
Enumeration and molecular characterization of circulating tumor cells enriched by microcavity array from stage III non-small cell lung cancer patients
Introduction
Circulating tumor cells (CTCs) are rare cells shed by solid tumors into the peripheral blood. Accurate quantification of CTCs may enable minimally invasive monitoring of disease status in a patient. Many approaches have been developed to capture and characterize CTCs in the blood. Currently, there are 2 general approaches to CTC enrichment and identification, exemplified by 2 of the longest-approved technologies: CELLSEARCH (Menarini Silicon Biosystems, Bologna, Italy) and AdnaTest (Qiagen, Hilden, Germany). The CELLSEARCH system is the first and only clinically validated U.S. Food and Drug Administration (FDA)-cleared blood test for enumerating CTCs in patients with metastatic breast, colorectal, or prostate cancer (1). This system is predictive of progression-free survival (PFS) and overall survival (OS) in patients with metastatic breast (2), colorectal (3), or prostate cancer (4). However, this method only captures CTCs of epithelial phenotype [leukocyte common antigen (CD45)−, EPCAM+, and cytokeratin 8+, cytokeratin 18+, and/or cytokeratin 19+]. The AdnaTest captures tumor cells using magnetic beads coated with a cocktail of antibodies against multiple cell surface receptors (5). In contrast to CELLSEARCH, mRNA is then extracted from the enriched cell populations and subjected to polymerase chain reaction (PCR) to detect gene transcripts for epithelial tumor cells, such as EPCAM, and tumor cells undergoing epithelial-to-mesenchymal transition (EMT), such as TWIST1 and ALDH1A1 (6). Therefore, AdnaTest cannot provide enumeration, and CELLSEARCH is not validated for molecular analyses. Integration of these 2 approaches into a single platform would allow both enumeration and downstream molecular characterization of enriched CTCs.
The Micro Cavity Array (MCA) system (Hitachi Chemical Co., Tokyo, Japan) is an integrated microfluidic device that enriches tumor cells on the basis of differences in size and membrane deformability between tumor cells and cells of hematopoietic origin (7-11). Owing to the agnostic nature of enrichment that is not reliant on cell capture based on surface protein expression, the MCA system has the potential to capture more CTCs than the EPCAM-based CELLSEARCH system (7) and may be able to provide both enumeration and molecular analysis of the captured cells.
Herein, we used a preclinical tumor cell spike-in model with healthy donor blood (HDB) to evaluate the capture efficiency of the MCA system. We then validated the MCA technology for CTC enumeration and downstream targeted gene expression profiling in patients with stage III non-small cell lung cancer (NSCLC) and demonstrated the ability of this technology to predict patient outcomes. We present the following article in accordance with the MDAR reporting checklist (available at
Methods
Overview of MCA system
A succinct overview of the MCA system (Hitachi Chemical Co., Ltd., Ibaraki, Japan) was recently provided by Yagi and colleagues (9). The MCA is a semi-automated filtration method with metal filters coated in nickel and gold. The filters consist of 10,000 fabricated 8 µm × 100 µm rectangular pores that have been optimized to trap tumor cells while permitting blood cells to flow through the microcavities during whole-blood filtration. Protocols for priming, washing, fixing, permeabilization, and staining are all automated with reagents added to cells captured in the filtration cartridge in situ.
Cell culture
Human lung cancer cell lines H358 and A549 were cultured in RPMI-1640 medium containing 10% fetal bovine serum and 10% penicillin-streptomycin in an incubator at 37 °C with a 5% CO2 atmosphere. Cells were harvested with 0.25% trypsin-EDTA when they reached 70% confluence, and cell viability was determined by trypan blue exclusion.
Tumor cell spiking by micromanipulation
A micromanipulation system (TransferMan 4r, Eppendorf, Hamburg, Germany) mounted on an inverted phase contrast microscope (Nikon, Tokyo, Japan) was used to obtain a known number of tumor cells (0, 10, 50, or 100 cells from each cell line), which were then deposited onto a 20-µL drop of tissue culture medium on the flat surface of a tissue culture dish. The culture medium containing a known number of tumor cells was transferred to 9.5 mL of normal donor blood. A sham spike condition was included as a control.
Study cohorts
This study was conducted at The University of Texas MD Anderson Cancer Center, was approved by the Institutional Review Board as Lab09-0307, and conforms to the provisions of the Declaration of Helsinki as revised in 2013. Study outcomes did not affect the clinical management of the enrolled patients. During November 2014 through October 2017, stage III NSCLC patients undergoing comprehensive treatment at MD Anderson were prospectively recruited for blood collection, and written informed consent from the recruits was obtained (Table 1). Peripheral blood was collected at the start of chemoradiation therapy. Since the study period preceded the FDA approval of durvalumab, no patients received durvalumab after chemoradiation therapy. Sex-matched individuals self-declared as cancer-free were recruited as healthy blood donors. Written informed consent was obtained from all healthy donors according to an Institutional Review Board-approved protocol (PA14-0063). HDB was collected in BD Vacutainer EDTA tubes (BD Biosciences, San Jose, CA).
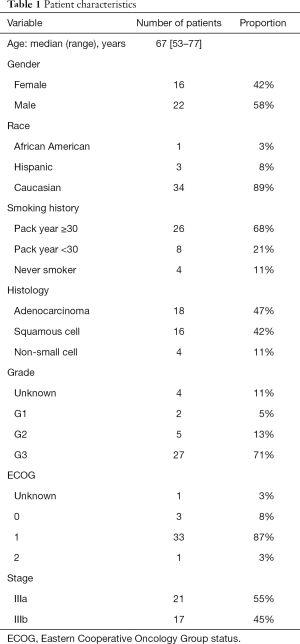
Full table
CTC capture
Enrichment procedures for enumeration and molecular characterization were performed in parallel on separate aliquots of the same blood sample. For HDB, contrived samples comprising known quantities of tumor cells in HDB, and NSCLC patient samples, ~9.5 mL of blood was loaded onto the MCA system for each aliquot. A flow rate of 200 µL/min was maintained with negative pressure through the MCA cartridge to capture tumor cells.
In situ staining and enumeration of CTCs
For imaging, the automated staining protocol of the MCA system sequentially applied BD FACS Lyse solution (BD Biosciences), BD FACS Permeabilizing Solution 2 (BD Biosciences), and an antibody cocktail, with intermittent and final automated washing steps. The antibody cocktail consisted of anti-pan-CK (AE1/AE3) conjugated with Alexa Fluor 488 (Thermo Fisher Scientific, Waltham, MA); anti-pan-CK (CK3-6H5) conjugated to fluorescein isothiocyanate (Miltenyi Biotec, Bergisch Gladbach, Germany); anti-CD45 antibody conjugated with Alexa Fluor 594 (BioLegend, San Diego, CA); and 4',6-diamidino-2-phenylindole (DAPI). The MCA cartridges containing captured tumor cells were imaged using an Olympus IX81-DSU system (Olympus, Tokyo, Japan; The Flow Cytometry and Cellular Imaging Core Facility at MD Anderson NCI Cancer Center Support Grant P30CA16672). Captured 10× images, along with bright-field morphology, were used to identify CTCs, defined as nucleated cells (DAPI+) that were CK+ and CD45−.
Leukocyte depletion and CTC capture for downstream molecular studies
To diminish the leukocyte background for samples subjected to downstream molecular studies, it was necessary to significantly deplete leukocytes from peripheral blood samples. To generate this leukocyte-poor fraction, we incubated peripheral blood samples with StraightFrom Whole Blood CD45 MicroBeads at 50 µL per mL of blood passed through a magnetic column (Miltenyi Biotec) before enrichment by the MCA system.
Following cell capture, samples were cleared of residual red blood cells using BD FACS Lyse solution (BD Biosciences). Enriched cells were harvested by applying a phenol/guanidine thiocyanate solution (QIAzol, Qiagen/Thermo Fisher Scientific, Inc.) to the open face of the capture filter to lyse the captured cells, and the lysate was archived at −80 °C for subsequent batch processing. RNA was extracted in batches with chloroform extraction and column enrichment (RNeasy mini, Qiagen). cDNA was prepared using random primers (ABI High-Capacity cDNA Reverse Transcription kit, Thermo Fisher Scientific).
Relative gene expression of captured CTCs
Gene expression profiling was performed by quantitative reverse transcription PCR (qRT-PCR) using customized Bio-Rad SYBR Green PrimePCR plates according to the manufacturer’s instructions (Bio-Rad Laboratories, Hercules, CA). In short, primers were designed following the principles established via the Minimum Information for Publication of Quantitative Real-Time PCR Experiments (MIQE) guidelines (12) and wet lab-validated by the manufacturer. Each PCR array plate was designed to accommodate 6 samples in triplicate and contained primers for 16 genes of interest and 3 housekeeping genes as well as quality control wells for RNA quality, reverse transcription quality, PCR quality, and DNA contamination for each sample. Cq analysis was performed by Bio-Rad CFX Manager software with a constant baseline adjustment of the relative fluorescent units for all array runs to allow accurate comparisons across samples. Data were normalized against the TBP, GAPDH, and HPRT housekeeping genes using the ΔΔCq method. Samples were considered positive based on the distribution of leukocyte-depleted enriched HDB samples (n=18) after removing HDB outliers with the ROUT method in GraphPad Prism Version 7 (GraphPad, San Diego, CA). A cut-off for positive expression was established at 2.5 standard deviations above the HDB median.
The panel of genes investigated included genes related to epithelial characteristics (EPCAM, HER2, EGFR), EMT (TWIST1, SNAI1), stem-like tumor cells (ALDH1A1, CD44), and signaling pathways commonly perturbed in NSCLC [ALK, BCL2, CD274 (PD-L1), FGFR1, KRAS, MET, RAD50, TERT and TP53]. PTPRC (CD45) was also included as a white blood cell control.
Statistical analysis
For time-to-event variables, the Kaplan-Meier method was used to estimate the survival functions. OS was defined as the time from the initiation of chemoradiation till death or most recent contact date. PFS was defined as the time from the initiation of chemoradiation until progression, death, or last follow-up. The log-rank test was used to compare survival times between groups. The Cox proportional hazards regression model was used to evaluate the effects of potential prognostic factors on OS or PFS, P values of <0.05 were considered statistically significant. Variables included in the multivariate survival models were selected using a backward model selection algorithm where CTC count (as a continuous variable) and BCL2 expression (considered positive if normalized expression was above the range of HDB) were pre-determined to be included in the final model. Other candidate variables (chosen on the basis of either the significance of their association with survival on univariate analysis or their clinical importance) were assessed in the initial model before selection. Owing to the limited sample size, covariates with P<0.4 were maintained in the model. S-PLUS software (TIBCO Software Inc., Palo Alto, CA), SAS 9.4 software (SAS Institute, Cary, NC), and R version 3.5 (R Foundation for Statistical Computing, Vienna, Austria) were used for statistical analysis.
Results
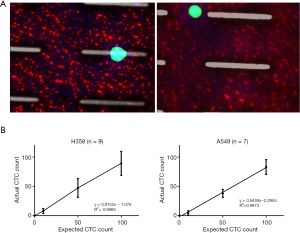
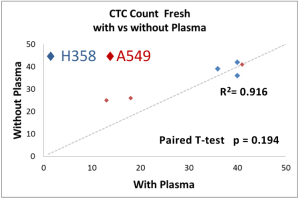
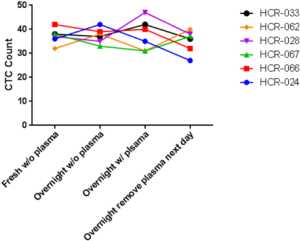
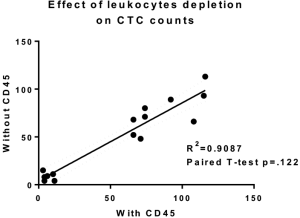
The performance of CTC enrichment by the MCA system was validated using precise numbers of cells from human lung cancer cell lines spiked into HDB as a model. This validation showed that the MCA cartridge is amenable to direct in situ imaging of captured cells. Using commercially available fluorophore-conjugated antibodies, CTCs defined as pan-CK+/DAPI+/CD45− cells in Figure 1A) were identified against a background of CD45+/DAPI+/pan-CK− leukocytes (Figure 1A). Plasma depletion before enrichment had minimal effects on CTC recovery (Figure S1). Likewise, storage of peripheral blood overnight with and without plasma did not make a significant difference in the recovery of CTCs (Figure S2). CTC recovery levels were also similar with and without prior leukocyte depletion (Figure S3), which greatly enhanced CTC purity (Figure S4) before molecular characterization. The CTC yield of the MCA system was assessed in HDB (n=13) spiked with 10, 50, or 100 individual cells from 2 lung cancer cell lines (H358 and A549). The recovery rates were 91.5% and 84.3% for the H358 and A549 cell lines, with a linearity of R2 of 0.9969 and 0.9973, respectively (Figure 1B).
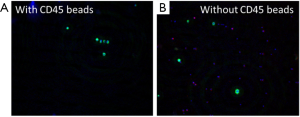
CTC enumeration in stage III NSCLC patients by manual counting
Samples from 38 patients with NSCLC were analyzed for CTCs after enrichment by MCA. CTC counts ranged from 0 to 33 CTCs per ~9.5-mL sample (Figure 2A). At least 1 CTC was detected in 30 of the 38 samples (79%), and the median was 2 CTCs (Figure 2B). There was no significant difference in CTC count by sex, stage, histology, or smoking status (data not shown).
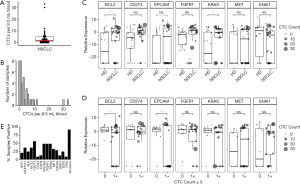
Gene expression profile in stage III NSCLC patients by qRT-PCR
Following CTC enrichment by MCA, samples from 25 NSCLC patients that underwent molecular characterization had significantly higher expression of BCL2 (Student’s t-test P=0.0240) and KRAS (P=0.0398) compared with samples from HDB (Figure 2C). Furthermore, samples with CTCs detectable by imaging had higher expression of BCL2 and EPCAM (P=0.046 and P=0.019, respectively, Figure 2D). Additional genes related to survival as described below are also presented in Figure 2. HDB similarly processed by MCA after leukocyte depletion (n=18) was used to establish cut-offs for positive gene expression of CTC-related genes as described in the Methods. Roughly half (52%) of the NSCLC patients’ samples were positive for the epithelial marker EPCAM. In contrast, 23 of 25 patients (92%) were positive for at least 1 gene from the full panel of 16 genes (Figure 2E). Furthermore, 11 patients with at least 1 CTC by imaging and 4 patients with at least 5 CTCs by imaging were nonetheless negative by the measure of EPCAM expression.
CTC count associated with survival
The 38 patients with NSCLC were followed for a median of 43.1 months. Median PFS was 16.4 months and median (Figure 3A,B,C) overall survival was 38.7 months (Figure 3D,E,F). In these baseline blood draws, there was no significant difference in CTC counts between patients whose tumors progressed during the study (Figure 3A) and those who died of their disease (Figure 3D). In receiver operating characteristic curve analysis (ROC) of CTC enumeration, the areas under the curve for PFS and OS were 0.52 and 0.53, respectively. However, all 7 patients with at least 7 CTCs experienced progression (Figure 3B) and eventually died (Figure 3E). When we applied a cut-off of ≥7 CTCs per sample, the baseline CTC count was able to predict OS with 100% specificity and positive predictive value, but with a sensitivity of 0.350 and negative predictive value of 0.581. For PFS, the sensitivity of this cut-off was 0.269, and the negative predictive value was 0.387. Patients with less than 7 CTCs had a significantly longer PFS compared with patients with 7 or more CTCs (median 24.7 vs. 3.8 months, P=0.0006, HR =4.24, 95% CI, 1.73–10.40, Figure 3C) and longer OS (median time not reached vs. 18.2 months, P<0.0001, HR =8.17, 95% CI, 2.87–23.26, Figure 3F). CTC count remained a significant prognostic factor for both PFS and OS in multivariate analysis that included standard staging (Figure S5).
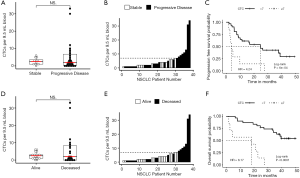
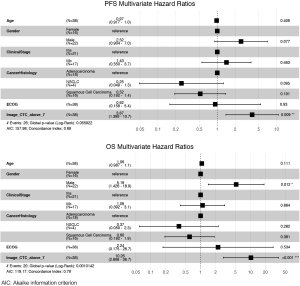
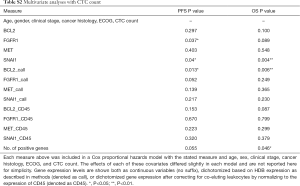
Gene expression predicts survival
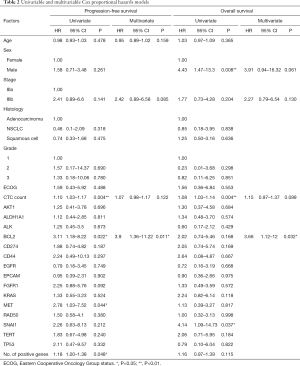
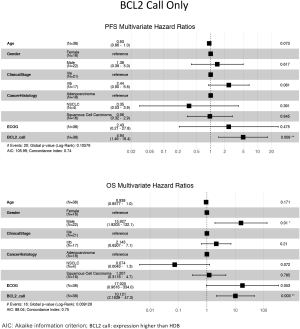
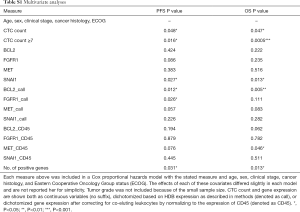
The ability of the positively expressed CTC-related genes to predict PFS and OS in 25 NSCLC patients is reported in Table 2. Positive expression of BCL2, positive expression of MET, and the total number of positive genes were each significantly associated with an increased risk of progression in the univariate Cox proportional hazards model. Similarly, positive expression of BLC2 and positive expression of SNAI1 were significantly associated with an increased risk of death. BCL2 remained a significant prognostic factor for both PFS and OS in multivariate analysis that included standard clinical variables in the initial model and required CTC count in the final model (Table 2), as well as in an unselected model that did not include CTC count (Figure S6). Additionally, FGFR1, MET, SNAI1, and the number of positive genes remained significant predictors in the unselected Cox proportional hazard models for PFS or OS (Tables S1 and S2).
Full table
Full table
Full table
Discussion
In this study, the Hitachi Chemical MCA system was evaluated for enrichment of CTCs from peripheral blood of patients with NSCLC. This report is the first empirical demonstration that blood samples enriched for CTCs by the MCA system are amenable to molecular characterization, specifically of gene expression by PCR. Furthermore, the gene expression profiles of these enriched CTCs are clinically relevant.
One of the challenges of enriching CTCs from peripheral blood is the rarity of CTCs among blood cellular elements. It has been suggested that for every CTC, there are 108 blood cellular elements including leukocytes and erythrocytes, as well as platelets (13). Depletion of leukocytes that co-elute with cancer cells in CTC enrichment systems is therefore crucial in improving the efficiency of these systems for molecular analysis (14). Although leukocyte contamination of CTCs may not affect enumeration, it still may play a significant role in downstream molecular analysis. Here, we have shown that leukocyte depletion with magnetic beads coated with anti-CD45 antibody before CTC enrichment can increase CTC purity with minimal effects on CTC recovery.
Removal of plasma prior to loading blood unto the MCA also did not affect the capture efficiency in contrived samples or, by extension, of the CTCs in patient samples. Since plasma is a critical component of liquid biopsy that may provide valuable clinical information, the extraction of plasma before processing blood through the MCA system would be a judicious use of patient samples and provide an excellent resource for additional interrogations.
Our basic development data showing high rates of CTC recovery demonstrated the MCA to be a stable and reliable system for conducting both translational and clinical research with patient samples processed either fresh or after overnight storage. This observation that the CTC enumeration is minimally affected by overnight storage of blood could be beneficial in clinical settings where samples may not be processed immediately.
Using the conditions established in our validation studies, we processed samples from NSCLC patients for CTC enumeration and molecular gene expression. For better statistical power, no CTC count threshold was used in survival modeling with Cox proportional hazards; instead, the HR for each unit increase in CTC was calculated. The resulting HR was small (per CTC) but highly significant.
CTC count thresholds for cancer diagnosis using the CELLSEARCH system have been well-validated for metastatic breast cancer (2) and prostate cancer (4) (for each, 5 CTCs/7.5 mL of blood) and colorectal cancer (3 CTCs/7.5 mL of blood) (3). Although several CTC cut-offs have been proposed for NSCLC, including 1 CTC (15), 2 CTCs (16,17), and 5 CTCs (17-19), none have gained the clinical fortitude of the cut-offs for the aforementioned neoplasms. Using the MCA system, Ichimura and colleagues (20) recently showed in a subset of 12 non-surgical lung cancer patients that a follow-up count of 3 or more CTCs in 3 mL of blood predicted poor survival. This result is similar to the poor OS and PFS predicted by 7 CTCs in 9.5 mL of blood that we observed here; however, Ichimura et al. observed no correlation between CTC count and survival in the larger cohort of 38 patients with stage I, II, III and IV lung cancer (20). Here, we found that a cut-off of 7 CTCs per 9.5 mL of blood had 100% prognostic specificity in this training set. We also observed that CTC count was independent of clinical stage and histology in NSCLC, along the lines of some previous reports (15,20). As the small sample size precludes drawing any generalizable conclusions, further investigation is warranted.
The agnostic nature of the CTC enrichment process using the MCA system can add tremendous value to the final product and, consequently, the feasibility of clinical applications. Surface protein expression of the epithelial cell adhesion molecule EPCAM is traditionally used to capture CTCs as well as to differentiate between CTCs and contaminating leukocytes (1). Here, we found only 48% of NSCLC patients have elevated EPCAM mRNA. Furthermore, using image analysis, we found a large portion of the samples containing CTCs, identified by CK+CD45− status, to have low levels of EPCAM mRNA, within the range of expression by healthy donors. As MCA captures CTC independent of EPCAM, it is not unexpected that the captured cells may or may not express EPCAM. Although EPCAM can both promote and inhibit metastasis, EMT processes can downregulate EPCAM as tumor cells disaggregate and disseminate (21), and inflammation may play a role in downregulating EPCAM expression (22). Therefore, relying on EPCAM for enrichment could potentially underestimate the number of CTC in patients (6,23,24). However, since the prognostic value of the EPCAM negative CTCs may be limited (25), better-powered studies are warranted.
Expression of BCL2 and KRAS transcripts was elevated in MCA-enriched NSCLC blood samples compared with those of healthy donors. In NSCLC samples with at least 1 CTC, BCL2 and EPCAM transcripts were expressed at higher levels. BCL2 is a critical inhibitor of apoptosis and is overexpressed in NSCLC (26). Furthermore, in contrast to metareviews of primary tumors (27-29), we found that high BCL2 expression in CTCs of NSCLC patients was correlated with poor survival and was an independent prognostic factor. This result is similar to those of other studies of BCL2+ CTCs in lung cancer patients (30) but contrasts studies in metastatic breast cancer where high BCL2 correlated with better survival (31).
We were also able to detect PD-L1 (CD274) transcripts in enriched CTCs. As studies have shown that patients may show a response to anti-PD-L1 therapy even when the protein is not detected in tissue samples (32), an additional measure that can be evaluated serially may be beneficial. The patients in this study were enrolled before immunotherapy was established as a standard of care, but it is encouraging that the MCA system may be able to track PD-L1 through easily accessible liquid biopsy for patients undergoing immune checkpoint inhibitor therapy.
To our knowledge, this is the first report that MCA-enriched CTCs are amenable to molecular characterization. Here, we demonstrate that these samples are not only conducive to gene expression analysis but also yield data that correlate well with clinical outcomes.
Analysis of CTC-specific biomarkers has the potential to stratify patients into prognostic groups. This approach may also open avenues for finding effective targets for new chemotherapeutic approaches. Importantly, biomarker testing in CTCs offers the key advantage of conducting these tests using peripheral blood, thereby avoiding the need for repeated invasive procedures to obtain primary tissue. The challenges that have precluded the widespread use of CTCs in in vitro diagnostic applications are thus being gradually addressed with recent advances in technology such as the Hitachi Chemical MCA system.
Acknowledgments
We wish to acknowledge Sanda Tin and Lin Han for obtaining healthy donor blood and processing samples, respectively and Jared Burks for microscopy support. This work was supported in part by the National Institutes of Health through MD Anderson’s Cancer Center Support Grant CA016672, the NCI’s Research Specialist 1 R50 CA243707-01A1, and by research support from Hitachi Chemical Co., Japan. The manuscript was edited by Sarah Bronson, ELS, Research Medical Library, MD Anderson.
Footnote
Provenance and Peer Review: This article was commissioned by the editorial office, Translational Lung Cancer Research for the series “New era of treatment for unresectable locally advanced non-small cell lung cancer”. The article has undergone external peer review.
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/tlcr-20-841
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tlcr-20-841
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tlcr-20-841). The series “New era of treatment for unresectable locally advanced non-small cell lung cancer” was commissioned by the editorial office without any funding or sponsorship. SHL served as the unpaid Guest Editor of the series and serves as an unpaid editorial board member of Translational Lung Cancer Research from Sep 2019 to Sep 2021. ENC reports grants from National Cancer Institute, grants from Hitachi Chemical Co., Japan, during the conduct of the study; grants from Angle, Plc, outside the submitted work. GJ reports grants from National Cancer Institute, grants from Hitachi Chemical Co., Japan, during the conduct of the study; grants from Angle, Plc, outside the submitted work. HG reports grants from National Cancer Institute, grants from Hitachi Chemical Co., Japan, during the conduct of the study; grants from Angle, Plc, outside the submitted work. WQ reports grants from National Cancer Institute, grants from Hitachi Chemical Co., Japan, during the conduct of the study; grants from Angle, Plc, outside the submitted work. SL reports grants from National Cancer Institute, grants from Hitachi Chemical Co., Japan, during the conduct of the study. JH reports grants from National Cancer Institute, grants from Hitachi Chemical Co., Japan, during the conduct of the study. YQ reports grants from National Cancer Institute, grants from Hitachi Chemical Co., Japan, during the conduct of the study. LY reports grants from National Cancer Institute, grants from Hitachi Chemical Co., Japan, during the conduct of the study. SHL reports grants from National Cancer Institute, grants from Hitachi Chemical Co., Japan, during the conduct of the study; grants from BeyondSpring Pharmaceuticals Inc., personal fees from AstraZeneca, personal fees from Varian Medical Systems, outside the submitted work. JMR reports grants from National Cancer Institute, grants from Hitachi Chemical Co., Japan, during the conduct of the study; grants and personal fees from Angle, Plc, outside the submitted work. JMR serves on the Scientific Advisory Board for Angle, Plc.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was conducted in compliance with the Declaration of Helsinki (as revised in 2013) and its subsequent amendments and was approved by the Institutional Review Board of the University of Texas MD Anderson Cancer Center (Lab09-0307) and informed consent was taken from all individual participants.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Allard WJ, Matera J, Miller MC, et al. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin Cancer Res 2004;10:6897-904. [Crossref] [PubMed]
- Cristofanilli M, Budd GT, Ellis MJ, et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med 2004;351:781-91. [Crossref] [PubMed]
- Cohen SJ, Punt CJ, Iannotti N, et al. Relationship of circulating tumor cells to tumor response, progression-free survival, and overall survival in patients with metastatic colorectal cancer. J Clin Oncol 2008;26:3213-21. [Crossref] [PubMed]
- de Bono JS, Scher HI, Montgomery RB, et al. Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin Cancer Res 2008;14:6302-9. [Crossref] [PubMed]
- Zieglschmid V, Hollmann C, Gutierrez B, et al. Combination of immunomagnetic enrichment with multiplex RT-PCR analysis for the detection of disseminated tumor cells. Anticancer Res 2005;25:1803-10. [PubMed]
- Mego M, Mani SA, Lee BN, et al. Expression of epithelial-mesenchymal transition-inducing transcription factors in primary breast cancer: The effect of neoadjuvant therapy. Int J Cancer 2012;130:808-16. [Crossref] [PubMed]
- Hosokawa M, Kenmotsu H, Koh Y, et al. Size-based isolation of circulating tumor cells in lung cancer patients using a microcavity array system. PLoS One 2013;8:e67466. [Crossref] [PubMed]
- Negishi R, Hosokawa M, Nakamura S, et al. Development of the automated circulating tumor cell recovery system with microcavity array. Biosens Bioelectron 2015;67:438-42. [Crossref] [PubMed]
- Yagi S, Koh Y, Akamatsu H, et al. Development of an automated size-based filtration system for isolation of circulating tumor cells in lung cancer patients. PLoS One 2017;12:e0179744. [Crossref] [PubMed]
- Yoshino T, Takai K, Negishi R, et al. Rapid imaging and detection of circulating tumor cells using a wide-field fluorescence imaging system. Anal Chim Acta 2017;969:1-7. [Crossref] [PubMed]
- Yoshino T, Tanaka T, Nakamura S, et al. Evaluation of cancer cell deformability by microcavity array. Anal Biochem 2017;520:16-21. [Crossref] [PubMed]
- Bustin SA, Beaulieu JF, Huggett J, et al. MIQE precis: Practical implementation of minimum standard guidelines for fluorescence-based quantitative real-time PCR experiments. BMC Mol Biol 2010;11:74. [Crossref] [PubMed]
- Yu M, Stott S, Toner M, et al. Circulating tumor cells: approaches to isolation and characterization. J Cell Biol 2011;192:373-82. [Crossref] [PubMed]
- Guo J, Xiao B, Zhang X, et al. Combined use of positive and negative immunomagnetic isolation followed by real-time RT-PCR for detection of the circulating tumor cells in patients with colorectal cancers. J Mol Med (Berl) 2004;82:768-74. [Crossref] [PubMed]
- Hofman V, Ilie MI, Long E, et al. Detection of circulating tumor cells as a prognostic factor in patients undergoing radical surgery for non-small-cell lung carcinoma: comparison of the efficacy of the CellSearch Assay and the isolation by size of epithelial tumor cell method. Int J Cancer 2011;129:1651-60. [Crossref] [PubMed]
- Juan O, Vidal J, Gisbert R, et al. Prognostic significance of circulating tumor cells in advanced non-small cell lung cancer patients treated with docetaxel and gemcitabine. Clin Transl Oncol 2014;16:637-43. [Crossref] [PubMed]
- Lindsay CR, Blackhall FH, Carmel A, et al. EPAC-lung: pooled analysis of circulating tumour cells in advanced non-small cell lung cancer. Eur J Cancer 2019;117:60-8. [Crossref] [PubMed]
- Muinelo-Romay L, Vieito M, Abalo A, et al. Evaluation of Circulating Tumor Cells and Related Events as Prognostic Factors and Surrogate Biomarkers in Advanced NSCLC Patients Receiving First-Line Systemic Treatment. Cancers (Basel) 2014;6:153-65. [Crossref] [PubMed]
- Wei T, Zhu D, Yang Y, et al. The application of nano-enrichment in CTC detection and the clinical significance of CTCs in non-small cell lung cancer (NSCLC) treatment. PLoS One 2019;14:e0219129. [Crossref] [PubMed]
- Ichimura H, Nawa T, Yamamoto Y, et al. Detection of circulating tumor cells in patients with lung cancer using metallic micro-cavity array filter: A pilot study. Mol Clin Oncol 2020;12:278-83. [Crossref] [PubMed]
- van der Gun BT, Melchers LJ, Ruiters MH, et al. EpCAM in carcinogenesis: the good, the bad or the ugly. Carcinogenesis 2010;31:1913-21. [Crossref] [PubMed]
- Cohen EN, Gao H, Anfossi S, et al. Inflammation Mediated Metastasis: Immune Induced Epithelial-To-Mesenchymal Transition in Inflammatory Breast Cancer Cells. PLoS One 2015;10:e0132710. [Crossref] [PubMed]
- Gorges TM, Tinhofer I, Drosch M, et al. Circulating tumour cells escape from EpCAM-based detection due to epithelial-to-mesenchymal transition. BMC Cancer 2012;12:178. [Crossref] [PubMed]
- Hyun KA, Koo GB, Han H, et al. Epithelial-to-mesenchymal transition leads to loss of EpCAM and different physical properties in circulating tumor cells from metastatic breast cancer. Oncotarget 2016;7:24677-87. [Crossref] [PubMed]
- de Wit S, van Dalum G, Lenferink AT, et al. The detection of EpCAM(+) and EpCAM(-) circulating tumor cells. Sci Rep 2015;5:12270. [Crossref] [PubMed]
- Sartorius UA, Krammer PH. Upregulation of Bcl-2 is involved in the mediation of chemotherapy resistance in human small cell lung cancer cell lines. Int J Cancer 2002;97:584-92. [Crossref] [PubMed]
- Martin B, Paesmans M, Berghmans T, et al. Role of Bcl-2 as a prognostic factor for survival in lung cancer: a systematic review of the literature with meta-analysis. Br J Cancer 2003;89:55-64. [Crossref] [PubMed]
- Zhao XD, He YY, Gao J, et al. High expression of Bcl-2 protein predicts favorable outcome in non-small cell lung cancer: evidence from a systematic review and meta-analysis. Asian Pac J Cancer Prev 2014;15:8861-9. [Crossref] [PubMed]
- Zhang J, Wang S, Wang L, et al. Prognostic value of Bcl-2 expression in patients with non-small-cell lung cancer: a meta-analysis and systemic review. Onco Targets Ther 2015;8:3361-9. [Crossref] [PubMed]
- Messaritakis I, Nikolaou M, Politaki E, et al. Bcl-2 expression in circulating tumor cells (CTCs) of patients with small cell lung cancer (SCLC) receiving front-line treatment. Lung Cancer 2018;124:270-8. [Crossref] [PubMed]
- Smerage JB, Budd GT, Doyle GV, et al. Monitoring apoptosis and Bcl-2 on circulating tumor cells in patients with metastatic breast cancer. Mol Oncol 2013;7:680-92. [Crossref] [PubMed]
- Brahmer J, Reckamp KL, Baas P, et al. Nivolumab versus Docetaxel in Advanced Squamous-Cell Non-Small-Cell Lung Cancer. N Engl J Med 2015;373:123-35. [Crossref] [PubMed]


