
Ex vivo culture of circulating tumour cells derived from non-small cell lung cancer
Introduction
Lung cancer (LC) is the leading cause of cancer-related deaths amongst men and women worldwide (1). In 2018, the incidence of LC was estimated to be 1.8 million new cases, accounting for 12.9% of all new cancers diagnosed globally (2,3). The 5-year survival rate is 18% for all stages of LC, with poor outcomes, largely due to late diagnosis, progression of disease and resistance to therapy (1,4,5). The majority of patients with LC present with locally advanced or metastatic disease with the annual mortality rate being higher than colon, breast and prostate cancers combined (4-6). Therefore, it is imperative that novel biomarkers are identified to assist in clinical decision making to improve outcomes for patients.
Metastasis is a highly complex process and is the main cause of cancer related deaths (7). Circulating tumour cells (CTCs) play a pivotal role in metastasis (8). CTCs disseminate into the bloodstream as single CTCs, clusters or as circulating tumour microemboli (CTM) (9-14). Cellular aggregates termed CTM include platelets, stromal cells and heamatopoietic cells which ‘protect’ tumour cells from undergoing apoptosis or being attacked by the immune system (12,15). CTC clusters have shown to have a higher metastatic capacity compared to single CTCs (9,16). Whilst CTCs were first described over a century ago, it is now that they have gained attention due to the rapid advancement of technologies to enrich for these rare cells from a large background of hematopoetic cells (17). Clinical trials and meta-analyses’ have demonstrated that the presence of CTCs in the blood correlates with poor overall survival in patients with metastatic prostate, breast, lung and colon cancers (18-23). Analysis of CTCs as a liquid biopsy has shown promise for the serial assessment of tumour evolution during disease progression, real-time monitoring of recurrences or relapses, as well as providing a snapshot of the tumour heterogeneity using a simple blood draw from a cancer patient (24,25).
Currently, therapeutic decisions rely on tumour biopsies, which are invasive and especially difficult to access due to the anatomical location of the tumour, resulting in limited tumour material. In order to make clinical decisions based on the tumour tissue activity, there is need for serial biopsies to evaluate the dynamics changes of tumour activities, which is extremely invasive and, in most cases, unattainable. In addition, when a cancer patient has undergone surgery, there is no availability of tumour tissue post resection. It is important to also note that the recurrence rates for NSCLC are between 30% to 50% and therefore, non-invasive means of assessing the disease are needed (26). Liquid biopsies have also shown great promise in detecting and characterizing minimal residual disease (MRD), leading to recurrences or relapse (27).
Studies have sought to expand CTCs outside a patients’ body for in depth characterization of metastasis initiating cells as well as to understand the biology of metastatic disease (28). Once expanded, ex vivo cultures also present the possibility of pre-clinical testing to determine drug susceptibility. CTC enrichment techniques have also impacted on the development of CTC culture models and therefore optimal culture conditions are yet to be established (28). The immortalization of stable long term cell lines hold vital clues when developing personalized treatments for cancer as well as testing of therapeutic effectiveness using drug screening (29). This approach could better assist in determining therapeutic regimen that is beneficial for patients, and therefore drive towards personalized oncology (30).
There is a rising interest in genomic profiling of CTCs in order to monitor the tumour’s response to therapy over the course of treatment, as CTCs may signify intact and functional cancer cells circulating in the peripheral blood with the capacity to metastasise and initiate secondary tumour growth (31). Previous studies have shown clinical value in sequencing individual CTCs to identify driver mutations and possible drug targets to prevent tumour resistance (32-35). In the current study, the spiral microfluidic chip technology was utilised to isolate CTCs in a cohort of NSCLC patients. Using this technology CTCs are separated using hydrodynamic forces for size-based sorting (36). In addition, isolated CTCs were assessed for clinically relevant markers EGFR exon 19 deletion and ALK translocations. CTCs were detected in 54.3% in bloods from patients with NSCLC. We found that NSCLC patients with >2 CTCs at diagnosis showed a poor outcome compared with the patients with <2 CTC cells in their bloods. In parallel, RosetteSep™ (antibody-based depletion) was used to isolate and expand CTCs. We were able to maintain CTC in culture (n=9) in most cases for up to 40 days. One CTC culture was confirmed by WES to be of NSCLC origin, providing evidence that the CTCs are derived from primary tumour tissue.
Methods
Patients and samples
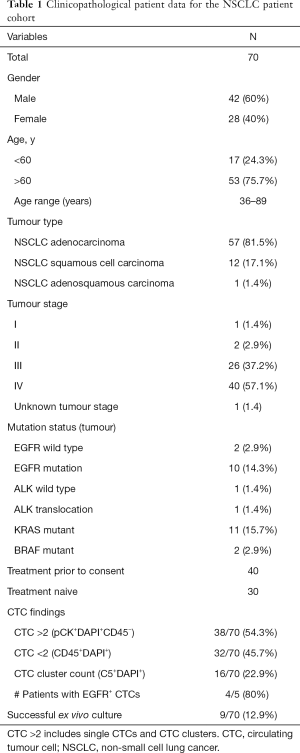
Ethics approval was obtained from the Metro South Health District Human Research Ethics Committee in accordance with the National Health and Medical Research Council’s guidelines (HREC/11/QPAH/331) to collect blood samples from the Princess Alexandra Hospital (PAH). All methods in this study were performed in accordance with these ethical guidelines and regulations and with the Helsinki Declaration (as revised in 2013). This study has Queensland University of Technology ethics approval (1100001420). Following written informed consent, 15–20 mL of blood samples were collected from 70 NSCLC patients. The patient demographic and clinicopathological findings are presented in Table 1.
Full table
CTC enrichment and detection
The workflow that we have adopted is shown in Figure 1.
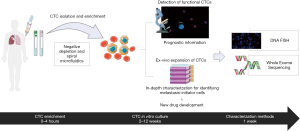
RosetteSep and ex vivo CTC culture
Patients whole blood (10 mL) was collected in heparin tubes (BD-Plymouth, UK) and incubated with RosetteSepTM (Human CD45 depletion cocktail, Stemcell Technologies™, Vancouver, Canada) at 50 µL/mL of whole blood for 20 minutes at room temperature. After incubation, cellular separation was achieved in SepMate™-50 mL tubes containing Lymphoprep™ (Stemcell Technologies™, Vancouver, Canada) density gradient medium and centrifugation at 1,200 g for 10 minutes. The enriched cellular suspension was resuspended in Clevers media and seeded in 96 well standard microplates (Greiner Bio-One, Austria) and 16 well Chamber Slide™ (Lab-Tek®, Australia).
Preparation of Clevers media
Clevers media is comprised of Advanced DMEM/F12 with the following additives: 50 ng/mL EGF (Sigma), 5% v/v R-spondin 1, 10% v/v Noggin, 10 ng/mL FGF10 (Peprotech), 1 ng/mL FGF2 (Peprotech), 10 nM Nicotinamide (Acros), 0.5 µM A83-01 (Tocris), 10 µM SB202190 (Sigma Aldrich), 10 µM Y-27632 (Selleck Chemical), 1X B27 Additive (Invitrogen), 1.25 mM N-Acetyl-L-cysteine (Sigma-Aldrich), 2 nM Glutamax (Invitrogen), 10 mM HEPES (Sigma Aldrich), 1:100 v/v Primocin (Invitrogen), 10% Foetal Bovine Serum as previously described (37). The enriched cells were cultured in Clevers media under hypoxic conditions (1–2% O2).
Microfluidic spiral chip
Isolation of CTCs using spiral microfluidic technology was performed as previously described (14,36). In brief, whole blood (10 mL) was collected in K2EDTA tubes (BD-Plymouth, UK), red blood cell lysed in 1:3 dilution (Astral Scientific, Australia), and run through a spiral chip using a syringe pump at 1.7 mL/min. The resulting CTC output was collected and spun down at 300 g for 5 min.
Characterization of CTCs
Spiral chip enriched samples were cyto-centrifuged onto glass slides and CTCs were identified by immunocytochemistry (ICC). Cells were fixed with 4% paraformaldehyde (PFA) and permeabilized with 0.25% Triton in PBS. The cells were then blocked with 10% FBS. Cells were immunostained with a combination of Cytokeratin Pan Antibody (Invitrogen), Vimentin Monoclonal Antibody (V9) (Invitrogen) and CD45 (AD). Subsequently, slides were stained with DAPI for nuclear staining. ProLong™ Gold Antifade mountant (Invitrogen) was used to prevent photobleaching and for the preservation of the fluorescent labelled molecules for long term storage. Slides were coverslipped and imaged using Zeiss Axio Imager Z2 microscope.
CTCs were identified as: (I) morphologically larger than background cells with intact nuclei; (II) high nucleus-cytoplasmic ratio; (III) pan-cytokeratin positive; (IV) DAPI positive; (V) CD45 negative; and (VI) cells larger than 14 µm. CTCs were additionally characterized for EGFR exon 19 deletion (1:200) (EGFR E746-A750, Cell Signaling, Beverly, MA, USA).
DNA fluorescence in situ hybridization
Enriched CTC samples were cyto-centrifuged, fixed in 4% PFA and dehydrated through an ethanol series of 70%, 85% and 96%. Slides were treated with RNAse (4 mg/mL) (Sigma, USA) and DNA FISH was then performed with EML4-ALK probe (Vysis LSI ALK break apart, Abbott, USA) as previously described (38). The cytospots were further counterstained with DAPI, coverslipped and imaged on a Zeiss Axio Imager Z2 microscope.
DNA extraction of CTCs from culture
DNA extraction was carried out using the QIAmp DNA Mini Kit (Qiagen, Germany) in concordance to the manufacturer’s instructions. DNA samples were evaluated for purity and quantified on a Qubit Fluorometer (Thermo Fisher Scientific, USA). In order to determine whether there was sufficient DNA prior to whole exome sequencing (WES) studies, DNA (50 ng) was used as a template for all polymerase chain reaction (PCR) amplifications. Human β-globin (forward: CAACTTCCACGGTTCACC; reverse: GAAGAGCCAAGGACAGGTAC) was used as a control to verify that cultured cells are of human origin. The qPCR used the following conditions: 50 °C for 2 min; 95 °C for 10 min; 40 cycles at 95 °C for 15 s and 60 °C for 1 min; and a final melting curve analysis with following conditions: 95 °C for 15 s, 60 °C for 1 min, and 95 °C for 15 s. qPCR was carried out for each sample on a QuantStudio™ 7 Flex Real-Time PCR System (Applied Biosystems™) using PowerUp™ SYBR® Green Master Mix (Applied Biosystems, Thermo Fisher Scientific, USA).
WES
Genomic DNA previously isolated was used for exome capture. The qualified genomic DNA sample was randomly fragmented by Covaris technology (Covaris Inc., Woburn, MA, USA), after which the sizes of the DNA fragments were distributed between 150–250 bp. End repair of DNA fragments was performed and an ‘A’ based was added at the 3' end of each stand, then ligated to both ends of the resulting fragments for amplification and sequencing. Extracted DNA was amplified by ligation-mediated PCR (LM-PCR) with hybridization, amplification and purification for enrichment. Each resulting qualified captured library was then sequenced on the BGISEQ-500 sequencing platform (Beijing Genomics Institute, Guangdong, China) and the desired average sequencing coverage for each sample was attained. Raw image files were processed using default parameters by BGISEQ base-calling software to generate pair-end reads for the data of each individual.
Variant analysis
Somatic variants were identified using the Australian Translational Genomics Centre’s clinical genomics bioinformatic pipeline v3.2.0. Briefly, FASTQ reads were aligned the GRCh37 human genome reference sequence using Novoalign v.3.09.00 (http://www.novocraft.com). Somatic variants were called from tumour-normal (cultured CTC versus blood from the same patient) pairs using MuTect2 as part of the Genome Analysis Toolkit v3.7 (39). In order to avoid spurious variant calls, filter PASS variants with variant allele frequencies (VAF) <0.1 were removed. Variants were subsequently annotated using variant effect predictor (VEP) v.86 (40) and converted to MAF format using vcf2maf.pl (https://github.com/mskcc/vcf2maf). Mutational signature analysis, tumour heterogeneity analysis and oncogenic pathway analysis was performed using Maftools v.1.9.20 (41) as implemented in R v.3.4.3 (42).
Statistical analysis
Correlation of CTC count at baseline with clinical stage was compared using Spearman’s ρ analysis. Associations of CTC baseline counts (>2 cells) and progression-free survival (PFS) were assessed using Kaplan-Meier analysis and log-rank tests. P<0.05 was considered to signify a statistically significant difference. Association with baseline CTC counts and short-term culture success was studied using the Mann-Whitney test.
Results
Patient demographic, CTC counts and ex vivo expansion
A total of 70 NSCLC patients were included in this study. Patient demographics are shown in Table 1. The median age was 66 years (range, 36–89 years); including n=42 men and n=28 women. The histological classifications were adenocarcinoma (n=57), squamous cell carcinoma (n=12) and adenosquamous carcinoma (n=1). Patient demographics are listed in Table 1.
CTC enrichment and characterization using spiral microfluidics
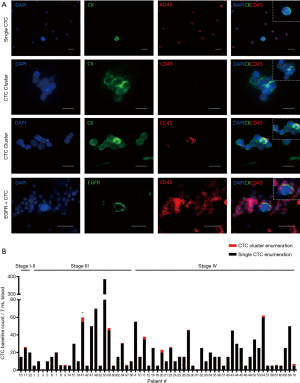
Peripheral blood samples from 70 patients were isolated using microfluidic spiral chip and RosetteSep as described previously (11,14,43). Putative CTCs and CTC clusters were defined as pan-cytokeratin-positive and CD45-negative cells (Figure 2A). Thirty-seven of 70 patients were positive for single CTCs (Figure 2A) with a CTC count of >2 while 16 patients had CTC clusters using spiral microfluidic technology (Table S1). Of the 16 CTC-cluster patients, 15 had single CTCs also detected. Identification of CTM were seen in one patient (Pt #41) who also had single CTCs. In four patients where EGFR exon 19 deletion in the primary tissue was identified by Queensland Pathology and corresponding bloods was taken for CTC evaluation, CTCs were identified in 4/5 patients with an EGFR exon 19 deletion specific antibody (EGFR E746-A750, Cell Signalling, USA) (Figure 2A). Pt #17 was positive for 25/30, Pt #23 was positive for 5/25, Pt #44 was positive for 27/30 and Pt #67 was positive for 1/1. The distribution of CTC types identified from the patient cohort is shown in Table 1. Single CTC and cluster count distribution for NSCLC clinical stages is illustrated in Figure 2B. CTCs were identified 3/3 stage I–II, 24/26 stage III, 37/40 stage IV patients. Higher CTC counts were observed in patients with advanced stage disease.

Full table
Association of CTC with PFS in NSCLC
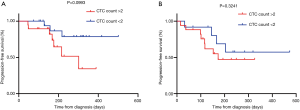
The CTC assessment at baseline was measured against patient survival curves using Kaplan-Meier curves. In total, 32 patients had <2 CTCs at baseline and 37 patients had >2 CTCs. Survival was evaluated for treatment naive and relapse/refractory patients. There was not a significant association between survival outcomes and treatment naive vs. relapse/refractory patients. Treatment naive patients with CTC counts of <2 did not have any significant PFS compared to those with >2 CTCs [hazard ratio (HR): 3.368; 95% confidence interval (CI): 0.9549–12.34; P=0.0993] (Figure 3A). This was also observed for relapse/refractory patients (HR: 1.630; 95% CI: 0.5227–5.085; P=0.3241) (Figure 3B).
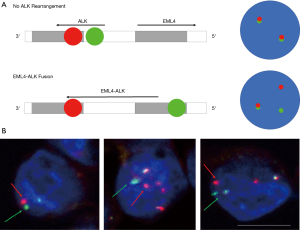
Analysis of ALK FISH using spiral enriched CTCs
Detection of ALK rearrangements from enriched CTCs using FISH by Vysis Break Apart FISH Probe kit (Abbott Molecular, Abbot Park, IL, USA). One patient was known to have an ALK-positive tumour, also had detectable CTC counts (Figure 4). EML4-ALK translocation was identified in in 8/9 (88.9%) CTCs (Figure 4).
Ex vivo culture of isolated CTCs from patients with non-small cell lung cancer (NSCLC)
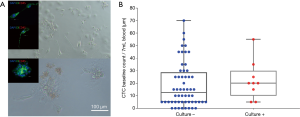
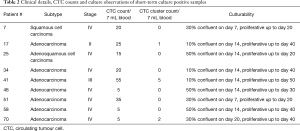
Short term cultures were established from 9 patients (13%) using RosetteSep (Stemcell Technologies™, Vancouver, Canada) enrichment for up to 40 days in Clevers media and hypoxic conditions (1–2% O2) (Figure 5A and Table 2). Once patient derived cultures were confluent (80–90%) the wells were collected for molecular/protein characterization assays. ICC on the cultured cells was showed that these cells were of epithelial origin and CD45 negative (Figure 5A). In patients 7, 25, 46, 51 and 58 high confluency was reached in 7–14 days and were proliferative for up to 50 days. Patients 17, 34, 41 and 66 reached a low cell confluency by day 14 and were proliferative for up to 40 days. All patients were CTC positive with clusters were detected in 2/7 cultured samples (Pt #17 and #41), additionally the presence of CTM was identified in patient #41 as shown in Table 2. Cultures were positive in samples with 5 or more CTCs in 7mls of blood as shown in Figure 5. It was also observed that the culturability wasn’t affected by an increased number of CTCs (Figure 5B).
Full table
WES and bioinformatics analysis of expanded CTCs
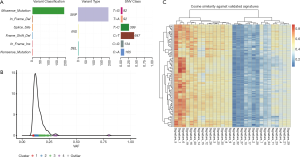
WES of cultured CTC and matched germline (blood) material from Patient #7 was conducted. A total of 1,633 somatic variants were detected in the CTCs, of which 1,430 were silent mutations. The remaining mutations were predominately missense mutations (n=191), as well as in-frame INDELs (n=5), splice site variants (n=3) nonsense mutations (n=2) and frameshift deletions (n=2) (Figure 6A). Density plotting of the variant allele frequency (VAF) distribution revealed four distinct clusters of VAFs with frequencies typically ranging from 0.1 to 0.25. These numerous, low VAF, clusters were indicative of a high degree of high tumour heterogeneity/polyclonality (Figure 6B). The mutational composition (a.k.a. signature) of the sample was compared to mutational signatures present in the COSMIC database (44) to determine the underlying mutational processes generating the somatic variants. Following 100 computational iterations, three distinct mutational signatures were noted: signature 1 (average cosine similarity 0.79) a common mutational signature across all cancer types which results from spontaneous deamination of 5-methylcytosine, signature 2 (average cosine similarity 0.75) a common mutational signature with unknown aetiology which is also observed across all cancer types, and signature 3 (average cosine similarity 0.74) a signature resulting from defective DNA mismatch repair which is noted to occur in LC as well as some other cancer types. Whilst the precise mutational process driving somatic mutation remains unclear, the observed mutational signature was consistent with that of a LC (Figure 6C). Three missense mutations were noted to occur in oncogenic pathways, two of which were predicted to be benign (TSC1-322Thr in the PI3K pathway and WNT8B-11Ser in the WNT pathway). The third mutation (FGFR4-388Arg in the RTK-RAS pathway) was predicted to be damaging and has previously been noted to promote LC progression (45). A full list of annotated variants is available at http://fp.amegroups.cn/cms/91cf48db5d4b8fef6b36eba0510a33ea/TLCR-20-521-1.pdf.
Discussion
Metastasis is the leading cause for cancer-related deaths. Over 30% of patients with NSCLC develop recurrences due to metastasis at diagnosis (46-48). One of the reasons for this is the lack of consideration for metastatic seed, CTCs. In our study, CTCs were identified in 38/70 (54.3%) NSCLC patient’s blood samples, with the inclusion of single CTCs, CTC clusters and CTM. Our CTC detection rates were comparative to that of Freidin et al. who detected 56% and 65% (49). We found no significant association between CTC counts in NSCLC cancer patients with early stage than with late stage disease. This is supported by previous studies by Freidin et al., Chudsama et al., Mascalchi et al., who reported no significant correlation between tumour stage and number of CTCs detected (49-51). Consistent with previous studies investigating the prognostic role of CTCs in LC (52-57), we confirmed that baseline CTC counts are significantly associated with PFS.
We were able to ex vivo culture CTCs derived from advanced stage patients’ blood samples. Cultures grown in hypoxic conditions were successful from CTC counts as low as 5 CTCs/7.5 mL of blood and were proliferative for up to 40 days. Hypoxia is a vital growth stimulus that promotes metastatic progression and is commonly used for CTC culture to ‘mimic’ conditions observed in vivo at distant sites where oxygen tensions are low (58-61). Another essential culture condition was the use of Clevers media, previously used for the culture of prostate derived CTCs (37). Clevers media promotes a stem cell phenotype. Additional growth factors in the media such as R-spondin-1 is used to sustain survival through Wnt signalling as a pivotal requirement for proliferation and growth of epithelial cells (30,37,62,63). We were able to confirm that the CTC culture from 1 patient showed a NSCLC genotype, which provided us with confidence that the developed culture methodology could be used in NSCLC. Further studies, using advance stage patients with higher burden-of-disease may be desirable to determine the effectiveness of the methodology.
In this study, we identified the presence of CTM in the blood of one patient, who also showed successful expansion short term (Pt #41). The presence of CTC clusters has been previously shown to have higher metastatic potential when compared with single CTCs (9). Clusters comprised of CTCs known as CTM, allow tumour cells within to be protected from apoptosis and have shown to drive cell cycle progression and increase metastatic potential of CTCs (12,15).
Clinically relevant biomarkers, EGFR del 19 mutation and ALK rearrangements were also analysed in a subset of patients. The presence of ALK rearrangement on CTCs from an ALK positive patient, validating previous studies (64,65). In 4/5 patients where tumours were identified as EGFR exon 19 del, EGFR mutation was detected in isolated CTCs, confirming that CTCs can be used as proxy to investigate tumour activities. This supports previous studies where EGFR mutation has been detected in CTCs which were concordant with mutation status in corresponding tumour (31,66-68). The mutation-specific antibody for E746-A750 selected has previously identified EGFR mutation detection in CTCs (11). This fast, low-cost approach shows promise for the detection of this mutation in NSCLC, particularly, where access to tumour tissue is limited, may enable early interventions and better treatment management (69). Further validation is necessary in order to confirm the clinical utility for screening EGFR del 19 mutation in NSCLC patients (70,71).
To determine the concordance between the primary tissue and CTC cultures, WES was performed in a subset of patients where sufficient DNA was available from the CTC cultures (n=9). Exome sequencing of CTC and white blood cells (as germline control) confirmed the presence of somatic mutations in the CTC culture of patient #7 with mutational signatures consistent with LC. Three missense mutations were observed to arise in oncogenic pathways. While two mutations (TSC1-322Thr in the PI3K pathway and WNT8B-11Ser in the WNT pathway) were predicted to be benign, the third mutation (FGFR4-388Arg in the RTK-RAS pathway) is predicted to be functionally relevant, having been reported to promote activation of pathways relative to cancer, and has been associated with the progression of LC (45). To better understand genomic heterogeneity, it is desirable to obtain genomic profiles throughout the course of the disease at multiple timepoints. Acquiring longitudinal samples was limited in the current study as the patient cohort had advanced stage with poor survival. Sufficient DNA yields were not available on the other eight CTC cultures at the timepoint at which the cultures were harvested.
Several efforts to propagate CTC ex vivo cultures have had limited success, particularly for NSCLC (72,73). Here we describe a method that allowed expansion of CTCs in culture in a subset of patients, which may provide a better understanding into CTC heterogeneity and mechanisms of metastasis. Further studies are necessary in the NSCLC field to establish CTC cell lines to model transient disease, in order to evaluate and discover new therapeutic approaches. Ideally, cancer cell lines would be routinely generated from each cancer patient but this is currently not realistic (15,28,30). This culture method may provide new spectrum of prospective functional analyses which in turn provide powerful ability for modelling cancer heterogeneity at clinically relevant timepoints.
Conclusions
With LC being the highest cause of cancer mortality worldwide, one of the biggest challenges for managing and treating patients is the lack of early screening/diagnostic methods (5,74). In order to monitor tumour evolution and understand mechanisms underlying resistance, genomic analyses of NSCLC are essential however, is currently limited by the accessibility to tumour specimens. The use of CTCs as a minimally-invasive liquid biopsy could accompany standard screening tests and also allow for molecular and genetic characterization during tumour progression in real time (75). Additionally, genomic analysis of CTCs provides important information for personalized therapy, however the difficulty of low DNA yield from the small number of captured CTCs remains a large hurdle. To meet this challenge, expansion of CTCs in cell culture systems allows for downstream functional analysis.
As demonstrated in our current study, the facilitation of liquid biopsies to capture CTCs for genomic analysis, has promise in non-invasive cancer genotyping and monitoring of disease. In addition, the expansion of CTCs captured through minimally invasive clinical samples provide valuable opportunity to study NSCLC biology. In the future, the authors envisage recruiting longitudinal samples from NSCLC patients for the monitoring of disease to determine the changes in CTCs over time.
Acknowledgments
The authors would like to thank Patricia Davidson and Jack Varghese (Clinical Trials, PAH) for their assistance.
Funding: This study was supported by the Cancer Australia (APP114567) and NHMRC Early Career Fellowship to AK (1157741). QUT postgraduate research scholarship for JK.
Footnote
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tlcr-20-521
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tlcr-20-521). KOB serves as an unpaid editorial board member of Translational Lung Cancer Research. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was conducted according to the principles of Declaration of Helsinki (as revised in 2013). Ethics approval was obtained by Metro South Health District Human Research Ethics Committee in accordance with the National Health and Medical Research Council’s guidelines (HREC/11/QPAH/331) and Queensland University of Technology (1100001420). All patients enrolled completed the informed consent form.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin 2017;67:7-30. [Crossref] [PubMed]
- Wong MCS, Lao XQ, Ho KF, et al. Incidence and mortality of lung cancer: global trends and association with socioeconomic status. Sci Rep 2017;7:14300. [Crossref] [PubMed]
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Insitute N-NC. Cancer Stat Facts: Lung and Bronchus Cancer. In: Surveillance, Epidemiology, and End Results Program. NIH- National Cancer Institute. 2017. Available online: https://seer.cancer.gov/statfacts/html/lungb.html. Accessed July 2017.
- Ellis PM, Vandermeer R. Delays in the diagnosis of lung cancer. J Thorac Dis 2011;3:183-8. [PubMed]
- Malik PS, Raina V. Lung cancer: Prevalent trends & emerging concepts. Indian J Med Res 2015;141:5-7. [Crossref] [PubMed]
- Massagué J, Obenauf AC. Metastatic colonization by circulating tumour cells. Nature 2016;529:298. [Crossref] [PubMed]
- Mehlen P, Puisieux A. Metastasis: a question of life or death. Nat Rev Cancer 2006;6:449-58. [Crossref] [PubMed]
- Aceto N, Bardia A, Miyamoto David T, et al. Circulating Tumor Cell Clusters Are Oligoclonal Precursors of Breast Cancer Metastasis. Cell 2014;158:1110-22. [Crossref] [PubMed]
- Au SH, Storey BD, Moore JC, et al. Clusters of circulating tumor cells traverse capillary-sized vessels. Proc Natl Acad Sci U S A 2016;113:4947-52. [Crossref] [PubMed]
- Kulasinghe A, Kapeleris J, Cooper C, et al. Phenotypic Characterization of Circulating Lung Cancer Cells for Clinically Actionable Targets. Cancers (Basel) 2019;11:380. [Crossref] [PubMed]
- Szczerba BM, Castro-Giner F, Vetter M, et al. Neutrophils escort circulating tumour cells to enable cell cycle progression. Nature 2019;566:553-7. [Crossref] [PubMed]
- Gkountela S, Castro-Giner F, Szczerba BM, et al. Circulating Tumor Cell Clustering Shapes DNA Methylation to Enable Metastasis Seeding. Cell 2019;176:98-112.e14. [Crossref] [PubMed]
- Kulasinghe A, Tran TH, Blick T, et al. Enrichment of circulating head and neck tumour cells using spiral microfluidic technology. Sci Rep 2017;7:42517. [Crossref] [PubMed]
- Dive C, Brady G. SnapShot: Circulating Tumor Cells. Cell 2017;168:742. [Crossref] [PubMed]
- Aceto N, Toner M, Maheswaran S, et al. En Route to Metastasis: Circulating Tumor Cell Clusters and Epithelial-to-Mesenchymal Transition. Trends Cancer 2015;1:44-52. [Crossref] [PubMed]
- Hong Y, Fang F, Zhang Q. Circulating tumor cell clusters: What we know and what we expect Int J Oncol 2016;49:2206-16. (Review). [Crossref] [PubMed]
- Cristofanilli M, Budd GT, Ellis MJ, et al. Circulating Tumor Cells, Disease Progression, and Survival in Metastatic Breast Cancer. N Engl J Med 2004;351:781-91. [Crossref] [PubMed]
- Scher HI, Jia X, de Bono JS, et al. Circulating tumour cells as prognostic markers in progressive, castration-resistant prostate cancer: a reanalysis of IMMC38 trial data. Lancet Oncol 2009;10:233-9. [Crossref] [PubMed]
- Stott SL, Hsu CH, Tsukrov DI, et al. Isolation of circulating tumor cells using a microvortex-generating herringbone-chip. Proc Natl Acad Sci U S A 2010;107:18392-7. [Crossref] [PubMed]
- Zhang L, Riethdorf S, Wu G, et al. Meta-analysis of the prognostic value of circulating tumor cells in breast cancer. Clin Cancer Res 2012;18:5701-10. [Crossref] [PubMed]
- Bidard FC, Peeters DJ, Fehm T, et al. Clinical validity of circulating tumour cells in patients with metastatic breast cancer: a pooled analysis of individual patient data. Lancet Oncol 2014;15:406-14. [Crossref] [PubMed]
- Lindsay CR, Blackhall FH, Carmel A, et al. EPAC-lung: pooled analysis of circulating tumour cells in advanced non-small cell lung cancer. Eur J Cancer 2019;117:60-8. [Crossref] [PubMed]
- Nurwidya F, Zaini J, Putra AC, et al. Circulating Tumor Cell and Cell-free Circulating Tumor DNA in Lung Cancer. Chonnam Med J 2016;52:151-8. [Crossref] [PubMed]
- Alix-Panabières C, Pantel K. Clinical Applications of Circulating Tumor Cells and Circulating Tumor DNA as Liquid Biopsy. Cancer Discov 2016;6:479-91. [Crossref] [PubMed]
- Kim L, Tsao MS. Tumour tissue sampling for lung cancer management in the era of personalised therapy: what is good enough for molecular testing? Eur Respir J 2014;44:1011-22. [Crossref] [PubMed]
- Pantel K, Alix-Panabieres C. Liquid biopsy and minimal residual disease - latest advances and implications for cure. Nat Rev Clin Oncol 2019;16:409-24. [Crossref] [PubMed]
- Maheswaran S, Haber DA. Ex Vivo Culture of CTCs: An Emerging Resource to Guide Cancer Therapy. Cancer Res 2015;75:2411-5. [Crossref] [PubMed]
- Khoo BL, Grenci G, Jing T, et al. Liquid biopsy and therapeutic response: Circulating tumor cell cultures for evaluation of anticancer treatment. Sci Adv 2016;2:e1600274. [Crossref] [PubMed]
- Wang R, Chu GCY, Mrdenovic S, et al. Cultured circulating tumor cells and their derived xenografts for personalized oncology. Asian J Urol 2016;3:240-53. [Crossref] [PubMed]
- Maheswaran S, Sequist LV, Nagrath S, et al. Detection of Mutations in EGFR in Circulating Lung-Cancer Cells. N Engl J Med 2008;359:366-77. [Crossref] [PubMed]
- Lohr JG, Adalsteinsson VA, Cibulskis K, et al. Whole-exome sequencing of circulating tumor cells provides a window into metastatic prostate cancer. Nat Biotechnol 2014;32:479-84. [Crossref] [PubMed]
- Zhu Z, Qiu S, Shao K, et al. Progress and challenges of sequencing and analyzing circulating tumor cells. Cell Biol Toxicol 2018;34:405-15. [Crossref] [PubMed]
- Carter L, Rothwell DG, Mesquita B, et al. Molecular analysis of circulating tumor cells identifies distinct copy-number profiles in patients with chemosensitive and chemorefractory small-cell lung cancer. Nat Med 2017;23:114-9. [Crossref] [PubMed]
- Faugeroux V, Lefebvre C, Pailler E, et al. An Accessible and Unique Insight into Metastasis Mutational Content Through Whole-exome Sequencing of Circulating Tumor Cells in Metastatic Prostate Cancer. Eur Urol Oncol 2019;S2588-9311(18)30213-X.
- Warkiani ME, Khoo BL, Wu L, et al. Ultra-fast, label-free isolation of circulating tumor cells from blood using spiral microfluidics. Nat Protoc 2016;11:134-48. [Crossref] [PubMed]
- Gao D, Vela I, Sboner A, et al. Organoid cultures derived from patients with advanced prostate cancer. Cell 2014;159:176-87. [Crossref] [PubMed]
- Khoo BL, Warkiani ME, Tan DSW, et al. Clinical Validation of an Ultra High-Throughput Spiral Microfluidics for the Detection and Enrichment of Viable Circulating Tumor Cells. PLoS One 2014;9:e99409. [Crossref] [PubMed]
- McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010;20:1297-303. [Crossref] [PubMed]
- McLaren W, Gil L, Hunt SE, et al. The Ensembl Variant Effect Predictor. Genome Biol 2016;17:122. [Crossref] [PubMed]
- Mayakonda A, Lin DC, Assenov Y, et al. Maftools: efficient and comprehensive analysis of somatic variants in cancer. Genome Res 2018;28:1747-56. [Crossref] [PubMed]
- Team RC. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. 2018. Available online: https://www.R-project.org/
- Kulasinghe A, Perry C, Warkiani ME, et al. Short term ex-vivo expansion of circulating head and neck tumour cells. Oncotarget 2016;7:60101-9. [Crossref] [PubMed]
- Tate JG, Bamford S, Jubb HC, et al. COSMIC: the Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res 2019;47:D941-7. [Crossref] [PubMed]
- Quintanal-Villalonga Á, Ojeda-Márquez L, Marrugal Á, et al. The FGFR4-388arg Variant Promotes Lung Cancer Progression by N-Cadherin Induction. Sci Rep 2018;8:2394. [Crossref] [PubMed]
- al-Kattan K, Sepsas E, Fountain SW, et al. Disease recurrence after resection for stage I lung cancer. Eur J Cardiothorac Surg 1997;12:380-4. [Crossref] [PubMed]
- Carnio S, Novello S, Papotti M, et al. Prognostic and predictive biomarkers in early stage non-small cell lung cancer: tumor based approaches including gene signatures. Transl Lung Cancer Res 2013;2:372-81. [PubMed]
- Uramoto H, Tanaka F. Recurrence after surgery in patients with NSCLC. Transl Lung Cancer Res 2014;3:242-9. [PubMed]
- Freidin MB, Tay A, Freydina DV, et al. An assessment of diagnostic performance of a filter-based antibody-independent peripheral blood circulating tumour cell capture paired with cytomorphologic criteria for the diagnosis of cancer. Lung Cancer 2014;85:182-5. [Crossref] [PubMed]
- Chudasama D, Barr J, Beeson J, et al. Detection of Circulating Tumour Cells and Survival of Patients with Non-small Cell Lung Cancer. Anticancer Res 2017;37:169-73. [Crossref] [PubMed]
- Mascalchi M, Falchini M, Maddau C, et al. Prevalence and number of circulating tumour cells and microemboli at diagnosis of advanced NSCLC. J Cancer Res Clin Oncol 2016;142:195-200. [Crossref] [PubMed]
- Tong B, Xu Y, Zhao J, et al. Prognostic significance of circulating tumor cells in non-small cell lung cancer patients undergoing chemotherapy. Oncotarget 2017;8:86615-24. [Crossref] [PubMed]
- Tanaka F, Yoneda K, Kondo N, et al. Circulating tumor cell as a diagnostic marker in primary lung cancer. Clin Cancer Res 2009;15:6980-6. [Crossref] [PubMed]
- Krebs MG, Sloane R, Priest L, et al. Evaluation and prognostic significance of circulating tumor cells in patients with non-small-cell lung cancer. J Clin Oncol 2011;29:1556-63. [Crossref] [PubMed]
- Zhang Z, Xiao Y, Zhao J, et al. Relationship between circulating tumour cell count and prognosis following chemotherapy in patients with advanced non-small-cell lung cancer. Respirology 2016;21:519-25. [Crossref] [PubMed]
- Hou JM, Krebs MG, Lancashire L, et al. Clinical significance and molecular characteristics of circulating tumor cells and circulating tumor microemboli in patients with small-cell lung cancer. J Clin Oncol 2012;30:525-32. [Crossref] [PubMed]
- Naito T, Tanaka F, Ono A, et al. Prognostic impact of circulating tumor cells in patients with small cell lung cancer. J Thorac Oncol 2012;7:512-9. [Crossref] [PubMed]
- Vaupel P. The role of hypoxia-induced factors in tumor progression. Oncologist 2004;9 Suppl 5:10-7. [Crossref] [PubMed]
- Yuan J, Glazer PM. Mutagenesis induced by the tumor microenvironment. Mutat Res 1998;400:439-46. [Crossref] [PubMed]
- Muz B, de la Puente P, Azab F, et al. The role of hypoxia in cancer progression, angiogenesis, metastasis, and resistance to therapy. Hypoxia (Auckl) 2015;3:83-92. [Crossref] [PubMed]
- Rankin EB, Giaccia AJ. Hypoxic control of metastasis. Science 2016;352:175-80. [Crossref] [PubMed]
- Tüysüz N, van Bloois L, van den Brink S, et al. Lipid-mediated Wnt protein stabilization enables serum-free culture of human organ stem cells. Nat Commun 2017;8:14578. [Crossref] [PubMed]
- Sato T, Stange DE, Ferrante M, et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology 2011;141:1762-72. [Crossref] [PubMed]
- Ilie M, Long E, Butori C, et al. ALK-gene rearrangement: a comparative analysis on circulating tumour cells and tumour tissue from patients with lung adenocarcinoma. Ann Oncol 2012;23:2907-13. [Crossref] [PubMed]
- Pailler E, Adam J, Barthélémy A, et al. Detection of Circulating Tumor Cells Harboring a Unique ALK Rearrangement in ALK-Positive Non-Small-Cell Lung Cancer. J Clin Oncol 2013;31:2273-81. [Crossref] [PubMed]
- Marchetti A, Del Grammastro M, Felicioni L, et al. Assessment of EGFR Mutations in Circulating Tumor Cell Preparations from NSCLC Patients by Next Generation Sequencing: Toward a Real-Time Liquid Biopsy for Treatment. PLoS One 2014;9:e103883. [Crossref] [PubMed]
- Punnoose EA, Atwal S, Liu W, et al. Evaluation of circulating tumor cells and circulating tumor DNA in non-small cell lung cancer: association with clinical endpoints in a phase II clinical trial of pertuzumab and erlotinib. Clin Cancer Res 2012;18:2391-401. [Crossref] [PubMed]
- Sundaresan TK, Sequist LV, Heymach JV, et al. Detection of T790M, the Acquired Resistance EGFR Mutation, by Tumor Biopsy versus Noninvasive Blood-Based Analyses. Clin Cancer Res 2016;22:1103-10. [Crossref] [PubMed]
- Saarenheimo J, Eigeliene N, Andersen H, et al. The Value of Liquid Biopsies for Guiding Therapy Decisions in Non-small Cell Lung Cancer. Front Oncol 2019;9:129. [Crossref] [PubMed]
- Simonetti S, Molina MA, Queralt C, et al. Detection of EGFR mutations with mutation-specific antibodies in stage IV non-small-cell lung cancer. J Transl Med 2010;8:135. [Crossref] [PubMed]
- Yu J, Kane S, Wu J, et al. Mutation-Specific Antibodies for the Detection of EGFR Mutations in Non-Small-Cell Lung Cancer. Clin Cancer Res 2009;15:3023. [Crossref] [PubMed]
- Khoo BL, Grenci G, Lim YB, et al. Expansion of patient-derived circulating tumor cells from liquid biopsies using a CTC microfluidic culture device. Nat Protoc 2018;13:34-58. [Crossref] [PubMed]
- Kapeleris J, Kulasinghe A, Warkiani ME, et al. The Prognostic Role of Circulating Tumor Cells (CTCs) in Lung Cancer. Front Oncol 2018;8:311. [Crossref] [PubMed]
- Price KA, Cohen EE. Current treatment options for metastatic head and neck cancer. Curr Treat Options Oncol 2012;13:35-46. [Crossref] [PubMed]
- Murlidhar V, Ramnath N, Nagrath S, et al. Optimizing the Detection of Circulating Markers to Aid in Early Lung Cancer Detection. Cancers (Basel) 2016;8:61. [Crossref] [PubMed]


