
The role of plasma genotyping in ALK- and ROS1-rearranged lung cancer
Introduction
Non-small cell lung cancer (NSCLC) is a heterogeneous disease that encompasses diverse molecular subsets, each of which is associated with unique therapeutic vulnerabilities and distinct disease outcomes (1,2). The genetic alterations that characterize these subsets are similarly broad and include point mutations, insertion/deletions, splicing alterations, amplifications, and rearrangements that activate oncogenic signaling (1). Given the therapeutic implications of identifying these important genetic alterations, it is critical that diagnostic approaches detect the various types of alterations with high sensitivity and specificity. Compared to activating mutations and insertion/deletions, detection of oncogenic rearrangements can be particularly challenging from a technical perspective due to structural complexity (3-5). Indeed, multiple breakpoints and diverse fusion partners can contribute to an oncogenic rearrangement.
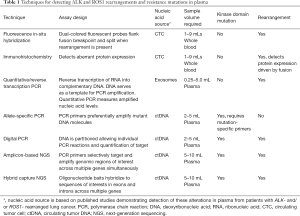
To overcome this diagnostic hurdle, initial strategies for detecting oncogenic rearrangements gravitated towards immunohistochemistry (IHC) or fluorescence in-situ hybridization (FISH) (6,7). The former is predicated on lack of expression of certain oncogenic proteins (which are constitutively expressed in tumors with activating rearrangements) in normal tissues whereas the latter relies on splitting of dual-colored fluorescent probes designed to flank common breakpoints (Table 1) (7,8). However, FISH is expensive and requires technical expertise and specialized microscopy which may not be readily available in many laboratories. Although more widely available and less challenging to implement, IHC performance can vary depending on the antibody used and background levels of expression of target proteins in normal tissues (9,10). While the sensitivity and specificity of both of these techniques can exceed 90% in the optimal context (11,12), practical considerations—specifically the ever-expanding number of important molecular alterations and need to conserve tissue—have shifted diagnostic approaches away from single-gene assays (i.e., FISH and IHC) towards more comprehensive strategies such as next-generation sequencing (NGS) that can interrogate DNA and RNA to simultaneously uncover relevant genetic alterations and fusion transcripts in hundreds of genes with a single assay (13).
Full table
To date, tissue biopsies have been the primary material used for molecular profiling, predominantly in the form of formalin-fixed paraffin-embedded (FFPE) tissues. In addition to FFPE tissues, cytology specimens can also serve as substrate for molecular genotyping (14). As FFPE is the most common tissue type that is processed in pathology laboratories for the majority of solid tumor specimens, most molecular diagnostic platforms have been validated using genetic material derived from this specimen source. However, several factors can adversely impact the utility of FFPE specimens for molecular testing, including inadequate tumor purity and nucleic acid degradation, the latter of which is dependent on several pre-analytic factors including but not limited to fixation, decalcification, and age of the block (15,16). In addition to these limitations, molecular testing may not be feasible due to insufficient tissue remaining within the block after prior sectioning for histologic studies and ancillary testing. Beyond the technical factors discussed above, disease-specific considerations such as high risk of procedural complications may preclude tissue sampling altogether.
These limitations of tissue-based molecular profiling coupled with significant advances in techniques for isolating and analyzing tumor-derived genetic material in the circulation have spurred rapid uptake of blood-based genotyping in recent years (17). In the bloodstream, tumor-derived genetic material can be free-floating (i.e., cell-free DNA) or contained in circulating tumor cells (CTCs) or extracellular vesicles (EV) (18,19). Circulating free-floating tumor DNA (ctDNA) is released passively into the bloodstream from necrotic or apoptotic cancer cells or actively from living cancer cells and comprises between 0.01% to 90% of all cell-free DNA in the circulation (18,19). The percentage of tumor DNA in cell-free DNA depends on tumor type, metastatic burden, and clinical context (20-22). Compared to ctDNA which can be analyzed using methods similar to tissue genotyping, unique methods are required for capture, enrichment, and detection of CTCs and EVs (23,24). As a result, the uptake of ctDNA analysis has outpaced adoption of CTC and EV profiling in clinical practice.
As with tissue-dependent molecular testing, plasma-based testing has inherent limitations that may impact ctDNA yield. In contrast to tissue-based biopsies which are often enriched for tumor (and may be dissected to further enhance tumor purity), plasma specimens are primarily comprised of DNA derived from non-tumor sources (25). As such, careful processing of plasma specimens is required to ensure that the lysis of normal white blood cells does not dilute the fraction of ctDNA. This is typically achieved through rapid processing of plasma or collection of plasma in specialized tubes that are designed to stabilize the specimen (26). Still, optimizing preanalytic steps to improve ctDNA recovery cannot overcome tumor-intrinsic factors such as minimal shedding of tumor-derived DNA into the circulation in the context of low disease burden or tumors confined to certain disease sites (e.g., thoracic cavity and central nervous system) (27). Furthermore, because of the underrepresentation of tumor cells relative to other cells in the plasma, mutations that originate in hematopoietic cells can be incorrectly attributed to solid tumors, thus confounding plasma-based analyses of the molecular makeup of solid tumors (28-30).
This review will discuss applications of plasma genotyping in NSCLC, with a particular emphasis on the unique considerations related to plasma genotyping in NSCLCs harboring ALK and ROS1 rearrangements.
Early applications of plasma genotyping in NSCLC
Initial applications of blood-based molecular analysis in metastatic NSCLC focused on detecting mutations in the epidermal growth factor receptor (EGFR) gene in ctDNA and CTCs (31-33). The focus on this molecular subset was likely fostered by favorable technical factors (e.g., ease of detecting mutations and insertions/deletions relative to more complex alterations) and clinical factors, specifically the early establishment of EGFR mutations as relevant actionable therapeutic targets and early adoption of serial molecular profiling for this group (34,35). The earliest studies in this area utilized allele-specific polymerase chain reaction, a technology that preferentially amplifies mutant DNA molecules to enhance detection of cancer-specific mutations in a background largely composed of signals from wild-type DNA (Table 1) (36,37). Although findings from these pioneering studies were promising, refinements of existing molecular genotyping techniques and introduction of highly sensitive PCR approaches (i.e., digital PCR) and broad-panel genotyping strategies that improved the performance of plasma assays in the decade that followed are primarily responsible for the increased prominence of plasma genoptying in clinical care of lung cancer patients (38-41).
Digital PCR enables absolute quantification of circulating nucleic acids by effectively diluting the genetic material into individual PCR reactions (Table 1) (40,42). In this technique, DNA is distributed or rather “partitioned”, with each partition serving as an independent PCR microreactor. Partitions generally consist of either droplets or chambers that function as physical dividers. Droplet-based platforms achieve individualization of PCR reactions via a variety of approaches, including emulsification and microfluidic droplet methods and BEAMing (Beads, Emulsion, Amplification, Magnets), a strategy where each magnetic bead captures a single molecule (38,40). Promising findings from studies utilizing digital droplet PCR (ddPCR) and BEAMing reignited interest in non-invasive genotyping of NSCLC (31). In multiple studies, high tissue-plasma concordance was observed for detecting lung cancer-specific point mutations (31,33,43). While the sensitivity of digital PCR is noteworthy, the coverage of only a single mutation and inability to reliably detect fusions have compromised its relevance in the era of comprehensive genotyping.
In recent years, NGS of cell-free DNA has emerged as a promising strategy for analyzing a broad spectrum of molecular alterations in plasma (17). Performance characteristics of various ctDNA assays has been extensively described in previous reviews (44). To date, most studies have used two genotyping techniques: amplicon-based NGS and hybrid-capture NGS. Amplicon-based NGS uses PCR technology and short primers to simultaneously identify mutated hotspots and regions of interest in exons of multiple genes (Table 1) (45,46). In retrospective analyses of patients with metastatic NSCLC, amplicon-based NGS detected a variety of alterations in plasma (including rearrangements) with sensitivity and specificity comparable to or exceeding that of ddPCR (47). In contrast to amplicon-based NGS, hybrid-capture NGS approaches rely on probes or “baits” consisting of barcoded DNA oligonucleotide adapters that span exonic and intronic regions of interest in multiple genes (Table 1) (21,48). In three prospective studies where tissue and plasma analysis were performed in parallel for patients with NSCLC (27,49,50), hybrid-capture plasma NGS demonstrated favorable performance characteristics across a wide range of driver alterations, including in a small group of tumors with oncogenic rearrangements. Analyzing ctDNA with this method uncovered relevant, actionable genetic alterations in a higher proportion of patients than tissue molecular profiling (49). “Bias-corrected” targeted NGS, a third method which improves upon traditional hybrid-capture methods by incorporating a primer extension step to reduce artifacts, using shorter probes to boost efficiency of capturing rearrangements, and tagging of DNA fragment to improve signal-to-noise ratio has proven to be a promising approach for broadly detecting targetable alterations with high specificity and sensitivity (51).
These early studies have made inroads towards establishing the clinical utility of comprehensive plasma genotyping in NSCLC. However, the plasma specimens included in these pivotal studies were predominantly derived from patients with NSCLCs that harbored activating mutations rather than oncogenic rearrangements. With the increasing number of actionable oncogenic rearrangements in NSCLC (1), it is essential to evaluate the performance of plasma genotyping in fusion-driven NSCLC. Therefore, the remainder of the review will summarize current understanding of plasma profiling—including ctDNA analysis and CTC and exosome profiling—in NSCLCs with two of the most common activating rearrangements: ALK and ROS1.
Anaplastic lymphoma kinase rearrangements
Approximately 5% of NSCLCs harbor chromosomal rearrangements of the anaplastic lymphoma kinase (ALK) gene (7,52). ALK rearrangements encode a constitutively active chimeric fusion protein that signals in a ligand-independent fashion (53). Multiple unique fusion partners have been described in ALK-rearranged (ALK+) NSCLC, but over 80% of ALK rearrangements juxtapose the ALK kinase domain with the promoter and oligomerization domain of EML4 (54). In the majority of ALK+ NSCLCs, the breakpoint in ALK occurs in intron 19 (exon 20) (54). However, there are >15 distinct EML4-ALK fusion variants which are distinguished by the location of the breakpoint in EML4 (55). The number of potential EML4-ALK fusion variants can pose a challenge if attempting to design unique PCR primers to cover all potential permutations. However, broadly detecting potential ALK fusions is not an insurmountable task for hybrid-capture assays as the conserved breakpoint in ALK and proclivity for EML4 as a fusion partner allow for dense bait coverage (also referred to as “tiling”) of introns and exons in these two genes.
Circulating tumor DNA
Several studies have explored the diagnostic utility of amplicon-based ctDNA NGS in newly diagnosed patients with ALK+ NSCLC and those relapsing on targeted therapy (47,56). For example, Guibert et al detected an EML4-ALK fusion in plasma from 6 (86%) of 7 treatment-naive patients known to have ALK+ NSCLC based on tissue analysis (47). The false negative result was from a patient with stage III NSCLC. Across platforms, sensitivity of ctDNA analysis in NSCLC patients with non-metastatic disease is notoriously inferior to sensitivity in patients with metastatic disease (22,57). In another study by Mezquita and colleagues which utilized primers that selectively targeted variants 1-3 of EML4-ALK in plasma, sensitivity for detecting ALK rearrangements was 78% for treatment-naïve patients with metastatic NSCLC (56). In comparison, an ALK rearrangement was only detected in 12% of patients responding to treatment, consistent with prior studies demonstrating that shedding of ctDNA decreases during periods of treatment response (56).
Compared to amplicon-based NGS, more studies have utilized hybrid-capture techniques to analyze plasma from patients with ALK+ NSCLC. In these studies, sensitivity for detecting ALK fusions was high and dependent on disease stage, with several studies reporting sensitivity approaching 80% (58-60). These studies suggest that capture-based techniques can identify a more diverse spectrum of fusion partners. For example, in a retrospective analysis which analyzed plasma from 88 patients with ALK+ NSCLC using a hybrid-capture NGS assay, seven unique fusion partners were observed (61). In addition, an analysis of 22 patients treated in the phase I/II ensartinib study who underwent paired plasma and tissue genotyping identified three different fusion partners in the cohort of patients: EML4, AKAP8L, PRKAR1A (62). Although the majority of tumors had EML4-ALK rearrangements, five unique EML4-ALK fusion variants were detected (62). Similarly, a separate study detected four distinct EML4-ALK fusion variants in a dataset comprised of 22 patients relapsing on ALK inhibitors who were treated at a single institution (60). In both of these studies and an analysis of 53 patients in the phase III ALEX study with paired tissue and plasma specimens (63), agreement between tissue and plasma fusion partner calls and EML4-ALK fusion variants ranged from 79.2–100% (60,62,63).
Based on the promising performance of capture-based plasma NGS, several studies have evaluated response to alectinib among patients with plasma-detected ALK rearrangements. In the pivotal phase III ALEX study which established superiority of alectinib over crizotinib as first-line treatment for ALK+ NSCLC, when progression-free survival (PFS) was compared in patients with plasma and/or tissue ALK rearrangements, the hazard ratio of investigator-assessed PFS was comparable between the plasma and tissue subgroups (63). In the BFAST study which prospectively enrolled patients with advanced NSCLC and assigned therapy based on alterations detected with the FoundationAct hybrid-capture plasma assay, the response rate to first-line alectinib among 87 patients with ALK+ NSCLC was 87.4% and the 12-month event-free survival rate was 78% (64). As these outcomes compare favorably with the ALEX study where tissue was used for eligibility (65), this study suggest that using plasma findings to select patients suitable for treatment with first-line alectinib is a valid approach. Beyond utility for identifying newly-diagnosed patients who might benefit from first-line ALK-directed therapy, monitoring the ALK rearrangement in plasma during treatment may serve as a readout of disease behavior. In support of this, a study of 121 patients enrolled in the phase II lorlatinib study observed a decrease in the allelic frequency of the ALK fusion at 6 weeks in most patients, a finding that has also been observed in smaller series of ALK+ patients treated with earlier-generation ALK TKIs (58,60,66). Notably, the group of patients with early complete clearance of the ALK rearrangement from plasma (i.e., undetectable plasma ALK rearrangement) had deeper responses and longer PFS on lorlatinib (66).
The clinical and diagnostic utility of detecting an ALK rearrangement in plasma depends on the rate of false positive and false negative results. Across studies, the positive predictive value of plasma-detected ALK rearrangements are high (58,59), suggesting that the risk of false positives is low. With hybrid-capture platforms, efficiency of capture of ctDNA fragments that span rearrangement breakpoints is a critical determinant of assay sensitivity. In a small study, the combination of shorter capture probes and primer extension improved sensitivity for detecting ALK fusions and enhanced the ability to detect ALK fusions with novel partners (67). Compared to probes that bind larger segments of DNA, shorter probes may overcome detection errors that result from misalignment to the fragmented ctDNA corresponding to breakpoints. Beyond capture efficiency, other disease-related factors can impact sensitivity. For example, sensitivity decreases when the disease burden is primarily concentrated in the central nervous system (68). In this setting, cerebrospinal fluid may be a higher yield source of ctDNA than plasma (19). Indeed, one study detected an ALK rearrangement in 82% of cerebrospinal fluid specimens compared to 46% of plasma specimens from 11 ALK+ patients with leptomeningeal disease who underwent paired plasma and cerebrospinal fluid analysis using hybrid-capture technology (69). Similar findings have been reported with amplicon-based NGS of cerebrospinal fluid (68).
Exosomes and platelets
In theory, analyzing expressed fusion transcripts may circumvent some of the challenges of DNA-based ALK fusion detection. In the circulation, cell-free tumor-derived RNA is rapidly degraded complicating its utility as a barometer of a tumor’s molecular composition (19). Tumor-derived nucleic acids that are incorporated into EVs are protected from degradation. EVs, therefore, represent a potential source of tumor-derived fusion transcripts. Several groups have attempted to identify ALK fusion transcripts in exosomes, a type of EV. Brinkmann and colleagues detected EML4-ALK rearrangements in half of the 52 patients with ALK+ NSCLC in their study using quantitative PCR (70). While it is possible that limitations of the assay led to failure to detect ALK rearrangements in the remaining cases, these NSCLCs may have expressed EML4-ALK variants or novel ALK fusions that were not targeted by the PCR primers. Interestingly, exosomes can serve as a conduit between cancer cells and platelets. This allows for transfer of ALK transcripts from cancer cells to platelets (71). In one study, reverse transcription PCR of platelet RNA from NSCLC patients demonstrated sensitivity and specificity of 65% and 100% for detecting EML4-ALK rearrangements, respectively (71). In longitudinal analyses, ALK fusions were undetectable in exosomes and platelets during response to ALK inhibitors and increased in abundance at relapse (70,71), suggesting that serial monitoring of these plasma components can provide insights into treatment response and disease biology.
CTCs
CTCs contain DNA, RNA, and proteins, allowing for detection of ALK+ NSCLCs using a variety of techniques. The handful of studies to date exploring feasibility of this genotyping approach have utilized FISH or IHC to distinguish CTCs from patients with ALK+ NSCLCs from those from patients with ALK wild-type NSCLCs (72,73). In these studies, rare CTCs with ALK rearrangements were detected in the circulation of patients who did not have ALK+ NSCLC, necessitating cutoffs to discriminate between “false-positive” and “true-positive” results. In a study by Pailler et al., the range of ALK-rearranged CTCs per milliliter among ALK+ cases was 4-34 prompting selection of 4 ALK-rearranged CTCs per milliliter as a cutoff for ALK+ NSCLC (73). Notably, at the cutoff of 4 ALK-rearranged CTCs per 1 milliliter of blood, sensitivity and specificity were each 100% (73). In a different study, a cutoff of 3 or more ALK-rearranged CTCs per milliliter was proposed based on analysis of a small cohort that included healthy controls and NSCLC patients with and without ALK rearrangements (74). Similar to findings from serial profiling of ctDNA and exosomes, CTC enumeration demonstrates reduction in the number of ALK-positive CTCs during response to treatment (73,75). In contrast to findings from ctDNA analysis (62), however, one study suggests that the pretreatment burden of ALK-rearranged CTCs and dynamics of these CTCs during treatment do not predict durability of response to ALK-directed therapy (75).
ROS1 rearrangements
Chromosomal rearrangements that promote constitutive activation of ROS1 are present in 1–2% of NSCLCs (6). Although there is considerable homology between the ALK and ROS1 kinases, there are notable differences between rearrangements involving these two genes. Unlike ALK rearrangements which generally harbor a conserved breakpoint in intron 19/exon 20 of ALK, breakpoints throughout introns 31-35 (exons 32-36) have been described in ROS1-rearranged (ROS1+) NSCLC (76). In addition, ROS1 fuses to more than 15 distinct partners in NSCLC (76-78). Although the frequency of certain fusion partners is higher than others (CD74, SLC34A2, SDC4, EZR), none approach the 80% seen with EML4-ALK (77,78). Indeed, the most prevalent partner CD74 only accounts for 50% of cases and CD74-ROS1 fusions can involve two different breakpoints in ROS1 (77,78). The inconsistency of fusion partners and spectrum of potential breakpoints introduce unique technical challenges for assay development and optimization.
Circulating Tumor DNA
Due to the low prevalence of ROS1 fusions in NSCLC (1,6), studies of ctDNA analysis in this molecular subset are limited. A small number of patients with this alteration have been included in studies that enrolled patients with heterogenous genotypes. For example, Guibert et al used an amplicon-based plasma NGS panel covering key intronic and exonic sequences corresponding to the breakpoints of four of the most common ROS1 rearrangements—CD74-ROS1, SLC34A2-ROS1, SDC4-ROS1, and EZR-ROS1—to correctly identify the 2 patients with ROS1 rearrangements in a group of 46 patients with metastatic NSCLC (47). Notably, both patients had CD74-ROS1 rearrangements. In another study by Mezquita et al. which used similar techniques (56), ROS1 fusions involving CD74 and SLC34A2 fusion partners were detected. Although only 6 patients with ROS1+ NSCLC were included in the study, sensitivity for detecting ROS1 fusions in treatment-naïve patients was 100% (56).
The number of patients evaluated for ROS1 rearrangements in plasma using hybrid-capture NGS in published studies is substantially larger than with amplicon-based NGS, but the actual number of patients with this alteration included in hybrid-capture studies is still quite small. For example, among 282 patients in the prospective NILE study (49), tissue-plasma concordance for detecting ROS1 rearrangements was 98.7% using a hybrid-capture NGS assay. However, there were only 2 patients with ROS1+ NSCLC in the NILE study who underwent paired plasma and tissue analysis. In both cases, the rearrangement was only detected in tissue (49). When the same assay was used to analyze plasma specimens from 20 patients known to have ROS1+ NSCLC in a separate study, sensitivity for detecting ROS1 rearrangements at relapse on targeted therapy was 50% (77). Similarly, in the lorlatinib phase I/II study, a ROS1 rearrangement was only detected in plasma from 23 (52%) of 44 patients with analyzable ctDNA (79). As many patients with ROS1+ NSCLC will not have extrathoracic, extracranial site involvement (80), the low sensitivity may be partially attributed to intrinsic disease characteristics (77), namely low ctDNA shedding from the thorax and brain (27,68).
Based on the limited number of ROS1+ patients in these studies and particularly the limited number patients with paired tissue and plasma profiling, data regarding the ability of ctDNA analysis to recapitulate the breadth of potential ROS1 rearrangements in NSCLC are scarce. In one study, paired analysis of plasma and tissue from 7 patients with NSCLC demonstrated complete concordance of fusion partners (77). Notably, there were four distinct fusion partners among the 7 patients, including a rare CCDC6-ROS1 rearrangement (77), suggesting that there should be confidence in ROS1 fusion calls based on plasma genotyping. The remaining data regarding prevalence of specific ROS1 fusion partners in plasma vs tissue are derived from a study which assessed fusion partners in two large non-overlapping datasets, including a group of 56 ROS1+ plasma specimens from a deidentified commercial database and 52 tissue specimens from ROS1+ NSCLC patients treated at a single institution cohort (77). In this analysis, the relative representation of common fusion partners was comparable in tissue and plasma (77). As the plasma cohort was generated from a deidentified database, the distribution of disease sites in the patients from whom plasma was collected is not known. These data therefore support utility of NGS for detecting diverse ROS1 rearrangements but do not provide insights into biological factors that may impact assay sensitivity. Larger well-annotated (including disease characteristics and sites of involvement) tissue-plasma concordance studies are necessary to robustly assess the performance characteristics (e.g., sensitivity, specificity, positive predictive value) of plasma NGS in ROS1+ NSCLC and evaluate how disease biology impacts assay performance. For now, the limited body of literature suggests that negative plasma results should be confirmed with tissue genotyping when feasible given the relatively low sensitivity of current assays for detecting ROS1 rearrangements.
CTCs
Given the limited sensitivity of current NGS assays for detecting ROS1 fusions, there is interest in exploring alternative non-invasive genotyping techniques. One case report suggests that CTC analysis may have advantages over plasma NGS. In this report, a patient with peritoneal carcinomatosis was found to have a ROS1 rearrangement in CTCs by FISH but was ROS1-negative by plasma NGS (81). The presence of the fusion was confirmed in tissue and the patient experienced durable disease control on crizotinib. Details regarding the plasma assay used were not included in the case report. While it is difficult to generalize findings from a single patient experience, there is evidence that ROS1 rearrangements can be readily detected in CTCs. In a study by Leong et al. (82), ROS1-rearranged CTCs were detected in 4 (67%) of 6 ROS1+ blood samples using ROS1 FISH and IHC, with up to 10 ROS1+ CTCs detected per 2 milliliters of blood. The disease characteristics of the patients included in the study were sparse. However, the findings suggest that there is potential for false negatives with CTC analysis as well. In the study (82), the frequency of ROS1-rearranged CTCs among healthy controls was sufficiently low that 1 ROS1+ cell per 2 milliliters of blood was proposed as a reliable cutoff for discriminating between ROS1+ and ROS1-negative cases.
An analysis by another group suggests that this cutoff may be too low. Specifically, a study by Pailler et al. detected a median number of ROS1 FISH-positive CTCs of 34.5 per 3 milliliters of blood from 4 patients with ROS1+ NSCLC compared to 7.5 per 3 milliliters of blood in ROS1-negative patients (83). The false positive rate in ROS1-negative patients was attributed to nonspecific background hybridization of FISH probes. Taken together, these two studies confirm that ROS1 rearrangements can be detected in CTCs but highlight the degree of variability across patients and assays. The studies conducted to date are not of sufficient size to ascertain whether variability in CTC yield is driven by disease biology or technical aspects of the assays. Therefore, standardization of assays and nomination of cutoffs informed by large groups of patients with ROS1+ NSCLC that represent the range of clinical presentations is needed before CTC profiling can be routinely applied to management of ROS1+ NSCLC.
Detecting acquired kinase domain mutations
In current practice, standard treatment of ALK+ and ROS1+ NSCLC most often involves sequential treatment with targeted therapies. Despite significant and noteworthy responses to tyrosine kinase inhibitors (TKIs) (65,84,85), relapse is inevitable and typically occurs within years of initiating of therapy. In ALK+ NSCLC, an array of kinase domain mutations can emerge at relapse (86,87). These mutations account for the majority of relapses on second-generation ALK TKIs (e.g., ceritinib, alectinib, brigatinib) (60,87). The complexity of ALK kinase domain mutations increases after treatment with the third-generation ALK TKI lorlatinib resulting in compound mutations in approximately one-third of patients (88). Current understanding of key molecular drivers of relapse in ROS1+ NSCLC is not as robust, but several studies suggest that secondary kinase domain mutations play a critical role in relapses and that next-generation ROS1 TKIs may be able to overcome certain acquired kinase domain mutations (80,89-91). Notably, the preliminary antitumor activity of next-generation ROS1 TKIs against acquired ROS1 kinase domain mutations is not as robust as the activity of the third-generation ALK TKI lorlatinib against analogous ALK kinase domain mutations (92). Given the implications of secondary resistance mutations, plasma diagnostics must detect fusions and mutations involving ALK and ROS1 with high confidence in order to meet the demands of clinical practice. Findings from plasma genotyping studies to date are summarized in Table 2.

Full table
ALK+ lung cancer
Several studies have confirmed that diverse ALK kinase domain mutations can be detected in plasma. The frequency of particular ALK mutations in plasma at relapse on specific ALK TKIs is comparable to the composition of ALK mutations identified through tissue-focused analyses (86,93). Furthermore, among patients who receive both methods of molecular profiling, ctDNA analysis is a highly sensitive approach for detecting tissue-identified ALK mutations, with reported sensitivity across studies of 82–90% (Table 2) (60,86,92). However, when tissue is considered the reference standard, specificity of plasma assays is seldom perfect as ALK mutations are often detected only in plasma (Table 2) (60,62,86). Indeed, one study found that while the proportion of patients without ALK mutations at relapse on ALK TKIs was comparable in tissue and plasma, the number of patients with multiple ALK mutations at progression was underestimated by tissue genotyping (86). As patients with plasma-only ALK mutations can achieve durable responses to ALK TKIs targeting those mutations (62), this occurrence likely reflects the ability of plasma to capture relevant alterations across multiple disease sites.
Consistent with findings from serial tissue profiling (87), longitudinal ctDNA analysis demonstrates temporal variation in ALK mutations in plasma during treatment with sequential ALK TKIs. Indeed, the plasma ALK mutation profile has been shown to be dynamic with clearance of ALK mutations that emerged on preceding ALK-directed therapies upon initiation of a different ALK TKI with activity against the specific mutation (60,62,86). Early data suggest that these ALK mutations in plasma can potentially provide insight into likelihood of responding to future targeted therapies. For example, in a correlative analysis of the lorlatinib phase II study, pretreatment plasma ALK mutation status was predictive of response to subsequent treatment with lorlatinib with ORR of 62% vs. 32% for ALK mutation-positive and ALK mutation-negative patients, respectively (92). Together, these studies support the potential utility of serial plasma analysis for guiding sequencing approaches in ALK+ NSCLC. This hypothesis is being formally assessed in the ongoing NCI-NRG ALK protocol (NCT03737994).
ROS1+ lung cancer
A handful of studies have evaluated utility of ctDNA analysis for detecting ROS1 kinase domain mutations. Consistent with tissue analyses (80), the solvent front G2032R mutation is the most commonly reported ROS1 mutation in plasma at progression on crizotinib. For example, an analysis of 18 patients who underwent plasma profiling at progression on crizotinib identified ROS1 kinase domain mutations in 6 (33%) specimens (77). Five specimens harbored G2032R whereas the remaining specimen demonstrated an L2026M gatekeeper mutation (77). Of the 4 cases in this group where both plasma and tissue were analyzed, three pairs were concordant, including one pair with the G2032R mutation in both plasma and tissue. In the final case, ROS1 G2032R was initially only identified in plasma. However, a biopsy several months later ultimately revealed G2032R in a different disease site than originally sampled (77). In the lorlatinib phase I/II study (79), 6 (15%) of 41 patients who had previously received another ROS1 TKI (mostly crizotinib) before initiating lorlatinib had detectable ROS1 mutations in ctDNA prior to treatment with lorlatinib, including G2032R (n=4), L2026M (n=1), and both L2026M and I2025I (n=1). Responses to lorlatinib were observed in 27% of patients without a plasma ROS1 mutation compared to 0% of the 6 patients with ROS1 kinase domain mutations (79). Acknowledging the small sample size, these data suggest that on-target mutation status may be less informative for predicting response to lorlatinib in ROS1+ NSCLC than ALK+ NSCLC, potentially reflecting differences in disease biology and disparate lorlatinib efficacy in these two subsets rather than limitations of plasma genotyping.
Conclusions
With a rapidly evolving therapeutic landscape that boasts an ever-expanding arsenal of approved targeted therapies, management of NSCLC is becoming increasingly complex and appropriately more personalized (2). As an accurate understanding of the molecular makeup of tumor cells forms the foundation of these therapeutic approaches, it is critical to nurture strategies that fully capture the molecular composition of NSCLC during a patient’s disease course. In recent years, plasma genotyping has emerged as a promising method for rapidly detecting relevant molecular alterations in NSCLC (49,50). In ROS1+ NSCLC, studies of plasma genotyping are sparse, but available data suggest that additional modifications will be necessary to enable plasma platforms to reliably capture the complex landscape of ROS1 rearrangements on a scale comparable to tissue genotyping. In contrast, early findings from studies using NGS-based plasma diagnostics to detect key alterations among patients with ALK+ NSCLC have been encouraging, with studies consistently demonstrating that NGS can identify ALK rearrangements and ALK resistance mutations with high sensitivity and specificity. Importantly, in prospective studies and retrospective correlative analyses from large trials, plasma-detected ALK alterations predict benefit from ALK-directed therapies (64,66), revealing the potential of using standalone plasma results to guide therapeutic strategies for ALK+ NSCLC. Beyond being an accurate representation of tissue findings and mirroring treatment outcomes seen with tissue biomarkers, plasma genotyping is ideally suited for the longitudinal analyses that are necessary for gaining early insights into long-term treatment outcomes and evolving resistance mechanisms.
As the number and complexity of relevant oncogenic fusions—including partners and breakpoints—is anticipated to continue to increase, diagnostic strategies that can overcome the limitations of current assays are necessary to both establish plasma analysis as a peer of tissue profiling and, most importantly, capitalize on all available therapeutic opportunities in a disease where availability and efficacy of targeted therapeutics has created stark differences in prognostic outcomes across molecular subsets. Validation strategies for novel plasma diagnostics should involve studies as robust as those used to vet novel therapeutic strategies, including prospective clinical trials. Finally, as disease biology influences performance of plasma diagnostics, it is critical that ongoing optimization of plasma assays does not adopt a myopic focus on improving assay performance in tumors with complex molecular alterations. Rather, as distribution of disease sites can handicap sensitivity of plasma platforms, future success depends on developing alternative approaches in these scenarios, including exploring analysis of tumor-derived molecular material in other biofluids (cerebrospinal fluid, pleural fluid) and piloting non-invasive diagnostic techniques that can enhance the yield of molecular profiling among patients with limited disease burden and limited sites of disease involvement (19,94).
Acknowledgments
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editors (Silvia Novello, Francesco Passiglia) for the series “Looking for Chimeras in NSCLC: Widen Therapeutic Options Targeting Oncogenic Fusions” published in Translational Lung Cancer Research. The article has undergone external peer review.
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tlcr-2019-cnsclc-09). The series “Looking for Chimeras in NSCLC: Widen Therapeutic Options Targeting Oncogenic Fusions” was commissioned by the editorial office without any funding or sponsorship. IDJ reports consulting fees from Boerhinger Ingelheim, AstraZeneca and Catalyst Pharmaceuticals, honoraria from Foundation Medicine, travel fees from Pfizer, Array American Lung Association and CEC, research support from Pfizer, Genentech/Roche and Array, outside the submitted work. The other author has no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Jordan EJ, Kim HR, Arcila ME, et al. Prospective Comprehensive Molecular Characterization of Lung Adenocarcinomas for Efficient Patient Matching to Approved and Emerging Therapies. Cancer Discov 2017;7:596-609. [Crossref] [PubMed]
- Arbour KC, Riely GJ. Systemic Therapy for Locally Advanced and Metastatic Non-Small Cell Lung Cancer: A Review. JAMA 2019;322:764-74. [Crossref] [PubMed]
- Zheng Z, Liebers M, Zhelyazkova B, et al. Anchored multiplex PCR for targeted next-generation sequencing. Nat Med 2014;20:1479-84. [Crossref] [PubMed]
- Benayed R, Offin M, Mullaney K, et al. High Yield of RNA Sequencing for Targetable Kinase Fusions in Lung Adenocarcinomas with No Mitogenic Driver Alteration Detected by DNA Sequencing and Low Tumor Mutation Burden. Clin Cancer Res 2019;25:4712-22. [Crossref] [PubMed]
- Davies KD, Le AT, Sheren J, et al. Comparison of Molecular Testing Modalities for Detection of ROS1 Rearrangements in a Cohort of Positive Patient Samples. J Thorac Oncol 2018;13:1474-82. [Crossref] [PubMed]
- Bergethon K, Shaw AT, Ou SH, et al. ROS1 rearrangements define a unique molecular class of lung cancers. J Clin Oncol 2012;30:863-70. [Crossref] [PubMed]
- Shaw AT, Yeap BY, Mino-Kenudson M, et al. Clinical features and outcome of patients with non-small-cell lung cancer who harbor EML4-ALK. J Clin Oncol 2009;27:4247-53. [Crossref] [PubMed]
- Mino-Kenudson M, Chirieac LR, Law K, et al. A novel, highly sensitive antibody allows for the routine detection of ALK-rearranged lung adenocarcinomas by standard immunohistochemistry. Clin Cancer Res 2010;16:1561-71. [Crossref] [PubMed]
- Shen Q, Wang X, Yu B, et al. Comparing four different ALK antibodies with manual immunohistochemistry (IHC) to screen for ALK-rearranged non-small cell lung cancer (NSCLC). Lung Cancer 2015;90:492-8. [Crossref] [PubMed]
- Hofman V, Rouquette I, Long-Mira E, et al. Multicenter Evaluation of a Novel ROS1 Immunohistochemistry Assay (SP384) for Detection of ROS1 Rearrangements in a Large Cohort of Lung Adenocarcinoma Patients. J Thorac Oncol 2019;14:1204-12. [Crossref] [PubMed]
- Wynes MW, Sholl LM, Dietel M, et al. An international interpretation study using the ALK IHC antibody D5F3 and a sensitive detection kit demonstrates high concordance between ALK IHC and ALK FISH and between evaluators. J Thorac Oncol 2014;9:631-8. [Crossref] [PubMed]
- Takeuchi K, Togashi Y, Kamihara Y, et al. Prospective and clinical validation of ALK immunohistochemistry: results from the phase I/II study of alectinib for ALK-positive lung cancer (AF-001JP study). Ann Oncol 2016;27:185-92. [Crossref] [PubMed]
- Kalemkerian GP, Narula N, Kennedy EB, et al. Molecular Testing Guideline for the Selection of Patients With Lung Cancer for Treatment With Targeted Tyrosine Kinase Inhibitors: American Society of Clinical Oncology Endorsement of the College of American Pathologists/International Association for the Study of Lung Cancer/Association for Molecular Pathology Clinical Practice Guideline Update. J Clin Oncol 2018;36:911-9. [Crossref] [PubMed]
- Roh MH. The Utilization of Cytologic Fine-Needle Aspirates of Lung Cancer for Molecular Diagnostic Testing. J Pathol Transl Med 2015;49:300-9. [Crossref] [PubMed]
- Araujo LH, Timmers C, Shilo K, et al. Impact of Pre-Analytical Variables on Cancer Targeted Gene Sequencing Efficiency. PLoS One 2015;10:e0143092. [Crossref] [PubMed]
- Lennon NJ, Adalsteinsson VA, Gabriel SB. Technological considerations for genome-guided diagnosis and management of cancer. Genome Med 2016;8:112. [Crossref] [PubMed]
- Merker JD, Oxnard GR, Compton C, et al. Circulating Tumor DNA Analysis in Patients With Cancer: American Society of Clinical Oncology and College of American Pathologists Joint Review. J Clin Oncol 2018;36:1631-41. [Crossref] [PubMed]
- Corcoran RB, Chabner BA. Application of Cell-free DNA Analysis to Cancer Treatment. N Engl J Med 2018;379:1754-65. [Crossref] [PubMed]
- Siravegna G, Marsoni S, Siena S, et al. Integrating liquid biopsies into the management of cancer. Nat Rev Clin Oncol 2017;14:531-48. [Crossref] [PubMed]
- Zill OA, Banks KC, Fairclough SR, et al. The Landscape of Actionable Genomic Alterations in Cell-Free Circulating Tumor DNA from 21,807 Advanced Cancer Patients. Clin Cancer Res 2018;24:3528-38. [Crossref] [PubMed]
- Newman AM, Bratman SV, To J, et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med 2014;20:548-54. [Crossref] [PubMed]
- Chaudhuri AA, Chabon JJ, Lovejoy AF, et al. Early Detection of Molecular Residual Disease in Localized Lung Cancer by Circulating Tumor DNA Profiling. Cancer Discov 2017;7:1394-403. [Crossref] [PubMed]
- Möhrmann L, Huang HJ, Hong DS, et al. Liquid Biopsies Using Plasma Exosomal Nucleic Acids and Plasma Cell-Free DNA Compared with Clinical Outcomes of Patients with Advanced Cancers. Clin Cancer Res 2018;24:181-8. [Crossref] [PubMed]
- Yu M, Bardia A, Wittner BS, et al. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science 2013;339:580-4. [Crossref] [PubMed]
- Diaz LA Jr, Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol 2014;32:579-86. [Crossref] [PubMed]
- Warton K, Yuwono NL, Cowley MJ, et al. Evaluation of Streck BCT and PAXgene Stabilised Blood Collection Tubes for Cell-Free Circulating DNA Studies in Plasma. Mol Diagn Ther 2017;21:563-70. [Crossref] [PubMed]
- Aggarwal C, Thompson JC, Black TA, et al. Clinical Implications of Plasma-Based Genotyping With the Delivery of Personalized Therapy in Metastatic Non-Small Cell Lung Cancer. JAMA Oncol 2019;5:173-80. [Crossref] [PubMed]
- Chabon JJ, Hamilton EG, Kurtz DM, et al. Integrating genomic features for non-invasive early lung cancer detection. Nature 2020;580:245-51. [Crossref] [PubMed]
- Razavi P, Li BT, Brown DN, et al. High-intensity sequencing reveals the sources of plasma circulating cell-free DNA variants. Nat Med 2019;25:1928-37. [Crossref] [PubMed]
- Hu Y, Ulrich BC, Supplee J, et al. False-Positive Plasma Genotyping Due to Clonal Hematopoiesis. Clin Cancer Res 2018;24:4437-43. [Crossref] [PubMed]
- Sacher AG, Paweletz C, Dahlberg SE, et al. Prospective Validation of Rapid Plasma Genotyping for the Detection of EGFR and KRAS Mutations in Advanced Lung Cancer. JAMA Oncol 2016;2:1014-22. [Crossref] [PubMed]
- Sundaresan TK, Sequist LV, Heymach JV, et al. Detection of T790M, the Acquired Resistance EGFR Mutation, by Tumor Biopsy versus Noninvasive Blood-Based Analyses. Clin Cancer Res 2016;22:1103-10. [Crossref] [PubMed]
- Reckamp KL, Melnikova VO, Karlovich C, et al. A Highly Sensitive and Quantitative Test Platform for Detection of NSCLC EGFR Mutations in Urine and Plasma. J Thorac Oncol 2016;11:1690-700. [Crossref] [PubMed]
- Yu HA, Arcila ME, Rekhtman N, et al. Analysis of tumor specimens at the time of acquired resistance to EGFR-TKI therapy in 155 patients with EGFR-mutant lung cancers. Clin Cancer Res 2013;19:2240-7. [Crossref] [PubMed]
- Sequist LV, Waltman BA, Dias-Santagata D, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med 2011;3:75ra26. [Crossref] [PubMed]
- Whitcombe D, Theaker J, Guy SP, et al. Detection of PCR products using self-probing amplicons and fluorescence. Nat Biotechnol 1999;17:804-7. [Crossref] [PubMed]
- Maheswaran S, Sequist LV, Nagrath S, et al. Detection of mutations in EGFR in circulating lung-cancer cells. N Engl J Med 2008;359:366-77. [Crossref] [PubMed]
- Diehl F, Li M, He Y, et al. BEAMing: single-molecule PCR on microparticles in water-in-oil emulsions. Nat Methods 2006;3:551-9. [Crossref] [PubMed]
- Diehl F, Schmidt K, Choti MA, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med 2008;14:985-90. [Crossref] [PubMed]
- Hindson BJ, Ness KD, Masquelier DA, et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal Chem 2011;83:8604-10. [Crossref] [PubMed]
- Pinheiro LB, Coleman VA, Hindson CM, et al. Evaluation of a droplet digital polymerase chain reaction format for DNA copy number quantification. Anal Chem 2012;84:1003-11. [Crossref] [PubMed]
- Hudecova I. Digital PCR analysis of circulating nucleic acids. Clin Biochem 2015;48:948-56. [Crossref] [PubMed]
- Oxnard GR, Thress KS, Alden RS, et al. Association Between Plasma Genotyping and Outcomes of Treatment With Osimertinib (AZD9291) in Advanced Non-Small-Cell Lung Cancer. J Clin Oncol 2016;34:3375-82. [Crossref] [PubMed]
- Ou SI, Nagasaka M, Zhu VW. Liquid Biopsy to Identify Actionable Genomic Alterations. Am Soc Clin Oncol Educ Book 2018;38:978-97. [Crossref] [PubMed]
- Kinde I, Wu J, Papadopoulos N, et al. Detection and quantification of rare mutations with massively parallel sequencing. Proc Natl Acad Sci U S A 2011;108:9530-5. [Crossref] [PubMed]
- Oxnard GR, Paweletz CP, Sholl LM. Genomic Analysis of Plasma Cell-Free DNA in Patients With Cancer. JAMA Oncol 2017;3:740-1. [Crossref] [PubMed]
- Guibert N, Hu Y, Feeney N, et al. Amplicon-based next-generation sequencing of plasma cell-free DNA for detection of driver and resistance mutations in advanced non-small cell lung cancer. Ann Oncol 2018;29:1049-55. [Crossref] [PubMed]
- Odegaard JI, Vincent JJ, Mortimer S, et al. Validation of a Plasma-Based Comprehensive Cancer Genotyping Assay Utilizing Orthogonal Tissue- and Plasma-Based Methodologies. Clin Cancer Res 2018;24:3539-49. [Crossref] [PubMed]
- Leighl NB, Page RD, Raymond VM, et al. Clinical Utility of Comprehensive Cell-free DNA Analysis to Identify Genomic Biomarkers in Patients with Newly Diagnosed Metastatic Non-small Cell Lung Cancer. Clin Cancer Res 2019;25:4691-700. [Crossref] [PubMed]
- Sabari JK, Offin M, Stephens D, et al. A Prospective Study of Circulating Tumor DNA to Guide Matched Targeted Therapy in Lung Cancers. J Natl Cancer Inst 2019;111:575-83. [Crossref] [PubMed]
- Paweletz CP, Sacher AG, Raymond CK, et al. Bias-Corrected Targeted Next-Generation Sequencing for Rapid, Multiplexed Detection of Actionable Alterations in Cell-Free DNA from Advanced Lung Cancer Patients. Clin Cancer Res 2016;22:915-22. [Crossref] [PubMed]
- Gainor JF, Varghese AM, Ou SH, et al. ALK rearrangements are mutually exclusive with mutations in EGFR or KRAS: an analysis of 1,683 patients with non-small cell lung cancer. Clin Cancer Res 2013;19:4273-81. [Crossref] [PubMed]
- Soda M, Choi YL, Enomoto M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature 2007;448:561-6. [Crossref] [PubMed]
- Ross JS, Ali SM, Fasan O, et al. ALK Fusions in a Wide Variety of Tumor Types Respond to Anti-ALK Targeted Therapy. Oncologist 2017;22:1444-50. [Crossref] [PubMed]
- Lin JJ, Riely GJ, Shaw AT. Targeting ALK: Precision Medicine Takes on Drug Resistance. Cancer Discov 2017;7:137-55. [Crossref] [PubMed]
- Mezquita L, Hu Y, Howarth K, et al. Abstract 4581: Feasibility of an amplicon-based liquid biopsy for ALK and ROS1 fusions in advanced non-small cell lung cancer (NSCLC) patients. Cancer Research 2018.4581.
- Jamal-Hanjani M, Wilson GA, McGranahan N, et al. Tracking the Evolution of Non-Small-Cell Lung Cancer. N Engl J Med 2017;376:2109-21. [Crossref] [PubMed]
- Wang Y, Tian PW, Wang WY, et al. Noninvasive genotyping and monitoring of anaplastic lymphoma kinase (ALK) rearranged non-small cell lung cancer by capture-based next-generation sequencing. Oncotarget 2016;7:65208-17. [Crossref] [PubMed]
- Cui S, Zhang W, Xiong L, et al. Use of capture-based next-generation sequencing to detect ALK fusion in plasma cell-free DNA of patients with non-small-cell lung cancer. Oncotarget 2017;8:2771-80. [Crossref] [PubMed]
- Dagogo-Jack I, Brannon AR, Ferris LA, et al. Tracking the Evolution of Resistance to ALK Tyrosine Kinase Inhibitors through Longitudinal Analysis of Circulating Tumor DNA. JCO Precis Oncol 2018;2018:PO.17.00160.
- McCoach CE, Blakely CM, Banks KC, et al. Clinical Utility of Cell-Free DNA for the Detection of ALK Fusions and Genomic Mechanisms of ALK Inhibitor Resistance in Non-Small Cell Lung Cancer. Clin Cancer Res 2018;24:2758-70. [Crossref] [PubMed]
- Horn L, Whisenant JG, Wakelee H, et al. Monitoring Therapeutic Response and Resistance: Analysis of Circulating Tumor DNA in Patients With ALK+ Lung Cancer. J Thorac Oncol 2019;14:1901-11. [Crossref] [PubMed]
- Camidge DR, Dziadziuszko R, Peters S, et al. Updated Efficacy and Safety Data and Impact of the EML4-ALK Fusion Variant on the Efficacy of Alectinib in Untreated ALK-Positive Advanced Non-Small Cell Lung Cancer in the Global Phase III ALEX Study. J Thorac Oncol 2019;14:1233-43. [Crossref] [PubMed]
- Gadgeel SM, Mok TSK, Peters S, et al. Phase II/III blood-first assay screening trial (BFAST) in treatment-naïve NSCLC: initial results from the ALK+ cohort. Ann Oncol 2019;30:v851-v934. [Crossref]
- Peters S, Camidge DR, Shaw AT, et al. Alectinib versus Crizotinib in Untreated ALK-Positive Non-Small-Cell Lung Cancer. N Engl J Med 2017;377:829-38. [Crossref] [PubMed]
- Shaw AT. Early circulating tumor (ct)DNA dynamics and efficacy of lorlatinib in patients (pts) with advanced ALK-positive non-small cell lung cancer (NSCLC). J Clin Oncol 2019;37:abstr 9019.
- Supplee JG, Milan MSD, Lim LP, et al. Sensitivity of next-generation sequencing assays detecting oncogenic fusions in plasma cell-free DNA. Lung Cancer 2019;134:96-9. [Crossref] [PubMed]
- Aldea M, Hendriks L, Mezquita L, et al. Circulating Tumor DNA Analysis for Patients with Oncogene-Addicted NSCLC With Isolated Central Nervous System Progression. J Thorac Oncol 2020;15:383-91. [Crossref] [PubMed]
- Zheng MM, Li YS, Jiang BY, et al. Clinical Utility of Cerebrospinal Fluid Cell-Free DNA as Liquid Biopsy for Leptomeningeal Metastases in ALK-Rearranged NSCLC. J Thorac Oncol 2019;14:924-32. [Crossref] [PubMed]
- Brinkmann K, Enderle D, Flinspach C, et al. Exosome liquid biopsies of NSCLC patients for longitudinal monitoring of ALK fusions and resistance mutations. J Thorac Oncol 2018;36:e24090.
- Nilsson RJ, Karachaliou N, Berenguer J, et al. Rearranged EML4-ALK fusion transcripts sequester in circulating blood platelets and enable blood-based crizotinib response monitoring in non-small-cell lung cancer. Oncotarget 2016;7:1066-75. [Crossref] [PubMed]
- Ilie M, Long E, Butori C, et al. ALK-gene rearrangement: a comparative analysis on circulating tumour cells and tumour tissue from patients with lung adenocarcinoma. Ann Oncol 2012;23:2907-13. [Crossref] [PubMed]
- Pailler E, Adam J, Barthélémy A, et al. Detection of circulating tumor cells harboring a unique ALK rearrangement in ALK-positive non-small-cell lung cancer. J Clin Oncol 2013;31:2273-81. [Crossref] [PubMed]
- Tan CL, Lim TH, Lim TKh, et al. Concordance of anaplastic lymphoma kinase (ALK) gene rearrangements between circulating tumor cells and tumor in non-small cell lung cancer. Oncotarget 2016;7:23251-62. [Crossref] [PubMed]
- Pailler E, Oulhen M, Borget I, et al. Circulating Tumor Cells with Aberrant. Cancer Res 2017;77:2222-30. [Crossref] [PubMed]
- Lin JJ, Shaw AT. Recent Advances in Targeting ROS1 in Lung Cancer. J Thorac Oncol 2017;12:1611-25. [Crossref] [PubMed]
- Dagogo-Jack I, Rooney M, Nagy RJ, et al. Molecular Analysis of Plasma From Patients With ROS1-Positive NSCLC. J Thorac Oncol 2019;14:816-24. [Crossref] [PubMed]
- Park S, Ahn BC, Lim SW, et al. Characteristics and Outcome of ROS1-Positive Non-Small Cell Lung Cancer Patients in Routine Clinical Practice. J Thorac Oncol 2018;13:1373-82. [Crossref] [PubMed]
- Shaw AT, Solomon BJ, Chiari R, et al. Lorlatinib in advanced ROS1-positive non-small-cell lung cancer: a multicentre, open-label, single-arm, phase 1-2 trial. Lancet Oncol 2019;20:1691-701. [Crossref] [PubMed]
- Gainor JF, Tseng D, Yoda S, et al. Patterns of Metastatic Spread and Mechanisms of Resistance to Crizotinib in ROS1-Positive Non-Small-Cell Lung Cancer. JCO Precis Oncol 2017. [Crossref] [PubMed]
- Raez LE, Manca P, Rolfo C, et al. ROS-1 Rearrangements in Circulating Tumor Cells. J Thorac Oncol 2018;13:e71-2. [Crossref] [PubMed]
- Leong MC, Lim TH, Chua YW, et al. Molecular detection of ROS1-rearranged circulating tumor cells (CTC) in non-small cell lung cancer (NSCLC) patients. J Clin Oncol 2016;34:e23086. [Crossref]
- Pailler E, Auger N, Lindsay CR, et al. High level of chromosomal instability in circulating tumor cells of ROS1-rearranged non-small-cell lung cancer. Ann Oncol 2015;26:1408-15. [Crossref] [PubMed]
- Shaw AT, Ou SH, Bang YJ, et al. Crizotinib in ROS1-rearranged non-small-cell lung cancer. N Engl J Med 2014;371:1963-71. [Crossref] [PubMed]
- Camidge DR, Kim HR, Ahn MJ, et al. Brigatinib versus Crizotinib in ALK-Positive Non-Small-Cell Lung Cancer. N Engl J Med 2018;379:2027-39. [Crossref] [PubMed]
- Dagogo-Jack I, Rooney M, Lin JJ, et al. Treatment with Next-Generation ALK Inhibitors Fuels Plasma. Clin Cancer Res 2019;25:6662-70. [Crossref] [PubMed]
- Gainor JF, Dardaei L, Yoda S, et al. Molecular Mechanisms of Resistance to First- and Second-Generation ALK Inhibitors in ALK-Rearranged Lung Cancer. Cancer Discov 2016;6:1118-33. [Crossref] [PubMed]
- Yoda S, Lin JJ, Lawrence MS, et al. Sequential ALK Inhibitors Can Select for Lorlatinib-Resistant Compound. Cancer Discov 2018;8:714-29. [Crossref] [PubMed]
- McCoach CE, Le AT, Gowan K, et al. Resistance Mechanisms to Targeted Therapies in ROS1(+) and ALK(+) Non-small Cell Lung Cancer. Clin Cancer Res 2018;24:3334-47. [Crossref] [PubMed]
- Facchinetti F, Loriot Y, Kuo MS, et al. Crizotinib-Resistant ROS1 Mutations Reveal a Predictive Kinase Inhibitor Sensitivity Model for ROS1- and ALK-Rearranged Lung Cancers. Clin Cancer Res 2016;22:5983-91. [Crossref] [PubMed]
- Drilon A, Ou SI, Cho BC, et al. Repotrectinib (TPX-0005) Is a Next-Generation ROS1/TRK/ALK Inhibitor That Potently Inhibits ROS1/TRK/ALK Solvent- Front Mutations. Cancer Discov 2018;8:1227-36. [Crossref] [PubMed]
- Shaw AT, Solomon BJ, Besse B, et al. ALK Resistance Mutations and Efficacy of Lorlatinib in Advanced Anaplastic Lymphoma Kinase-Positive Non-Small-Cell Lung Cancer. J Clin Oncol 2019;37:1370-9. [Crossref] [PubMed]
- Noé J, Lovejoy A, Ou SI, et al. ALK Mutation Status Before and After Alectinib Treatment in Locally Advanced or Metastatic ALK-Positive NSCLC: Pooled Analysis of Two Prospective Trials. J Thorac Oncol 2020;15:601-8. [Crossref] [PubMed]
- Krug AK, Enderle D, Karlovich C, et al. Improved EGFR mutation detection using combined exosomal RNA and circulating tumor DNA in NSCLC patient plasma. Ann Oncol 2018;29:700-6. [Crossref] [PubMed]


