
SRC and PIM1 as potential co-targets to overcome resistance in MET deregulated non-small cell lung cancer
Introduction
The hepatocyte growth factor receptor (MET) is a transmembrane receptor tyrosine kinase, identified as dysregulated in lung cancer in the 1990s (1). Firstly studied as mechanisms of innate or acquired resistance to epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors (TKIs) in EGFR mutant non-small cell lung cancer (NSCLC), MET alterations have also been identified as de novo oncogenic drivers in NSCLC, representing a potential new therapeutic target (2-4).
In lung cancers the MET receptor can be altered mostly as a result of gene amplification or gene mutation. MET amplification may result in protein overexpression and constitutive kinase activation and is present in about 4% of non-small cell lung cancer (5,6). Nevertheless, different scoring systems have been used to define amplification by using fluorescence in situ hybridization (FISH) and the real role of MET amplification as a driver is still debated (6,7). Pathogenic MET gene mutations usually occur in the juxtamembrane domain and determine aberrant splicing of exon 14, leading to the production of a protein lacking the Y1003 c-Cbl binding site and consequent decreased ubiquitination and protein degradation (5,8). This kind of alteration is found in about 2.7% of NSCLC, up to 8% in adenosquamous and 22% in sarcomatoid histology (9,10). Recently, an intronic deletion involving the MET extracellular domain has been identified in high grade gliomas, determining the loss of exons 7 and 8 and the fusion of Immunoglobulin-plexin-transcription 1-2 (IPT1-2), resulting in defective furin cleavage, intracellular retention, and auto-activation of the uncleaved preform (11). No data on the prevalence and role of this alteration in NSCLC are available.
The first drugs used to inhibit MET were multi-target inhibitors, such as, crizotinib and cabozantinib (7,12-16). More selective MET inhibitors, tepotinib, savolitinib and capmatinib, look more promising: they demonstrated activity in MET exon 14 mutated NSCLC and are currently under clinical evaluation in a molecularly selected population (17-19). Very recently, capmatinib received approval from Food and Drug Administration (FDA) for the treatment of MET exon 14 mutated NSCLC. Among the antibody compounds triggering MET, data recently presented on safety and efficacy of Sym015, show promising activity of this novel drug (20). Unfortunately, virtually all patients develop resistance to these drugs and the acquired resistance mechanisms have not yet been studied in depth.
Proviral integration site for Moloney murine leukemia virus (PIM) is a serine/threonine kinase, involved in cell cycle progression, cell growth, cell survival and treatment resistance. In a MET amplified NSCLC cell line, treatment with a MET inhibitor led to upregulation of PIM-1 as a mechanism of resistance, while treatment with PIM inhibitors restored sensitivity to MET inhibition in the MET inhibitor-resistant cell line (21). Moreover, in a prostate cancer model, PIM-1 signaling was associated with resistance to AKT inhibitors through the upregulation of MET and other receptor tyrosine kinases (RTKs) (22). Therefore, dual PIM-1 and MET inhibition might prevent the occurrence of resistance mechanisms in MET dependent cell lines.
Src homology 2-containing protein tyrosine phosphatase 2 (SHP2), a non-receptor protein tyrosine phosphatase, is central in RTK signaling and in Src activation. We have previously shown that the overexpression of RTKs, like AXL and the transmembrane protein CUB domain-containing protein-1 (CDCP1), as well as Src activation, are mechanisms of intrinsic resistance to EGFR inhibition in EGFR mutant lung cancer (23). Also, in MET amplified EGFR mutant lung cancer cell lines resistant to EGFR TKIs, Src expression was increased, compared to EGFR TKI sensitive counterpart. In this model, Src inhibition led to cell growth arrest and apoptosis (24).
The present study aims to test the role of PIM and Src in resistance to MET inhiabition in MET addicted tumors, and the potential activity of combination treatments in this context.
We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/tlcr-20-681).
Methods
Chemical and reagents
Tepotinib, AZD1208 and dasatinib were purchased from Selleck Chemicals (Houston, TX, U.S.). Savolitinib was purchased from Med Chem Express (Monmouth Junction, NJ, U.S.). PIM447 (LGH447), crizotinib and cabozantinib were purchased from Selleck Chemicals (Houston, TX, U.S.). Drugs were prepared in dimethyl sulfoxide (DMSO) at a concentration of 10–100 mM stock solutions and stored at −20 °C. Further dilutions were made in culture medium to final concentration before use. Primary antibodies and secondary antibodies used are in Table S1.
Full table
Cell viability assay
The EBC-1 human squamous lung cancer cell line with MET amplification, the Hs746T human gastric cancer cell line with MET exon 14 skipping mutation, and the H1993 human lung adenocarcinoma cell line harboring MET amplification were purchased from American Type Culture Collection (ATCC). The E98 human glioblastoma cell line with MET exon 7–8 skipping variant (11) was provided by Dr William Leenders, from the Department of Biochemistry, Radboud Institute for Molecular Life Sciences, Nijmegen, The Netherlands. Cells were seeded on 96-well plates at the following densities: 5×103, 8×103, 8×103, 16×103 cells/well, respectively EBC-1, H1993, Hs746T, E98, and incubated for 24 h, as previously described (25). Cells were treated with serial dilutions of the drugs administrated at indicated doses. After 72 h of incubation, 0.5 mg/mL of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) reagent (Sigma-Aldrich, St. Louis, MO, U.S.) was added to the medium in the wells for 2 h at 37 °C and formazan crystals in viable cells were solubilized with 100 µL DMSO and spectrophotometrically quantified using a microplate reader (Infinite 200 PRO, Tecan Group Ltd, Mannedorf, CH) at 565 nm of absorbance. Fractional survival was then calculated as percentage to control cells. Half maximal inhibitory concentrations (IC50s) in nM range identified drug sensitivity, while those in µM range identified drug resistance. Data of combined drug effects were subsequently analyzed by the Chou and Talalay method (26,27). Combination index (CoI) values <1, =1 and >1 indicated synergism, additive effect and antagonism, respectively. High or slight synergism was further identified within values <1, according to previous works (23,25).
Western blot analyses
Cells were washed with cold phosphate-buffered saline (PBS) and re-suspended in ice-cold radioimmunoprecipitation assay buffer containing protease inhibitor mixture (RIPA buffer, Cell Signaling Technology, Leiden, NL) as previously described (23). Centrifugation at 18,620 ×g for 15 min at 4 °C was used to obtain cell lysis and the resulting supernatant was collected as the total cell lysate. Briefly, the lysates containing 30 µg proteins were electrophoresed on 10% sodium dodecyl sulphate (SDS)-polyacrylamide gel electrophoresis (Life Technologies, Carlsbad, CA, U.S.) and transferred to polyvinylidene difluoride membranes (Bio-Rad laboratories Inc., Hercules, CA, U.S.). Membranes were blocked in Odyssey blocking buffer (Li-Cor Biosciences, Lincoln, NE, U.S.) and phosphoBLOCKER blocking reagent (Cell Biolabs INC., San Diego, CA, U.S.), according to manufacturer instructions, to identify non-phosphorylated and phosphorylated proteins, respectively. All target proteins were immunoblotted with appropriate primary and horseradish peroxidase (HPR)- conjugated secondary antibodies. Chemiluminescent (HRP-conjugated) bands were detected in a ChemiDoc MP Imaging System (Bio-Rad laboratories Inc.). β-actin was used as an internal control to confirm equal gel loading.
Patients
Pretreatment tumor specimens from advanced NSCLC patients with squamous cell carcinoma, sarcomatoid histology, and EGFR mutant adenocarcinoma, consecutively treated at Istituto Oncologico Veneto (Italy) from 2012 to 2016, were retrospectively collected. Inclusion criteria were the availability of histological specimen collected before starting any systemic treatment, and the availability of complete clinical and radiological information and adequate follow-up. The clinical data was assessed in accordance with the protocol approved by the institutional review board of Istituto Oncologico Veneto (IOV; protocol number MET-2017; date of approval: 11th December 2017) and de-identified for patient confidentiality. The study was conducted in accordance with the precepts of the Helsinki declaration (as revised in 2013). Signed informed consent, approved by the Ethics Committee of IOV, was obtained for collection, analysis and publication of data, according to the Italian data protection authority dispositions.
FISH and immunohistochemistry (IHC)
FISH for MET was performed with the ZytoLight® SPEC MET/CEP7 Dual Color Probe (ZytoVision, Bremerhaven, Germany) according to manufacturer’s instructions. At least 50 tumor cells per sample were counted. Three positivity criteria for MET amplification were used, namely a MET/CEP7 ratio ≥2; average ≥6 MET copies per tumor cell; and ≥5 MET copies in >50% of tumor cells or ≥15 copies in >10% tumor cells.
Immunostaining was performed with MET SP44 clone (Roche, Mannheim, Germany) on a BenchMark ULTRA automated tissue staining system (Ventana Medical Systems, Tucson, Arizona, USA). MET membrane staining was graded as: 0, absent; 1+, weak/faint; 2+ moderate and 3+, strong. A minimum of 50 tumor cells were evaluated per sample and the positivity criterion was 3+ in ≥50% of tumor cells.
Real-time PCR analyses
Formalin-fixed, paraffin-embedded (FFPE) tumor blocks and slides were obtained by standard procedures. The percentage of tumor cell infiltration, stroma, necrosis and inflammatory cell infiltration was evaluated by a pathologist. For tumor infiltration rate <85%, the selected tumor area was captured by laser microdissection (Zeiss-Palm, Oberlensheim, Germany); otherwise, manual macrodissection was performed. RNA was isolated from the tumor tissue specimens in accordance with a proprietary procedure (European patent number EP1945764-B1).
Samples and cell lines were lysed in a tris, sodium chloride, ethylenediaminetetraacetic (EDTA), SDS and proteinase K containing buffer. RNA was then extracted with phenol-chloroform-isoamyl alcohol, followed by precipitation with isopropanol in the presence of glycogen and sodium acetate. RNA was re-suspended in water and treated with DNAse I to avoid DNA contamination. Complementary DNA (cDNA) was synthesized using M-MLV retrotranscriptase enzyme.
Template cDNA was added to Taqman Universal Master Mix (Applied Biosystems) in a 12.5 µL reaction with specific primers and probes for each gene under analysis. Quantification of gene expression was performed using the ABI Prism 7900HT Sequence Detection System (Applied Biosystems) and was calculated according to the comparative Ct method as previously described (Karachaliou et al., EBioMed 2018). Commercial RNAs from lung and liver were used as internal controls (Liver and Lung; Stratagene, La Jolla, CA, USA). In all quantitative experiments, a sample was considered not evaluable when the standard deviation of the Ct values was >0.30 in two independent analyses. As a result, the number of evaluable samples varied among the several genes analysed.
A tissue-based RT-PCR assay was also used for the detection of MET exon 14 and 7–8 skipping mutations in tumor samples. RNA was converted to cDNA using M-MLV retrotranscriptase (ThermoFisher Scientific) and oligo-dT primers, and METΔex14 was amplified using HotStart Taq polymerase (Qiagen) in a 20 µL reaction and visualized in agarose gels. Primers used were located in exons 13 and 15, sequences were: forward (exon 13) 5'-TTTTCCTGTGGCTGAAAAAGA-3' and reverse (exon 15) 5'-GGGGACATGTCTGTCAGAGG-3'. Amplification generated a 246bp band for wild type (wt) MET RNA and a 106-bp band for METΔex14. Positive samples were confirmed by bidirectional Sanger sequencing of RT-PCR products, using the big-dye 3.1 sequencing kit (Applied Biosystems).
Next generation sequencing
MET copy number gain was also explored in tumor samples by next generation sequencing (NGS), and the results were correlated with FISH MET. NGS was performed with the GeneRead® QIAact Lung DNA UMI Panel (Qiagen), according to the manufacturer’s instructions. The panel targets genes frequently altered in lung cancer tumors and can also detect amplifications in MET and other genes.
For the GeneRead panel, up to 40 ng of purified DNA were used as a template. Clonal amplification was performed on 625 pg of pooled libraries and, following bead enrichment, the GeneReader instrument was used for sequencing. Qiagen Clinical Insight Analyze (QCI-A) software was employed to align the read data and call sequence variants, which were imported into the Qiagen Clinical Insight Interpret (QCI-I) web interface for data interpretation and generation of final report.
Statistical analyses
The preclinical data were analyzed with GraphPad Prism 7.0 statistical software. Variables were presented by using median value for continuous variables and percentages (numbers) for categorical variables. The median overall survival (mOS) from diagnosis of advanced NSCLC was estimated by using Kaplan-Meier method and measures were provided with their 95% CI. The log-rank test was used to compare survival between groups. A two-sided 5% significance level was set for all tests. The statistical analyses were performed using R software.
Results
MET addicted cell lines are sensitive to different classes of MET-TKIs
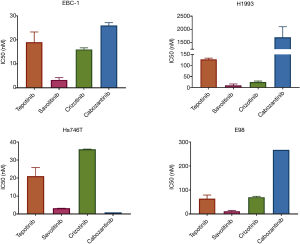
In cell viability experiments, all of the adenocarcinoma cell lines tested, were sensitive to single MET inhibition with tepotinib, savolitinib, crizotinib and cabozantinib, with half maximal inhibitory concentrations (IC50s) in nM range, with the exception of MET amplified lung adenocarcinoma H1993 cell line, which was resistant to cabozantinib (Figure 1). The MET amplified lung squamous carcinoma cell line EBC-1 showed high sensitivity (low nM range) to all compounds tested. All cell lines were highly sensitive to the selective MET inhibitors, savolitinib and tepotinib, while only Hs746T cell line (MET exon 14 gastric adenocarcinoma) was very highly sensitive to cabozantinib. Crizotinib showed an intermediate sensitivity profile compared to the other compounds in E98 (MET exon 7–8 glioblastoma), EBC-1 and H1993 cell lines.
Single PIM inhibition or single SRC inhibition had no activity in MET addicted cell lines
All cell lines tested were resistant to the pan-PIM inhibitors AZD1208 and PIM447, displaying IC50s in µM range in cell viability experiments. Similar results were obtained when using the SRC inhibitor, dasatinib, alone in the MET altered cell lines (Table S2).

Full table
Co-treatment with MET and PIM inhibitors showed synergism in MET amplified lung squamous cell carcinoma cell line EBC-1
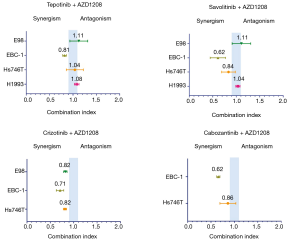
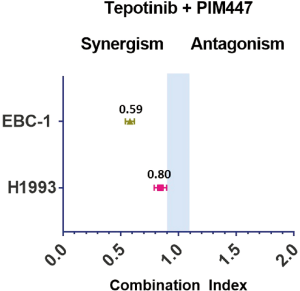
When combining the MET inhibitors with the PIM inhibitors in cell viability experiments, we obtained synergistic or additive effect in all the cell lines. The combination of AZD1208 with either, savolitinib, crizotinib or cabozantinib, was highly synergistic in the EBC-1 cell line, while the AZD1208 plus tepotinib combination was only slightly synergistic (Figure 2, Table S3). The tepotinib plus PIM447 combination was highly synergistic in the EBC-1 cell line (CoI 0.59), while it was slightly synergistic in the H1993 cell line (CoI 0.80) (Figure S1). In the H1993, E98 and Hs746T cell lines, the combination of AZD1208 with the MET inhibitors was slightly synergistic, or showed additive effect (Figure 2, Table S3).

Full table
Co-treatment with MET and SRC inhibitors showed strong synergism in MET addicted cell lines
In cell viability experiments, the combination of dasatinib with savolitinib was highly synergistic in all cell lines (CoI 0.51, 0.62, 0.70, 0.77 in EBC-1, H1993, Hs746T and E98, respectively). Similar results were found with the dasatinib plus tepotinib combination, with the exception of the H1993 cell line showing only slight synergism (Figure 3). The dasatinib plus crizotinib or cabozantinib combinations showed synergism as well (Table S4).
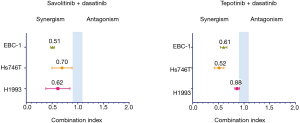

Full table
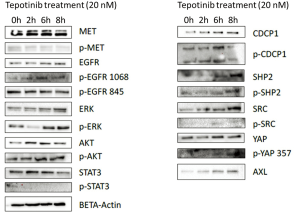
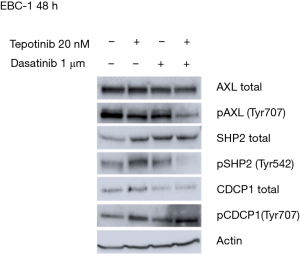
Co-treatment with MET and SRC inhibitors prevents the tepotinib-driven activation of p-AXL, p-SHP2 and total CDCP1
To understand the potential mechanisms of resistance to single MET inhibition, we evaluated the effects of tepotinib alone and in combination with dasatinib in the EBC-1 cell line, by analyzing the expression of proteins potentially involved in resistance mechanisms using immunoblotting experiments. In vitro experiments showed that single MET inhibition with tepotinib led to upregulation of the membrane proteins, AXL and CDCP1, the intracellular proteins, SRC, SHP2 and ERK, and an increase of their phosphorylated forms in the EBC-1 cell line (Figure S2). The addition of dasatinib could partially reverse these effects, causing decrease of activation of p-AXL, p-SHP2 and down-regulation of total CDCP1 (Figure 4).
RNA expression of genes potentially related to resistance to MET inhibition
Baseline RNA expression of MET, PIM, SRC, and other genes involved in resistance development pathways was explored in the four cell lines. EBC-1 and Hs746T cell lines showed higher baseline expression of PIM1, CDCP1, AXL, SHP2 and lower expression of SRC, compared to the H1993 and E98 (Figure S3).

MET alterations in advanced NSCLC samples
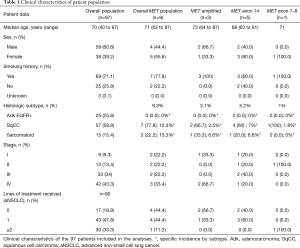
Clinical features of patients included in the analyses are summarized in Table 1.
Full table
Among the 127 NSCLC patients collected with complete clinical information and tumor tissue available, 97 tumor samples were evaluable for MET screening. Overall, MET alterations were found in 9 out of 97 (9.3%) NSCLC samples. MET IHC was positive in five samples, FISH analyses confirmed MET amplification in 3 of them (3.1%). MET amplified patients were confirmed by NGS. MET exon 14 skipping variant was found by RT-PCR in five samples (5.2%): four out of 57 (7%) squamous cell carcinoma samples and one out of 15 (6.6%) sarcomatoid carcinoma histology. MET exon 7–8 was found in one patient (1%) with squamous cell carcinoma. None of the MET mutations were associated with MET positive IHC. No MET alterations were found in EGFR mutant adenocarcinoma samples.
Median age at diagnosis was 71 years (63–87 years). Among the MET altered patients, three had metastatic disease at diagnosis. Six patients had resectable disease at diagnosis, they were radically treated and received adjuvant treatment according to pathological TNM stage. All of them experienced disease relapse, median time to relapsing disease was 10 months (2–38 months). Median age at diagnosis of advanced NSCLC (aNSCLC) was 73 years (64–88 years).
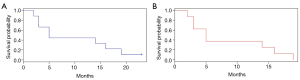
None of the patients received specific MET TKI treatment. Only five out of 9 MET altered patients (55.6%) received at least one line of platinum-based systemic treatment for aNSCLC, only the patient with MET exon 7–8 mutation received further treatment lines (Table 1). The remaining three patients died before starting first line treatment. Median overall survival (mOS) in MET altered patients was 5 months (95% CI, 3 to NA) (Figure 5). In the overall aNSCLC evaluated population (n=90), mOS was 10 months (95% CI, 8–15), with a trend of lower OS for MET positive compared to MET negative patients, but not reaching statistical significance, probably due to the small sample size (5 vs. 11 months, P=0.3). The trend was confirmed once EGFR mutated patients were excluded, with a mOS of 8 months (95% CI: 5–10) for MET negative, and 5 months (95% CI: 3 m–NA) for MET positive advanced stage sqCC and sarcomatoid patients (n=64, P=0.8).
Discussion
An increasing amount of data is available on the potential role of MET inhibitors in patients carrying exon 14 mutations, and acquired resistance mechanisms are under evaluation (19). In addition, MET acquired amplification is clearly established as an acquired resistance mechanism in EGFR mutated NSCLC treated with EGFR TKI (4).
The present manuscript depicts in vitro analyses on resistance mechanisms to MET inhibition and the potential role of new combination treatment.
Through in vitro experiments, we found that single inhibition of MET with tepotinib activates AXL, CDCP1, SHP2, and SRC in MET addicted cells. The combination of tepotinib with the SRC inhibitor, dasatinib, could partially prevent the activation of p-AXL, CDCP1, and p-SHP2 in the EBC-1 cell line. Our findings demonstrate that single MET inhibition might not be enough in MET addicted cell lines, while the combination of a MET inhibitor with dasatinib, or with a PIM inhibitor, could potentially prevent, or delay, the occurrence of treatment resistance. The role of Src was previously investigated in the setting of EGFR mutated NSCLC. In cell lines and NSCLC models showing resistance to EGFR or ALK inhibitors, MET and Src coactivation was demonstrated (24,28). The hyperactivation of RTKs and non-receptor TKs (non-RTKs) has been widely demonstrated as a mechanism of resistance to EGFR TKIs in EGFR mutation positive lung cancer (23). In primary MET addicted models, few reports are available on resistance mechanisms to MET inhibitors. In addition to MET secondary resistance mutations (29-31), MAPK and PI3K signaling have been shown to mediate resistance to MET TKIs in in vitro and in vivo models of NSCLC with MET exon 14 skipping or MET amplification (32-35), and same mechanisms were confirmed in clinical samples as well (36). Until now, no evidence has been available on the possible role of AXL, CDCP1, and Src in MET TKI resistance in primary MET altered NSCLC. Although a PIM dependent resistance mechanism to MET TKIs was described in MET amplified NSCLC models (21,34) and in EGFR mutation positive NSCLC (37), to the best of our knowledge, this is the first report to also identify a role of PIM in MET exon 14 skipping NSCLC.
Although clinical results achieved by using MET TKI monotherapy have not been fully satisfactory till now (14-16,38-40), the possibility of combining MET and Src inhibitors, or PIM inhibitors, in the clinical setting might raise some concern. Recently, a phase I trial was conducted with crizotinib plus dasatinib across multiple cancer types and the toxicity profile of the combination was worrying, with over 20% of patients experiencing dose limiting toxicities in the expansion cohort (41). PIM kinase inhibitors are being investigated in clinical trials, mainly in hematological malignancies. The development of several PIM inhibitors, including AZD1208, has been interrupted due to high toxicity rate (42).
In our study, we also included a screening in tumor samples. Overall, almost 10% of the tumor samples were positive for MET alterations. The proportion of MET mutant patients in our series is quite comparable with the previously reported data, identifying higher prevalence of MET alterations in lung squamous cell carcinoma (sqCC) and sarcomatoid histology (MET exon 14 mutations in 7% and 6.6%, respectively). In the largest series by Schrock et al., 7.7% of sarcomatoid tumors, 2.1% of squamous cell carcinoma, and 8.2% of adenosquamous carcinoma harbored MET exon 14 mutation (9). They also identified the MET mutation in 2.9% of lung adenocarcinomas (9). In our series, we did not find any MET alteration in lung adenocarcinoma patients with baseline EGFR mutations. This finding, although based on a small number of cases, may suggest that MET alterations are more likely to occur as a mechanism of acquired resistance to EGFR TKIs, rather than being present at baseline as a co-mutation driving primary resistance. Additionally, we have identified for the first time the presence of the MET exon 7–8 skipping mutation in a lung squamous cell carcinoma patient. This variant was previously identified in the E98 glioblastoma cell line and xenograft models, and in glioma samples (11,43). The pathogenetic mechanism of MET exon 7–8 resulting in intracellular retention and auto-activation, together with our preliminary evidence of MET TKI activity on this alteration, make it appear as a good molecular target candidate. Therefore, much effort should be made in the future to investigate the prevalence of this novel MET alteration in lung cancer.
The main limitations of this study are related to the retrospective nature of the tumor sample analysis, since the included patients were selected according to availability of evaluable tumor material and complete medical records. Patients with non-EGFR mutant adenocarcinoma histology were excluded from our analysis and, therefore, are not represented. Because of the inclusion timeframe of our sample collection, none of the included patients were screened for MET alterations in clinical practice, nor received MET TKIs within clinical trials.
Conclusions
In conclusion, based on our preclinical results, further investigation is required on the role of RTKs, SRC, SHP2, CDCP1, and PIM as potential co-targets that may be modulated by treatment with single agent MET inhibitors.
Acknowledgments
This work was supported by La Caixa.
Funding: None.
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/tlcr-20-681
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tlcr-20-681
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tlcr-20-681). NK and RR serve as unpaid editorial board members of Translational Lung Cancer Research. AP reports personal fees AstraZeneca, Roche, Eli Lilly, BMS, MSD, Dako/Agilent, Pfizer, outside the submitted work. FM reports personal fees from AstraZeneca, Roche, Novartis, BMS, MSD, Takeda, Pfizer outside the submitted work. GP reports personal fees from Astrazeneca, BMS, Boehringer Ing, MSD, Roche, Eli Lilly & Co., outside the submitted work. VG reports personal fees from Astrazeneca, Novartis, Roche, Eli Lilly & Co., outside the submitted work. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the precepts of the Helsinki declaration (as revised in 2013) and approved by the institutional review board of Istituto Oncologico Veneto (IOV; protocol number MET-2017; date of approval: 11th December 2017). Signed informed consent, approved by the Ethics Committee of IOV, was obtained for collection, analysis and publication of data, according to the Italian data protection authority dispositions.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Ichimura E, Maeshima A, Nakajima T, et al. Expression of c-met/HGF receptor in human non-small cell lung carcinomas in vitro and in vivo and its prognostic significance. Jpn J Cancer Res 1996;87:1063-9. [Crossref] [PubMed]
- Shi P, Oh YT, Zhang G, et al. Met gene amplification and protein hyperactivation is a mechanism of resistance to both first and third generation EGFR inhibitors in lung cancer treatment. Cancer Lett 2016;380:494-504. [Crossref] [PubMed]
- Chabon JJ, Simmons AD, Lovejoy AF, et al. Circulating tumour DNA profiling reveals heterogeneity of EGFR inhibitor resistance mechanisms in lung cancer patients. Nat Commun 2016;7:11815. [Crossref] [PubMed]
- Sequist LV, Han JY, Ahn MJ, et al. Osimertinib plus savolitinib in patients with EGFR mutation-positive, MET-amplified, non-small-cell lung cancer after progression on EGFR tyrosine kinase inhibitors: interim results from a multicentre, open-label, phase 1b study. Lancet Oncol 2020;21:373-86. [Crossref] [PubMed]
- Drilon A, Cappuzzo F, Ou SI, et al. Targeting MET in Lung Cancer: Will Expectations Finally Be MET? J Thorac Oncol 2017;12:15-26. [Crossref] [PubMed]
- Cappuzzo F, Marchetti A, Skokan M, et al. Increased MET gene copy number negatively affects survival of surgically resected non-small-cell lung cancer patients. J Clin Oncol 2009;27:1667-74. [Crossref] [PubMed]
- Camidge DR, Ou SHI, Shapiro G, et al. Efficacy and safety of crizotinib in patients with advanced c-MET-amplified non-small cell lung cancer (NSCLC). J Clin Oncol 2014;32:8001. [Crossref]
- Van Der Steen N, Giovannetti E, Pauwels P, et al. cMET Exon 14 Skipping: From the Structure to the Clinic. J Thorac Oncol 2016;11:1423-32. [Crossref] [PubMed]
- Schrock AB, Frampton GM, Suh J, et al. Characterization of 298 Patients with Lung Cancer Harboring MET Exon 14 Skipping Alterations. J Thorac Oncol 2016;11:1493-502. [Crossref] [PubMed]
- Liu X, Jia Y, Stoopler MB, et al. Next-Generation Sequencing of Pulmonary Sarcomatoid Carcinoma Reveals High Frequency of Actionable MET Gene Mutations. J Clin Oncol 2016;34:794-802. [Crossref] [PubMed]
- Navis AC, van Lith SA, van Duijnhoven SM, et al. Identification of a novel MET mutation in high-grade glioma resulting in an auto-active intracellular protein. Acta Neuropathol 2015;130:131-44. [Crossref] [PubMed]
- Camidge DR, Otterson GA, Clark JW, et al. Crizotinib in patients (pts) with MET-amplified non-small cell lung cancer (NSCLC): Updated safety and efficacy findings from a phase 1 trial. J Clin Oncol 2018;36:9062. [Crossref]
- Wang SXY, Zhang BM, Wakelee HA, et al. Case series of MET exon 14 skipping mutation-positive non-small-cell lung cancers with response to crizotinib and cabozantinib. Anticancer Drugs 2019;30:537-41. [Crossref] [PubMed]
- Drilon A, Clark J, Weiss J, et al. OA12.02 Updated Antitumor Activity of Crizotinib in Patients with MET Exon 14-Altered Advanced Non-Small Cell Lung Cancer. J Thorac Oncol 2018;13:S348. [Crossref]
- Landi L, Chiari R, Tiseo M, et al. Crizotinib in MET-Deregulated or ROS1-Rearranged Pretreated Non-Small Cell Lung Cancer (METROS): A Phase II, Prospective, Multicenter, Two-Arms Trial. Clin Cancer Res 2019;25:7312-9. [Crossref] [PubMed]
- Drilon A, Clark JW, Weiss J, et al. Antitumor activity of crizotinib in lung cancers harboring a MET exon 14 alteration. Nat Med 2020;26:47-51. [Crossref] [PubMed]
- Paik PK, Veillon R, Cortot AB, et al. Phase II study of tepotinib in NSCLC patients with METex14 mutations. J Clin Oncol 2019;37:9005. [Crossref]
- Wolf J, Seto T, Han JY, et al. Capmatinib (INC280) in METΔex14-mutated advanced non-small cell lung cancer (NSCLC): Efficacy data from the phase II GEOMETRY mono-1 study. J Clin Oncol 2019;37:9004. [Crossref]
- Fujino T, Kobayashi Y, Suda K, et al. Sensitivity and Resistance of MET Exon 14 Mutations in Lung Cancer to Eight MET Tyrosine Kinase Inhibitors In Vitro. J Thorac Oncol 2019;14:1753-65. [Crossref] [PubMed]
- Camidge DR, Janku F, Martinez-Bueno A, et al. Safety and preliminary clinical activity of the MET antibody mixture, Sym015 in advanced non-small cell lung cancer (NSCLC) patients with MET amplification/exon 14 deletion (METAmp/Ex14∆). J Clin Oncol 2020;38:9510. [Crossref]
- An N, Xiong Y, LaRue AC, et al. Activation of Pim Kinases Is Sufficient to Promote Resistance to MET Small-Molecule Inhibitors. Cancer Res 2015;75:5318-28. [Crossref] [PubMed]
- Cen B, Mahajan S, Wang W, et al. Elevation of receptor tyrosine kinases by small molecule AKT inhibitors in prostate cancer is mediated by Pim-1. Cancer Res 2013;73:3402-11. [Crossref] [PubMed]
- Karachaliou N, Chaib I, Cardona AF, et al. Common Co-activation of AXL and CDCP1 in EGFR-mutation-positive Non-smallcell Lung Cancer Associated With Poor Prognosis. EBioMedicine 2018;29:112-27. [Crossref] [PubMed]
- Yoshida T, Okamoto I, Okamoto W, et al. Effects of Src inhibitors on cell growth and epidermal growth factor receptor and MET signaling in gefitinib-resistant non-small cell lung cancer cells with acquired MET amplification. Cancer Sci 2010;101:167-72. [Crossref] [PubMed]
- Chaib I, Karachaliou N, Pilotto S, et al. Co-activation of STAT3 and YES-Associated Protein 1 (YAP1) Pathway in EGFR-Mutant NSCLC. J Natl Cancer Inst 2017.109. [Crossref] [PubMed]
- Chou TC. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res 2010;70:440-6. [Crossref] [PubMed]
- Narayan RS, Fedrigo CA, Brands E, et al. The allosteric AKT inhibitor MK2206 shows a synergistic interaction with chemotherapy and radiotherapy in glioblastoma spheroid cultures. BMC Cancer 2017;17:204. [Crossref] [PubMed]
- Tsuji T, Ozasa H, Aoki W, et al. Alectinib Resistance in ALK-Rearranged Lung Cancer by Dual Salvage Signaling in a Clinically Paired Resistance Model. Mol Cancer Res 2019;17:212-24. [Crossref] [PubMed]
- Ou SI, Young L, Schrock AB, et al. Emergence of Preexisting MET Y1230C Mutation as a Resistance Mechanism to Crizotinib in NSCLC with MET Exon 14 Skipping. J Thorac Oncol 2017;12:137-40. [Crossref] [PubMed]
- Bahcall M, Sim T, Paweletz CP, et al. Acquired METD1228V Mutation and Resistance to MET Inhibition in Lung Cancer. Cancer Discov 2016;6:1334-41. [Crossref] [PubMed]
- Li A, Yang JJ, Zhang XC, et al. Acquired MET Y1248H and D1246N Mutations Mediate Resistance to MET Inhibitors in Non-Small Cell Lung Cancer. Clin Cancer Res 2017;23:4929-37. [Crossref] [PubMed]
- Kim S, Kim TM, Kim DW, et al. Acquired Resistance of MET-Amplified Non-small Cell Lung Cancer Cells to the MET Inhibitor Capmatinib. Cancer Res Treat 2019;51:951-62. [Crossref] [PubMed]
- Bahcall M, Awad MM, Sholl LM, et al. Amplification of Wild-type KRAS Imparts Resistance to Crizotinib in MET Exon 14 Mutant Non-Small Cell Lung Cancer. Clin Cancer Res 2018;24:5963-76. [Crossref] [PubMed]
- Henry RE, Barry ER, Castriotta L, et al. Acquired savolitinib resistance in non-small cell lung cancer arises via multiple mechanisms that converge on MET-independent mTOR and MYC activation. Oncotarget 2016;7:57651-70. [Crossref] [PubMed]
- Gimenez-Xavier P, Pros E, Bonastre E, et al. Genomic and Molecular Screenings Identify Different Mechanisms for Acquired Resistance to MET Inhibitors in Lung Cancer Cells. Mol Cancer Ther 2017;16:1366-76. [Crossref] [PubMed]
- Awad MM, Lee JK, Madison R, et al. Characterization of 1,387 NSCLCs with MET exon 14 (METex14) skipping alterations (SA) and potential acquired resistance (AR) mechanisms. J Clin Oncol 2020;38:9511. [Crossref]
- Bracht JWP, Karachaliou N, Berenguer J, et al. PIM-1 inhibition with AZD1208 to prevent osimertinib-induced resistance in EGFR-mutation positive non-small cell lung cancer. J Cancer Metastasis Treat 2019;5:22. [Crossref]
- Park K, Felip E, Veillon R, et al. Tepotinib in NSCLC patients harboring METex14 skipping: Cohort A of phase II VISION study. Ann Oncol 2019;30:ix22-3. [Crossref]
- Heist RS, Wolf J, Seto T, et al. OA01.07 Capmatinib (INC280) in METΔEX14-Mutated Advanced NSCLC: Efficacy Data from the Phase 2 Geometry MONO-1 Study. J Thorac Oncol 2019;14:S1126. [Crossref]
- Wolf J, Overbeck TR, Han JY, et al. Capmatinib in patients with high-level MET-amplified advanced non–small cell lung cancer (NSCLC): results from the phase 2 GEOMETRY mono-1 study. J Clin Oncol 2020;38:9509. [Crossref]
- Kato S, Jardim DL, Johnson FM, et al. Phase I study of the combination of crizotinib (as a MET inhibitor) and dasatinib (as a c-SRC inhibitor) in patients with advanced cancer. Invest New Drugs 2018;36:416-23. [Crossref] [PubMed]
- Luszczak S, Kumar C, Sathyadevan VK, et al. PIM kinase inhibition: co-targeted therapeutic approaches in prostate cancer. Signal Transduct Target Ther 2020;5:7. [Crossref] [PubMed]
- van den Heuvel C, Das AI, de Bitter T, et al. Quantification and localization of oncogenic receptor tyrosine kinase variant transcripts using molecular inversion probes. Sci Rep 2018;8:7072. [Crossref] [PubMed]


