiority Diagnostic Value, but Also Economic and Turnaround Time Advantages From Digital Pathology. Am J Surg Pathol 2018;42:841-2. [Crossref] [PubMed]
- Têtu B, Paré G, Trudel M-C, et al. Whole-slide imaging-based telepathology in geographically dispersed Healthcare Networks. The Eastern Québec Telepathology project. Diagnostic Histopathology 2014;20:462-9. [Crossref]
- Fujisawa T, Mori K, Mikamo M, et al. Nationwide cloud-based integrated database of idiopathic interstitial pneumonias for multidisciplinary discussion. Eur Respir J 2019.53. [Crossref] [PubMed]
- Jack Copeland HAB. The Essential Turing: The Ideas that Gave Birth to the Computer Age. Oxford: Clarendon Press, 2004. Pp. viii+613. ISBN 0-19-825079-7. £50.00 (hardback). ISBN 0-19-825080-0. £14.99 (paperback). The British Journal for the History of Science 2006;39:470-1. [Crossref]
- McCarthy J, Minsky ML, Rochester N, et al. A Proposal for the Dartmouth Summer Research Project on Artificial Intelligence. August 31, 1955. Available online: http://jmc.stanford.edu/articles/dartmouth/dartmouth.pdf
- Russell S, Norvig P. Artificial Intelligence: A Modern Approach. Prentice Hall Press, 2009.
- Deng J, Dong W, Socher R, et al. editors. ImageNet: A large-scale hierarchical image database. 2009 IEEE Conference on Computer Vision and Pattern Recognition; 2009 20-25 June 2009.
- Goodfellow I, Pouget-Abadie J, Mirza M, et al. Generative Adversarial Networks. ArXiv e-prints (June 2014). arXiv preprint arXiv:14062661 2014;1406.
- Quiros AC, Murray-Smith R, Yuan K. Pathology GAN: Learning deep representations of cancer tissue. arXiv e-prints 2019.
- Mahmood F, Borders D, Chen R, et al. Deep Adversarial Training for Multi-Organ Nuclei Seg-mentation in Histopathology Images. arXiv e-prints 2018.
- Hou L, Agarwal A, Samaras D, et al. Unsupervised Histopathology Image Synthesis. arXiv e-prints 2017.
- Bera K, Schalper KA, Rimm DL, et al. Artificial intelligence in digital pathology - new tools for diagnosis and precision oncology. Nat Rev Clin Oncol 2019;16:703-15. [Crossref] [PubMed]
- Stang A, Pohlabeln H, Muller KM, et al. Diagnostic agreement in the histopathological evaluation of lung cancer tissue in a population-based case-control study. Lung Cancer 2006;52:29-36. [Crossref] [PubMed]
- Grilley-Olson JE, Hayes DN, Moore DT, et al. Validation of Interobserver Agreement in Lung Cancer Assessment: Hematoxylin-Eosin Diagnostic Reproducibility for Non–Small Cell Lung Cancer: The 2004 World Health Organization Classification and Therapeutically Relevant Sub-sets. Arch Pathol Lab Med 2013;137:32-40. [Crossref] [PubMed]
- Zhu Y, Li Y, Jung CK, et al. Histopathologic Assessment of Capsular Invasion in Follicular Thyroid Neoplasms-an Observer Variation Study. Endocr Pathol 2020;31:132-40. [Crossref] [PubMed]
- Liu Z, Bychkov A, Jung CK, et al. Interobserver and intraobserver variation in the morphological evaluation of noninvasive follicular thyroid neoplasm with papillary-like nuclear features in Asian practice. Pathol Int 2019;69:202-10. [Crossref] [PubMed]
- Nagtegaal ID, Odze RD, Klimstra D, et al. The 2019 WHO classification of tumours of the diges-tive system. Histopathology 2020;76:182-8. [Crossref] [PubMed]
- Dowsett M, Nielsen TO, A'Hern R, et al. Assessment of Ki67 in breast cancer: recommendations from the International Ki67 in Breast Cancer working group. J Natl Cancer Inst 2011;103:1656-64. [Crossref] [PubMed]
- Bui MM, Riben MW, Allison KH, et al. Quantitative Image Analysis of Human Epidermal Growth Factor Receptor 2 Immunohistochemistry for Breast Cancer: Guideline From the Col-lege of American Pathologists. Arch Pathol Lab Med 2019;143:1180-95. [Crossref] [PubMed]
- Šarić M, Russo M, Stella M, et al., editors. CNN-based Method for Lung Cancer Detection in Whole Slide Histopathology Images. 2019 4th International Conference on Smart and Sustainable Technologies (SpliTech); 2019: IEEE.
- Ehteshami Bejnordi B, Veta M, Johannes van Diest P, et al. Diagnostic Assessment of Deep Learning Algorithms for Detection of Lymph Node Metastases in Women With Breast Cancer. JAMA 2017;318:2199-210. [Crossref] [PubMed]
- Pham HHN, Futakuchi M, Bychkov A, et al. Detection of lung cancer lymph node metastases from whole-slide histopathological images using a two-step deep learning approach. Am J Pathol 2019;189:2428-39. [Crossref] [PubMed]
- Wei JW, Tafe LJ, Linnik YA, et al. Pathologist-level classification of histologic patterns on resected lung adenocarcinoma slides with deep neural networks. Sci Rep 2019;9:3358. [Crossref] [PubMed]
- Yu KH, Wang F, Berry GJ, et al. Classifying Non-Small Cell Lung Cancer Histopathology Types and Transcriptomic Subtypes using Convolutional Neural Networks. bioRxiv 2019:530360.
- Gertych A, Swiderska-Chadaj Z, Ma Z, et al. Convolutional neural networks can accurately distinguish four histologic growth patterns of lung adenocarcinoma in digital slides. Sci Rep 2019;9:1483. [Crossref] [PubMed]
- Yu KH, Zhang C, Berry GJ, et al. Predicting non-small cell lung cancer prognosis by fully auto-mated microscopic pathology image features. Nat Commun 2016;7:12474. [Crossref] [PubMed]
- Wang X, Janowczyk A, Zhou Y, et al. Prediction of recurrence in early stage non-small cell lung cancer using computer extracted nuclear features from digital H&E images. Sci Rep 2017;7:13543. [Crossref] [PubMed]
- Wang S, Chen A, Yang L, et al. Comprehensive analysis of lung cancer pathology images to dis-cover tumor shape and boundary features that predict survival outcome. Sci Rep 2018;8:10393. [Crossref] [PubMed]
- Bychkov D, Linder N, Turkki R, et al. Deep learning based tissue analysis predicts outcome in colorectal cancer. Sci Rep 2018;8:3395. [Crossref] [PubMed]
- Coudray N, Ocampo PS, Sakellaropoulos T, et al. Classification and mutation prediction from non-small cell lung cancer histopathology images using deep learning. Nat Med 2018;24:1559-67. [Crossref] [PubMed]
- Schaumberg AJ, Rubin MA, Fuchs TJ. H&E-stained Whole Slide Image Deep Learning Predicts SPOP Mutation State in Prostate Cancer. bioRxiv 2018:064279.
- Kim RH, Nomikou S, Dawood Z, et al. A Deep Learning Approach for Rapid Mutational Screening in Melanoma. bioRxiv 2019:610311.
- Hegde N, Hipp JD, Liu Y, et al. Similar image search for histopathology: SMILY. NPJ Digit Med 2019;2:56. [Crossref] [PubMed]
- Aprupe L, Litjens G, Brinker TJ, et al. Robust and accurate quantification of biomarkers of immune cells in lung cancer micro-environment using deep convolutional neural networks. PeerJ 2019;7:e6335. [Crossref] [PubMed]
- Furukawa T, Kuroda K, Bychkov A, et al. Verification of deep learning model to measure tumor cellularity in transbronchial biopsies of lung adenocarcinoma. In “Abstracts from USCAP 2019: Pulmonary Pathology.”. Modern Pathology 2019;32:1828.
- Yamamoto Y, Tsuzuki T, Akatsuka J, et al. Automated acquisition of explainable knowledge from unannotated histopathology images. Nat Commun 2019;10:5642. [Crossref] [PubMed]
- Wang S, Wang T, Yang L, et al. ConvPath: A software tool for lung adenocarcinoma digital pathological image analysis aided by a convolutional neural network. EBioMedicine 2019;50:103-10. [Crossref] [PubMed]
- Sha L, Osinski BL, Ho IY, et al. Multi-Field-of-View Deep Learning Model Predicts Nonsmall Cell Lung Cancer Programmed Death-Ligand 1 Status from Whole-Slide Hematoxylin and Eosin Images. J Pathol Inform 2019;10:24. [Crossref] [PubMed]
- Althammer S, Tan TH, Spitzmuller A, et al. Automated image analysis of NSCLC biopsies to pre-dict response to anti-PD-L1 therapy. J Immunother Cancer 2019;7:121. [Crossref] [PubMed]
- Wang X, Chen H, Gan C, et al. Weakly Supervised Deep Learning for Whole Slide Lung Cancer Image Analysis. IEEE Trans Cybern 2020;50:3950-62. [Crossref] [PubMed]
- Yi F, Yang L, Wang S, et al. Microvessel prediction in H&E Stained Pathology Images using fully convolutional neural networks. BMC Bioinformatics 2018;19:64. [Crossref] [PubMed]
- Kapil A, Meier A, Zuraw A, et al. Deep Semi Supervised Generative Learning for Automated Tumor Proportion Scoring on NSCLC Tissue Needle Biopsies. Sci Rep 2018;8:17343. [Crossref] [PubMed]
- Campanella G, Hanna MG, Geneslaw L, et al. Clinical-grade computational pathology using weakly supervised deep learning on whole slide images. Nat Med 2019;25:1301-9. [Crossref] [PubMed]
- Sica G, Yoshizawa A, Sima CS, et al. A grading system of lung adenocarcinomas based on histologic pattern is predictive of disease recurrence in stage I tumors. Am J Surg Pathol 2010;34:1155-62. [Crossref] [PubMed]
- Bándi P, Geessink O, Manson Q, et al. From Detection of Individual Metastases to Classification of Lymph Node Status at the Patient Level: The CAMELYON17 Challenge. IEEE Trans Med Imaging 2019;38:550-60. [Crossref] [PubMed]
- Smits AJ, Kummer JA, de Bruin PC, et al. The estimation of tumor cell percentage for molecular testing by pathologists is not accurate. Mod Pathol 2014;27:168-74. [Crossref] [PubMed]
- Tsou P, Wu CJ. Mapping Driver Mutations to Histopathological Subtypes in Papillary Thyroid Carcinoma: Applying a Deep Convolutional Neural Network. J Clin Med 2019;8:1675. [Crossref] [PubMed]
- Kather JN, Pearson AT, Halama N, et al. Deep learning can predict microsatellite instability directly from histology in gastrointestinal cancer. Nat Med 2019;25:1054-6. [Crossref] [PubMed]
- Steele KE, Brown C. Multiplex Immunohistochemistry for Image Analysis of Tertiary Lymphoid Structures in Cancer. Methods Mol Biol 2018;1845:87-98. [Crossref] [PubMed]
- Parra ER, Villalobos P, Behrens C, et al. Effect of neoadjuvant chemotherapy on the immune microenvironment in non-small cell lung carcinomas as determined by multiplex immunofluorescence and image analysis approaches. J Immunother Cancer 2018;6:48. [Crossref] [PubMed]
- Lopes N, Bergsland CH, Bjornslett M, et al. Digital image analysis of multiplex fluorescence IHC in colorectal cancer recognizes the prognostic value of CDX2 and its negative correlation with SOX2. Lab Invest 2020;100:120-34. [Crossref] [PubMed]
- Lee CW, Ren YJ, Marella M, et al. Multiplex immunofluorescence staining and image analysis assay for diffuse large B cell lymphoma. J Immunol Methods 2020;478:112714. [Crossref] [PubMed]
- Mobadersany P, Yousefi S, Amgad M, et al. Predicting cancer outcomes from histology and genomics using convolutional networks. Proc Natl Acad Sci U S A 2018;115:E2970-9. [Crossref] [PubMed]
- Hao J, Kosaraju SC, Tsaku NZ, et al. PAGE-Net: Interpretable and Integrative Deep Learning for Survival Analysis Using Histopathological Images and Genomic Data. Pac Symp Biocomput 2020;25:355-66. [PubMed]
- Qu H, Wu P, Huang Q, et al. Weakly Supervised Deep Nuclei Segmentation using Points Annotation in Histopathology Images. In: Cardoso MJ, Aasa F, Ben G, et al. editors. Proceedings of The 2nd International Conference on Medical Imaging with Deep Learning; Proceedings of Machine Learning Research: PMLR; 2019:390-400.
- Singh RV, Agashe SR, Gosavi AV, et al. Interobserver reproducibility of Gleason grading of prostatic adenocarcinoma among general pathologists. Indian J Cancer 2011;48:488-95. [Crossref] [PubMed]
- McKenney JK, Simko J, Bonham M, et al. The potential impact of reproducibility of Gleason grading in men with early stage prostate cancer managed by active surveillance: a multi-institutional study. J Urol 2011;186:465-9. [Crossref] [PubMed]
- Wright J, Churg A, Kitaichi M, et al. Reproducibility of visual estimation of lung adenocarcinoma subtype proportions. Mod Pathol 2019;32:1587-92. [Crossref] [PubMed]
- Shih AR, Uruga H, Bozkurtlar E, et al. Problems in the reproducibility of classification of small lung adenocarcinoma: an international interobserver study. Histopathology 2019;75:649-59. [Crossref] [PubMed]
- Selvaraju RR, Cogswell M, Das A, et al. Grad-CAM: Visual Explanations from Deep Networks via Gradient-Based Localization. International Journal of Computer Vision 2020;128:336-59. [Crossref]


