
Metformin reverses chemoresistance in non-small cell lung cancer via accelerating ubiquitination-mediated degradation of Nrf2
Introduction
Lung cancer is responsible for more deaths worldwide than any other malignancy (1). Non-small cell lung cancer (NSCLC) accounts for approximately 85% of lung cancer cases (2). In recent years, the tumor responses and clinical outcomes of patients with NSCLC have been dramatically improved by targeted therapy and immunotherapy (3,4). However, platinum-based systemic chemotherapy is still the preferred first-line therapy for the remaining patients and is the cornerstone in the treatment of resectable patients who undergo neoadjuvant chemotherapy (5). However, various factors can result in the development of resistance to chemotherapy (including reduced drug accumulation, enhanced drug-mediated detoxification of oxidative stress, and increased DNA adduct repair), which is a major obstacle that limits our ability to treat advanced NSCLC (6-8).
With the increasing number of combination strategies to attenuate tumor resistance and decrease chemotherapeutic toxicity (9), metformin, which is widely prescribed for the treatment of type II diabetes, has shown important anti-lung cancer effects in both in vitro and in vivo preclinical studies. The effect of metformin in combination with other treatment strategies has also been studied (10). Metformin was demonstrated to sensitize different cancer cell types to cisplatin cytotoxicity, and various mechanisms have been described, from mitochondrial apoptosis to the inhibition of DNA synthesis (11). Although the signal transduction mechanisms by which the combination of metformin with cisplatin potentiates cytotoxicity in lung cancer are evidenced by a large body of research (12-14), fewer studies have focused on the detoxification of reactive oxygen species (ROS) under cisplatin-induced oxidative stress. Notably, mutagenic ROS is involved during carcinogenesis and chemotherapy resistance (15). Conversely, high levels of ROS can further form DNA double-strand breaks, resulting in a DNA catastrophe and subsequently inducing apoptosis (16). Therefore, the increased cellular antioxidant capacity may play a vital role in lung cancer cellular adaptation to cisplatin-induced oxidative stress. ROS are generated in mitochondria. As a drug regulating glucose metabolism, metformin also regulates mitochondrial function. However, its effect on cellular ROS has not yet been fully elucidated.
The transcription factor nuclear factor erythoid-2-related factor 2 (NFE2L2/Nrf2), a master regulator of the antioxidant response, plays a role in the most important endogenous defense mechanism by which ROS are maintained at low physiological levels. Nrf2 is essential to redox homeostasis, especially after cells have been exposed to chemotherapeutic agents (17,18). Nrf2 exerts its detoxifying effect by binding to the antioxidant response element (ARE) and transactivating various cytoprotective genes, especially, heme oxygenase 1 (HO-1), which is one of the strongest antioxidant phase II detoxifying enzymes.
“Nrf2 addiction” refers to hyperactivation of the Nrf2 pathway in lung cancer cells, which promotes the development of NSCLC and can also enhance chemoresistance (19,20). Emerging evidence has shown that targeting Nrf2 is a potential therapeutic strategy for overcoming cisplatin resistance (21). Intriguingly, Truong Do M revealed that metformin suppresses the expression of Nrf2 at the transcriptional level by inhibiting Sirtuin 1 (Sirt1) (22), while another study reported the opposite result, with metformin also upregulating Sirt1 expression for decreasing the acetylation of Nrf2 and preventing its nuclear distribution (23). Metformin somehow negatively modulates Nrf2 expression in lung cancer, but there is complete lack of understanding of the underlying mechanisms. Some Nrf2-ECH homology (Neh) domains in Nrf2 are tightly regulated by various posttranslational modifications (PTMs), such as phosphorylation and ubiquitylation (24), which effectively confer changes in Nrf2 expression. Effective PTMs in Nrf2 can change its location or expression level (17). Extracellular signal-regulated kinases 1/2 (ERK1/2) were shown to be involved in the regulation of Nrf2 by metformin treatment (25). Butylated hydroxyanisole was reported to increase phosphorylation of the ERK1/2, thus promoting Nrf2 translocation into the nucleus (26). However, the relationship between ERK1/2 and Nrf2-related PTMs still remains unclear and few studies have explored the effect of clinical agents on the PTM status of Nrf2, which in turn affects Nrf2 activation.
In the current study, we investigated the role of Nrf2 in the regulation of cisplatin-induced ROS production and chemoresistance in NSCLC cells with a more comprehensive analysis. Our data elucidate, for the first time, that the promotion effect of metformin on mitochondrial ROS production plays a critical role in chemoresistance reversal in lung cancer. We explored the mechanism of metformin-mediated Nrf2 degradation through attenuating ERK-mediated Nrf2 phosphorylation to restore its polyubiquitination, which has a weakening effect on the ROS defense system in NSCLC. Importantly, for the first time, we evaluated the difference in Nrf2 levels in NSCLC patients before and after neoadjuvant chemotherapy as well as the correlation of this change with chemotherapeutic response and patient survival. We present the following article in accordance with the ARRIVE reporting checklist (available at http://dx.doi.org/10.21037/tlcr-20-1072).
Methods
Cell culture and transfection
Human lung cancer cells A549/DDP (Bogoo Biotechnology, Shanghai, China), a cisplatin-resistant A549 cell line, and H838 (Cell Bank of China Science Academy, Shanghai, China) were maintained in RPMI 1640 medium (CORNING, Mediatech, Inc., Manassas, VA, USA) supplemented with 10% fetal bovine serum (FBS, PAN-Biotech, Aidenbach, Germany) in a humidified atmosphere with 5% CO2. Cisplatin (800 ng/mL) was added in fresh medium for A549/DDP to maintain its resistance. Cell lines were authenticated by Biowing Applied Biotechnology Co., Ltd and Cinoasia Institude (Shanghai, China) via short tandem repeat profiling.
Commercial plasmids pcDNA3.1-mNrf2 and pcDNA3.1-mHO-1 were supplied by Genechem Co., Ltd (Shanghai, China). The plasmids were transfected into NSCLC cells using Lipofectamine 3000 reagent (Invitrogen, CA, USA) according to the manufacturer’s instructions. After 6 h, the original medium was replaced with fresh medium. At 48 h after transfection, the cells were harvested or treated with drugs.
For knockdown of Nrf2 expression, lentivirus containing an shRNA sequence targeting Nrf2 were supplied by Genechem Co., Ltd (Shanghai, China). NSCLC cells were seeded at 30% confluence in 24-well plates overnight before infection, and the media were replaced with a medium (250 µL) containing lentivirus and polybrene (8 µg/mL). After infection for 4 h, another 250 µL of fresh medium containing polybrene (8 µg/mL) was added into wells. The original medium was replaced with fresh medium 24 h later. And the infected cells were selected with puromycin (2 µg/mL) 72 h after infection. The mature antisense sequences targeting Nrf2 are as follows: 5'-CACCAGAACACTCAGTGGAAT-3' (shNrf2 #1), 5'-GAGCAGTTCAATGAAGCTCAA-3' (shNrf2 #2), 5'-AGCCATTCACTCTCTGAACTT-3' (shNrf2 #3).
Reagents and antibodies
Metformin (1,1-Dimethylbiguanide hydrochloride, #D150959-5G), Protoporphyrin IX zinc (ZnPPIX, #282820), N-Acetyl-L-cysteine (NAC, #A7250) and 2',7'-Dichlorofluorescein diacetate (DCFH-DA, #D6883) were purchased from Sigma-Aldrich (St. Louis, MO, USA). Cycloheximide (CHX, CAS 66-81-9) was purchased from MedChemExpress LLC (Monmouth Junction, NJ, USA). Cisplatin and proteasome inhibitor MG132 was purchased from Selleck Chemicals (Houston, TX, USA). The Annexin V-PE/7-AAD Apoptosis Detection Kit (AP104) was purchased from Multi Sciences (Hangzhou, China). U0126 (#9903). The antibodies used for immunoblotting and co-immunoprecipitation, β-actin (#58169), Nrf2 (#12721), caspase-3 (#9662), poly ADP-ribose polymerase (PARP) (#9532), Phospho-c-Raf (#9431), p44/42 MAPK (Erk1/2, #4695), Phospho-p44/42 MAPK (#8544), and ubiquitin (#3936), along with goat anti-rabbit immunoglobulin G (IgG, #7074) and goat anti-mouse (#7076) HRP-linked secondary antibodies, were purchased from Cell Signaling Technology, Inc. (Danvers, MA, USA). Antibodies against Keap1 (AF5266) and HO-1 (AF5393) were obtained from Affinity Biosciences (Cincinnati, OH, USA).
Cell viability assay
Cell viability was measured with a Cell Counting Kit-8 (CCK-8) assay (Dojindo, Japan). NSCLC cells were seeded in 96-well plates overnight at a density of 5,000 cells per well, and then treated with drugs under the indicated conditions and times. Next, 10% of the final volume of CCK-8 solution was added to each well and incubated at 37 °C for 1 h. A SpectraMax M4 microplate reader (Molecular Devices, LLC, Sunnyvale, CA, USA) was used to measure the absorbance at a wavelength of 450 nm.
The IC50 curves were calculated using variable slope for log (inhibitor) vs. normalized response (Graphpad Prism 8.3, La Jolla, CA, USA). The combination index (CI) was calculated using CompuSyn software (ComboSyn, Inc., Paramus, NJ, USA), with CI <0.9, 0.9–1.1, and >1.1 indicating synergism, additive effect, and antagonism, respectively.
Cell apoptosis assay
The distribution of apoptotic cells was evaluated using Annexin V-PE/7-AAD staining. After indicated treatments, NSCLC cells were trypsinized in 0.25% trypsin with Ethylenediaminetetraacetic acid (EDTA)-free, washed with cold phosphate-buffered saline (PBS), and then suspended in 100 µL PBS. Subsequently, the cells were stained with 5 µL of Annexin V-PE and 10 µL of 7-AAD, before being incubated at room temperature for 30 min in the dark. The stained cells were visualized by flow cytometry (FACS CantoII, BD Biosciences, USA) and analyzed using Flowjo (FlowJo 10.4, LLC, USA).
Drug toxicity was assessed by Hoechst 33342/PI staining. NSCLC cells were seeded into 6-well plates at a density of 4×105/well. After incubation for 24 h, the cells were treated with metformin and/or cisplatin under the indicated conditions. Next, the cells were trypsinized, resuspended in PBS with Hoechst 33342 10 µg/mL and incubated for 15 min at 37 °C. The cells were then washed twice with PBS, and stained with PI (10 µg/mL) for 15 min at room temperature. Finally, the cell suspension was dripped onto the glass slides and observed by fluorescence microscopy.
ROS detection
After the indicated treatments, adherent cells were harvested with trypsin-EDTA solution and washed once with PBS. For total cellular ROS detection, the cells were incubated with 10 µM ROS-sensitive probe (DCFH-DA) in PBS for 30 min at 37 °C in darkness. For mitochondrial ROS accumulation detection, NSCLC cells were incubated with 5 µM mitochondrial superoxide indicator (MitoSOX Red, Thermo Scientific, M36008) in PBS for 10 min at 37 °C in darkness. The cells were then washed twice with PBS and suspended in a fresh medium for immediate analysis by flow cytometer.
Western blot and immunoprecipitation (IP) assay
Cells were first washed with ice-cold PBS and then lysed in RIPA lysis buffer (Beyotime, Shanghai, China). Sonication was used to reduce the viscosity of the lysate, and a BCA assay (Solarbio, China) was carried out to measure the protein concentration. The lysates were then mixed with 3X reducing SDS loading buffer (#7723, Cell Signaling Technology, Inc., USA) and denatured at 100 °C for 5 min. The lysates were separated with 10–12% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) gels and then transferred onto a polyvinylidene difluoride (PVDF) membrane (Millipore, Billerica, MA). The membrane was blocked with 5% nonfat milk for 1 h at room temperature and incubated overnight at 4 °C with primary antibodies. After that, the membrane was washed 3 times with TBST (tris-buffered saline, 0.1% Tween-20) and then probed with a secondary antibody for 1 h. After washing with TBST, the blots were incubated with a high-sensitivity chemiluminescence detection system (Millipore) and visualized using the Amersham Biosciences ECL Detection System (Amersham plc, GE Healthcare, Chicago, IL, USA).
For the IP assay, the Pierce™ Direct Magnetic IP/Co-IP Kit (Thermo Scientific, #88828) was used. The main procedure was carried out in accordance with the manufacturer’s instructions. Briefly, Nrf2 antibody (5 µg) was incubated with magnetic beads (25 µL) for 60 min. Cell lysates were mixed with antibody-coupled magnetic beads and incubated for 2 h at room temperature on a rotator. Precipitated proteins (60 µL) were eluted from beads and prepared for western blot analysis.
Phos-tag analysis
For Phos-tag SDS-PAGE, 20 µM phosbind acrylamide (F4002, APExBIO, Houston, TX, USA) and 80 µM MnCl2 were added to 8% SDS-polyacrylamide gel before polymerization. The gel was electrophoresed at 25 mA for 2 h. Before transfer, the gel was first immersed three times in transfer buffer containing 10 mM EDTA for 10 min and then in transfer buffer without EDTA twice for 10 min. Protein transfer was performed for 3 h at 350 mA at 0 °C, and then the membrane was analyzed by western blotting with anti-Nrf2 antibody.
Dephosphorylation with lambda phosphatase
NSCLC cells were seeded in 10-cm culture dishes overnight and collected using 400 µL IP Lysis Buffer. The collected cells were incubated with 5 µL of lambda protein phosphatase (New England BioLabs, Ipswich, MA, USA) diluted in 50 µL of 10× Protein MetalloPhosphatases (PMP) and 50 µL of 10 mM MnCl2 for 30 min at 30 °C. After incubation, the samples were analyzed by western blot or IP as described above.
Quantitative real-time polymerase chain reaction
Total RNA was extracted using RNA-Quick Purification Kit (Yishan Biotech, Shanghai, China). cDNA was synthesized using HiScript II Q RT SuperMix (Vazyme Biotech, Nanjing, China). Real-time polymerase chain reaction (PCR) was performed on an ABI Prism 7500 Fast Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) using ChamQ SYBR qPCR Master Mix (Vazyme). The 2−ΔΔCt method was used to calculate the relative expression levels of Nrf2 with β-actin used for normalization. The sequences of the primers used for quantitative real-time PCR (qRT-PCR) were as follows:
- Nrf2: Forward 5'-TCAGCGACGGAAAGAGTATGA-3'; Reverse 5'-CCACTGGTTTCTGACTGGATGT-3'.
- β-actin: Forward 5'-TGGCACCCAGCACAATGAA-3'; Reverse 5'-CTAAGTCATAGTCCGCCTAGAAGCA-3'.
Xenograft model and treatments
All animal experiments were approved by the Committee of Animal Experimental Ethical Inspection of the First Affiliated Hospital, College of Medicine, Zhejiang University (reference No. 2019-1232), and followed by the institutional Guidelines for animal care and use.
Five-week-old male nude mice (Shanghai Sippr-bk Laboratory Animal Co., Ltd, China; weight, 16 to 20 g) were injected subcutaneously in the left flanks with 5×106 A549/DDP cells mixed 1:1 with 100% Matrigel. On day 8, when tumors had reached 40–80 mm3, the mice were separated into four groups (5 mice per group) and given water (50 µL/10 g; qd) + intraperitoneal saline (100 µL/10 g; qod; control group) by oral gavage, metformin (200 mg/kg; qd) + intraperitoneal saline (100 µL/10 g; qod; metformin group) by oral gavage, water (50 µL/10g; qd) + intraperitoneal cisplatin (2 mg/kg; qod; cisplatin group) orally, or metformin (200 mg/kg; qd) + intraperitoneal cisplatin (2 mg/kg; qod; combination group) orally. The tumor volume was calculated using the following formula: volume = 1/2(length × width2). The animals were euthanized by cervical dislocation according to the American Veterinary Medical Association Guidelines (Schaumburg, IL, USA).
Terminal deoxynucleotidyl transferase dUTP nick-end labeling assay
Detection of apoptotic cells was performed with a terminal deoxynucleotidyl transferase dUTP nick-end labeling (TUNEL) assay on paraffin-embedded sections of xenograft tumor tissue using an In Situ Cell Death Detection Kit, POD (Roche) according to the manufacturer’s instructions. Images were captured using a light microscope.
Immunohistochemical analysis
Paraffin-embedded tumor tissue sections (4 µM) were deparaffinized and subjected to heated-induced epitope retrieval using citrate buffer (pH 6.0). The tissues were incubated with rabbit anti-Nrf2 (1:200 dilution, Abcam, #ab31163) or anti-Ki67 (1:200 dilution, Abcam, #ab15580) antibody overnight at 4 °C. Following that, the tissues were incubated with the corresponding secondary antibodies. Peroxidase activity was visualized with 3,3'-diaminobenzidine (DAB; Darko, CA, USA) and then counterstaining with hematoxylin was performed.
The sections were examined by two senior pathologists in a double-blinded manner. The intensity of Nrf2 staining was quantified using a four-score grading system (0=0–10% of stained cells; 1=11–24% of stained cells; 2=25–75% of stained cells; 3=>75% of stained cells).
Tumor specimens and patients
All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). Patients were informed that the specimens were stored by the hospital and potentially used for scientific research, and signed informed consent to participants was waived by the Ethics Committee. This study was approved by the Clinical Research Ethics Committee of the First Affiliated Hospital, College of Medicine, Zhejiang University (Approval No.: 2020-13). Clinical samples were obtained from 50 primary lung cancer patients who received platinum-containing neoadjuvant chemotherapy followed by surgical resection. To be enrolled in this study, patients must have been pathologically confirmed before neoadjuvant chemotherapy and obtained their final pathological report after surgery. Matched biopsy and surgical resection tissue samples from eligible patients were embedded in formalin-fixed paraffin, and IHC staining was used to confirm chemotherapy-related changes in Nrf2 status. Patients were regularly followed and the chemotherapeutic response was clinically evaluated using the Response Evaluation Criteria in Solid Tumors (RECIST 1.1). The objective response rate (ORR) used in this cohort was defined as follows: (I) complete response (CR): disappearance of all known lesions; (II) partial response (PR), ≥30% regression in the sum of the measurable diameters of target lesions; (III) progressive disease (PD), ≥20% increase in the sum of the measurable diameters of target lesions; (IV) stable disease (SD), any response between PR and PD.
Statistical analysis
The two-tailed Student’s t-test was used for comparisons between two groups, and one-way or two-way analysis of variance (ANOVA) followed by Tukey’s test was used for multiple group comparisons. Categorical data were compared using Fisher’s exact text. For multi-classified variables, binary logistic regression analysis was adopted. Survival curves were estimated using the Kaplan-Meier method and analyzed using the log-rank test. Prism 8.3 (GraphPad, La Jolla, CA) and SPSS software 25.0 (IBM statistics, SPSS Inc., Chicago, IL, USA) were used for statistical analyses. A P value <0.05 was considered statistically significant.
Results
Metformin synergistically augments cisplatin-induced cytotoxicity with Nrf2 suppression
The effects of metformin combination therapy on the normal NSCLC cell line H838 and the cisplatin-resistant cell line A549/DDP were assessed. The CCK-8 assay revealed that metformin significantly enhanced the inhibitory effect of cisplatin on cell proliferation (Figure 1A). Then, single or combination treatment groups administered high concentrations of cisplatin were assessed to generate accurate IC50 curves. As shown in Figure 1B, compared to single cisplatin treatment, combination treatment with metformin clearly decreased the IC50 values of A549/DDP cells (from 24.87 to 17.53 µM) and H838 cells (from 3.18 to 1.56 µM). The combination index (CI) was calculated to determine the synergistic effects of combination therapy with metformin and cisplatin. The CI was <1, which showed that metformin and cisplatin synergistically suppressed NSCLC proliferation (Figure 1C).

Next, the effect of metformin on cisplatin-induced NSCLC cell death was evaluated. Cells in the late stage of apoptosis were stained with both Hoechst 33342 and PI (pink). Combination treatment with metformin and cisplatin significantly enhanced the induction of apoptosis (Figure 1D). Flow cytometry analysis showed that combination treatment significantly increased the number of apoptotic NSCLC cells (the total percentage of cells in quadrants Q2 + Q3) compared to that following treatment with cisplatin alone, while single treatment with metformin (3.2 and 12.8 mM for A549/DDP and H838 cells, respectively) did not clearly promote cell apoptosis (Figure 1E). The protein levels of caspase-3 (cleaved caspase-3), a mediator of apoptosis signaling, and PARP (cleaved PARP) were also evaluated, which revealed similar results in NSCLC cells treated with metformin and/or cisplatin (Figure 1F). Intriguingly, metformin augmented cisplatin-induced apoptosis by strongly inhibiting the expression of Nrf2. With cisplatin stimulation, the levels of Nrf2 and HO-1 dose-dependently increased. Metformin strongly inhibited cisplatin-mediated upregulation of Nrf2 and HO-1 in both chemoresistant and normal NSCLC cells (Figure 1G). These results therefore demonstrated that metformin synergistically enhanced cisplatin-mediated cytotoxicity and suppressed cisplatin-induced Nrf2/HO-1 axis upregulation in both chemoresistant and normal NSCLC cells.
Metformin eliminates the detoxification ability of the Nrf2/HO-1 axis and promotes ROS-mediated cisplatin-induced apoptosis in chemoresistance NSCLC cells
We hypothesized that the downregulation of Nrf2 expression and accumulation of ROS are associated with increased apoptosis in NSCLC cells treated with combination therapy. To explore the critical role of the Nrf2/HO-1 axis in the metformin-mediated reversal of resistance in NSCLC cells, Nrf2 and HO-1 were overexpressed in A549/DDP and H838 cells. As shown in Figure 2A, metformin did not block the protein expression of Nrf2 or HO-1 in cells overexpressing Nrf2 or HO-1. Under combination treatment, Nrf2 and HO-1 overexpression markedly reduced intrinsic apoptosis markers. Similarly, the CCK-8 assay and flow cytometry analysis also showed that the synergistic effect of the combination treatment was rescued by Nrf2 and HO-1 overexpression (Figure 2B, Figure S1A). Furthermore, NSCLC cells were transfected with lentivirus containing an shRNA sequence targeting Nrf2, and verified the inhibition efficiency (Figure S1B). Accordingly, the sensitivity and cytotoxicity of the NSCLC cells to cisplatin was remarkably enhanced upon Nrf2 knockdown in NSCLC cells (Figure 2C,D). The stable knockdown of Nrf2 showed a similar effect of chemoresistance reversal to the combination therapy. These results suggested that the detoxification ability of Nrf2/HO-1 axis toward cisplatin is critical for NSCLC cells to develop drug resistance.
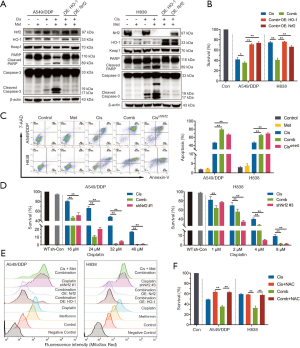
Next, the intracellular ROS level was detected in Nrf2-knockdown and Nrf2/HO-1-overexpressing NSCLC cells using MitoSOX Red (Figure 2E). Evaluation of mitochondrial superoxide levels revealed that metformin markedly increased the cisplatin-induced production of ROS. Nrf2-knockdown NSCLC cells also exhibited remarkably increased mitochondrial production of ROS in response to cisplatin treatment. With overexpression of Nrf2 and HO-1, the curve indicating ROS levels moved substantially to the left, showing decreased intracellular ROS accumulation under metformin and cisplatin treatment. The total ROS detection by 2',7'-Dichlorofluorescein diacetate (DCFH-DA) probe produces the similar results (Figure S1C). Then, the cells were treated with the antioxidant N-acetyl cysteine (NAC) for 48 h and cisplatin or the combination of metformin and cisplatin. NAC, a scavenger of ROS, inhibited the synergistic effects of the combination therapy on cell death (Figure S1D) and proliferation (Figure 2F), indicating that the synergistic effects of metformin and cisplatin depend on intracellular ROS levels. Moreover, metformin and specific HO-1 inhibitor ZnPPIX treatment (10 µM) significantly decreased the proliferation of A549/DDP and H838 cells compared to that following treatment with cisplatin alone (Figure S1E); this finding confirmed that HO-1 suppression is responsible for the antiproliferative effect of metformin. Overall, these results suggest that ROS-induced NSCLC cell death is regulated by the Nrf2/HO-1 signaling axis in chemotherapy and that metformin weakens the Nrf2/HO-1 axis and the ROS-detoxification system to further reverse chemoresistance.
Metformin inhibits the cisplatin-activated Nrf2/HO-1 axis via Raf-ERK inactivation
ERK1/2 have been reported to regulate Nrf2 expression via a keap1-independent mechanism (25). To evaluate the kinases upstream of metformin-abolished Nrf2 activation, the phosphorylation status of MAPKs with combination treatment with increasing concentrations of cisplatin was evaluated first. Cisplatin activated the phosphorylation of Raf and ERK1/2 in a dose-dependent manner, while metformin at the indicated concentration strongly suppressed the Nrf2/HO-1 axis and the phosphorylation of Raf-ERK1/2, which was activated by cisplatin (Figure 3A). Metformin caused a dose-dependent reduction in the protein levels of Nrf2 and HO-1 in A549/DDP and H838 cells. Similarly, the phosphorylation of Raf and ERK1/2 was dose-dependently reduced by metformin. However, the Keap1 protein level was not affected by metformin (Figure 3B).
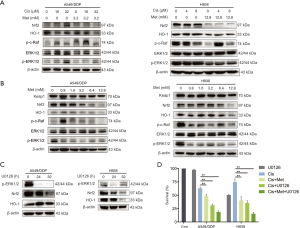
To further confirm the role of Raf-ERK signaling in the regulation of Nrf2, the two cell lines were treated with 30 µM U0126, a highly selective inhibitor of mitogen-activated protein kinase (MEK) 1 and MEK 2 that functions upstream of ERK1/2, for 24 and 32 h. U0126 strongly suppressed the expression of Nrf2 in a time-dependent manner (Figure 3C). Further, the combination treatment with U0126 and cisplatin for 48 h was found to have synergistic inhibitory effects on NSCLC cell proliferation (Figure 3D). Single treatment with U0126 (30 µM) had almost no inhibitory effect on the chemoresistant cell line (average survival rate, 97%), although it strongly inhibited cell proliferation in a combination treatment with 16 µM cisplatin (average survival rate, 31%). Taken together, these results indicate that inactivation of the Raf-ERK signaling pathway is involved in the metformin-mediated downregulation of Nrf2 and HO-1.
Metformin accelerates Nrf2 degradation by increased ubiquitin modification from its dephosphorylation
Nrf2 is mainly controlled through the regulation of protein turnover by ubiquitin-mediated proteasomal degradation and exhibits a half-life of approximately 30 min (27). Given that metformin strongly suppressed the protein level of Nrf2, even under cisplatin-activated conditions, we hypothesized that metformin can inhibit the mRNA expression of Nrf2, thus reducing the protein synthesis of Nrf2, and/or regulate Nrf2 protein at the PTM level. On the basis that a mechanism must underlie metformin-mediated Nrf2 suppression, we investigated the possibilities. A549/DDP and H838 cells were maintained in medium supplemented with cisplatin (8 and 1 µM, respectively) to stably activate Nrf2 expression and simulate combination therapy. Then, the two cell lines were treated with metformin (12.8 mM) and harvested at various time points. Western blotting revealed that the expression levels of Nrf2 and HO-1 were remarkably inhibited within 8 h of metformin treatment (Figure 4A). Conversely, qRT-PCR showed that metformin increased Nrf2 mRNA expression in both cell lines at all time points (Figure 4B). However, the results of qRT-PCR alone cannot explain the dramatic decrease in Nrf2 at the protein level. Therefore, we investigated whether the downregulation of Nrf2 by metformin could result from increased Nrf2 degradation. The degradation of Nrf2 was accelerated by co-treatment with metformin and CHX, an inhibitor of de novo protein synthesis (Figure 4C). When the Nrf2 protein levels with or without pretreatment with the proteasome inhibitor MG132 were compared in each group, a large gap in protein expression was found, which suggested that Nrf2 degradation in A549/DDP and H838 cells is dependent on the proteasome and that both metformin and U0126 have a strong ability to promote the degradation of enormous amounts of Nrf2 protein (Figure 4D).
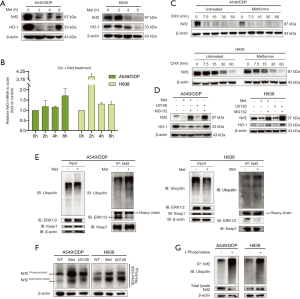
To analyze the ubiquitination status of Nrf2 in NSCLC cells with and without metformin treatment, a co-immunoprecipitation (co-IP) assay was performed. As Raf-ERK signaling is involved in metformin-mediated regulation of Nrf2, the amount of ERK1/2 that binds to Nrf2 was also analyzed. Nrf2 polyubiquitination was found to be increased in NSCLC cells treated with metformin compared with the control cells. Intriguingly, metformin had no effect on the binding of Keap1 and Nrf2, although it significantly reduced protein-protein interaction between ERK1/2 and Nrf2 (Figure 4E). Further, metformin and U0126 promoted the dephosphorylation of Nrf2 in NSCLC cells (Figure 4F). Next, fresh cell extract from NSCLC cells was treated with lambda phosphatase for 30 min, a protein phosphatase with activity towards phosphorylated serine, threonine, and tyrosine residues. Lambda phosphatase strongly increased ubiquitin modification of Nrf2 (Figure 4G), revealing that phosphorylated Nrf2 prevented its polyubiquitination. Overall, these results clearly demonstrate that metformin accelerates the ubiquitin-mediated proteolysis of Nrf2 in a Keap1-independent mechanism through preventing ERK1/2 phosphorylating Nrf2 extensively to restore polyubiquitination of Nrf2.
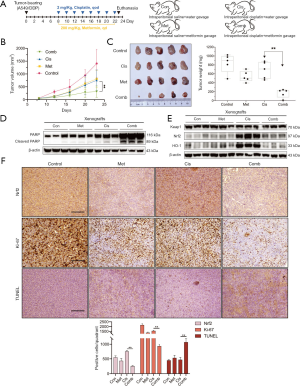
Combination therapy overcomes resistance to cisplatin in vivo
A chemotherapeutic resistance model was established using BALB/c mice. Figure 5A shows the experimental protocol and drug treatment protocols. The average tumor volume of the cisplatin group showed a considerable increase within the 23 days following tumor implantation, reaching 756 mm3, whereas that in the combination group exhibited a substantial decrease, with an average tumor volume of 312 mm3 (P<0.001; Figure 5B). The mice were weighed twice a week, and no weight loss was observed (Figure S2A,B). The median tumor weight in the combination group was remarkably reduced compared with that in the cisplatin group (708 vs. 220 mg; P<0.001; Figure 5C). Xenograft tumor tissues were randomly selected from three mice in each group. In the combination group, the levels of cleaved PARP were dramatically elevated and cisplatin-induced activation of the Nrf2/HO-1 signaling axis was inhibited (Figure 5D,E). The results of IHC staining for Nrf2 in xenograft tissues correlated with those of western blot analysis. Moreover, Ki-67 and TUNEL staining confirmed that combination treatment inhibited proliferation and increased apoptosis in NSCLC cells in vivo (Figure 5F). The findings from our in vivo experiments recapitulated the results obtained through our in vitro experiments.
Non-decreased Nrf2 expression in patients after neoadjuvant chemotherapy is associated with poor survival and chemoresistance in NSCLC
To elucidate whether the tumor Nrf2 status is related to neoadjuvant chemotherapeutic response and survival in NSCLC patients, Nrf2 expression was detected and scored in matched tumor tissues using IHC staining. The matched tumor samples were obtained from 50 NSCLC patients who underwent biopsy prior to receiving cisplatin-based neoadjuvant chemotherapy followed by surgical resection. The baseline characteristics of the patients with different responses to chemotherapy are shown in Table S1. The patients were divided into two groups according to their Nrf2 IHC staining scores (Figure 6A): the Nrf2 low group (score of 0 or 1) and the Nrf2 high group (score of 2 or 3). Patients with decreased Nrf2 scores after neoadjuvant chemotherapy were classified into the decreased Nrf2 group, and those with increased or unchanged Nrf2 scores were classified into the non-decreased Nrf2 group (Figure 6B). Low Nrf2 expression after chemotherapy was associated with an improved chemotherapeutic response (P<0.0001; Figure 6C, Fisher’s exact test).
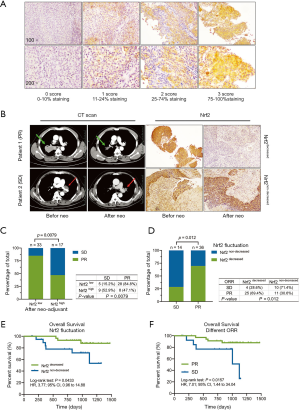
Next, the differences in Nrf2 expression before and after neoadjuvant chemotherapy, as well as the corresponding response to chemotherapy, were studied further. A higher proportion of patients in the non-decreased Nrf2 group had SD (71.4%) than PR (30.6%) (P=0.012; Figure 6D, Fisher’s exact test). Furthermore, fluctuation in the level of Nrf2 was significantly correlated with overall survival in this cohort (P=0.0433; Figure 6E, log-rank test). Patients who were less sensitive to chemotherapy had significantly poorer outcomes than PR patients (P=0.0157; Figure 6F, log-rank test). Collectively, these data demonstrated that variation in Nrf2 expression may be a crucial clinical marker for predicting the prognosis of patients who undergo cisplatin-based neoadjuvant chemotherapy before surgery.
Discussion
The biguanide metformin has been successfully used to treat millions of patients with type II diabetes worldwide. In addition to normalizing blood glucose levels, metformin has shown antitumor properties, which has led to many preclinical and clinical studies seeking to repurpose metformin as an anticancer agent. Data obtained so far indicate that metformin monotherapy shows strong anticancer effects in various cancers, including lung cancer (28-32). Recent studies have tended to combine metformin with chemotherapeutic drugs to enhance their cytotoxicity, reduce side effects, and reverse tumor resistance to these drugs (11,33). Combination strategies allow metformin to be used at a lower concentration and simultaneously improve the treatment efficacy of other agents.
Combinations of metformin and chemotherapeutic agents exert their effects through various mechanisms (10,34). Lin et al. reported that metformin inhibited STAT3 phosphorylation and mammalian target of rapamycin (mTOR) activity, thus enhancing sensitivity to cisplatin (12). They also found that metformin inhibited cisplatin-induced ROS production. Interestingly, our research showed that metformin augmented the cisplatin-induced increase in the ROS level, which was associated with decreased Nrf2 protein expression. The substantial accumulation of ROS leads to an imbalance in “redox capacity”, which disrupts redox homeostasis (35). Cisplatin remains a classic anticancer drug because it causes DNA crosslinking and is one of the strongest ROS generators. Elevated ROS levels can damage cellular components, resulting in a further increase in ROS levels, which induces cell cycle arrest and apoptosis (36). However, lung cancer cells have developed a strong antioxidant system to manage endogenous or exogenous ROS overproduction (37,38). As the core element of this antioxidant defense system, Nrf2 was observed to be sensitive to cisplatin and elevated by cisplatin in a dose-dependent manner. Metformin completely eliminated the detoxification ability of cisplatin and induced ROS-mediated oxidative burst during cellular processes. We demonstrated that the mechanism of metformin to reverse chemoresistance is based on oxidative damage caused by the rapid accumulation of ROS and a defective antioxidant system in lung cancer cell lines.
Metformin was reported to suppress Nrf2 protein levels in cancer cells via a Keap1-independent mechanism. Raf-ERK signaling attenuation was shown to be involved in the regulation of Nrf2 by metformin in the treatment of lung cancer (25). However, the p38 MAPK and JNK signaling pathways were not found to affect Nrf2 protein expression (21). Our study produced similar results. Accordingly, the mechanism underlying Nrf2 regulation by metformin is complicated. Although metformin has been shown to reduce Nrf2 mRNA levels in human hepatic carcinoma (25), no study to date has examined metformin-induced changes in Nrf2 mRNA expression in lung cancer cells. Intriguingly, we found that metformin elevated Nrf2 mRNA expression in a time-dependent manner, which was consistent with the sensitivity of lung cancer to foreign agents. However, metformin had entirely the opposite effect on Nrf2 protein and mRNA levels. PTMs, such as ubiquitylation, often result in rapid changes in Nrf2 protein expression.
Accumulating evidence suggests that several Nrf2 statuses, such as Nrf2 activation and subcellular localization, are tightly regulated by some types of PTM (17). Unphosphorylated Nrf2 stays in the cytoplasm, whereas the phosphorylated form is preferentially localized in the nucleus (39). Protein kinase C can phosphorylate Nrf2 at Ser-40 in its Neh2 domain and disrupt the association between Nrf2 and Keap1, thus resulting in the nuclear localization of Nrf2 (40). However, even when Nrf2 was phosphorylated at Ser-40, the Nrf2 protein was degraded by metformin or U0126 treatment (41). Ser-40 has been reported to not be necessary for the nuclear accumulation and increased stability of Nrf2 (42). Unlike metformin, butylated hydroxyanisole was shown to increase phosphorylation of the ERK1/2 signaling pathway, which is associated with Nrf2 release from Keap1 (26). Curiously, glycogen synthase kinase 3-beta was shown to phosphorylate the Neh6 region of Nrf2, facilitating its ubiquitination and subsequent proteasomal degradation (43).
Based on our results (Figure 7), the specific phosphorylation of Nrf2 at one or two phosphorylation sites does not affect the ubiquitination and subsequent degradation of Nrf2. Substantial phosphorylation of Nrf2 by ERK prevents the polyubiquitination of Nrf2, even though it binds to Keap1. When the remaining Keap1 is saturated by Nrf2, the phosphorylated Nrf2 becomes more resistant to degradation and accumulates freely in the cytoplasm, thus indirectly promoting the translocation of Nrf2 into the nucleus. Nrf2 contains over 100 serine, threonine, and tyrosine residues (44). Nrf2 contains many putative PTM sites, which complicates the identification of phosphorylation events. Changes to Nrf2 PTMs alter the protein levels and intracellular location of Nrf2, which should be carefully examined in lung cancer.
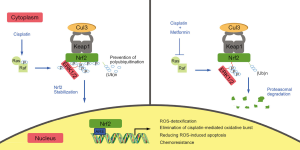
Nrf2 activation is common in cancer, and the transcription of its downstream genes promotes various cancer markers (24). Continuous activation of Nrf2 promotes malignant transformation, cancer progression and resistance to chemotherapy and is considered a poor prognostic factor (20). These malignant phenotypes have been observed in lung cancer, leading to the definition of Nrf2 as an oncogene (19,45-47). Solis et al.’s study evaluated Nrf2 expression in 63 NSCLC patients who received adjuvant treatment and reported that nuclear Nrf2 expression was associated with worse progression-free survival (48). Another study found a marginal but significant trend toward the benefit of chemotherapy in lung squamous carcinoma in a group with low expression of Nrf2 and its associated genes (49).
Our study demonstrates, for the first time, that variation in Nrf2 expression before and after neoadjuvant chemotherapy is strongly associated with poor patient prognosis in NSCLC. As our research showed that low Nrf2 expression in tumor tissues after neoadjuvant was correlated with a better ORR, fluctuations in Nrf2 expression should be an effective clinical biomarker for predicting the ORR and prognosis of patient who receive cisplatin-based chemotherapy, in view of the dynamic changes in Nrf2 responses to foreign stimulation. The emerging role of Nrf2 in NSCLC suggests that a strategy in which Nrf2 activation is inhibited could help to overcome chemoresistance and enhance the sensitivity of NSCLC to chemotherapeutic drugs or even chemotherapy combined with immunotherapy (50,51).
Conclusions
Combination therapy with metformin and cisplatin strongly decreased the protein levels of Nrf2 by inhibiting the Ras/Raf/ERK pathway in A549/DDP and H838 NSCLC cells, thereby eliminating the detoxification effect of Nrf2 and promoting ROS-mediated apoptosis in NSCLC. Mechanistically, metformin increased Nrf2 polyubiquitination by attenuating ERK-mediated phosphorylation of Nrf2, subsequently facilitating its proteasomal degradation. Additionally, fluctuating levels of Nrf2 were discovered to confer a strong predictive ability for chemotherapeutic response and survival outcomes in NSCLC patients who underwent neoadjuvant chemotherapy.
Acknowledgments
The authors thank Dr. Qiaonan Shan and Dr. Sunbin Ling for technical assistance.
Funding: We gratefully acknowledge funding from the National Natural Science Foundation of China (No. 81900099), the National Key R&D Program of China (No. 2017YFC0113500), the Key Subject of Zhejiang Province Traditional Chinese Medicine (No. 2017-XK-A33), the Special Project for Major Science and Technology of Zhejiang Province (No. 2020C03058), the Research Center for Diagnosis and Treatment of Lung Neoplasms of Zhejiang Province (JBZX-202007), and the Wu Jieping Medical Foundation (320.320.2730.1869).
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at http://dx.doi.org/10.21037/tlcr-20-1072
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tlcr-20-1072
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tlcr-20-1072). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). Patients were informed that the specimens were stored by the hospital and potentially used for scientific research, and signed informed consent to participants was waived by the Ethics Committee. This study was approved by the Clinical Research Ethics Committee of the First Affiliated Hospital, College of Medicine, Zhejiang University (Approval number: 2020-13). All animal experiments were approved by the Committee of Animal Experimental Ethical Inspection of the First Affiliated Hospital, College of Medicine, Zhejiang University (Reference number: 2019-1232), and followed by the institutional guidelines for animal care and use.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin 2020;70:7-30. [Crossref] [PubMed]
- Torre LA, Siegel RL, Jemal A. Lung Cancer Statistics. Adv Exp Med Biol 2016;893:1-19. [Crossref] [PubMed]
- Facchinetti F, Tiseo M. Digging into lorlatinib resistance in ALK-positive lung cancer: an editorial. Chin Clin Oncol 2019;8:S2. [Crossref] [PubMed]
- Hu Z, Li M, Chen Z, et al. Advances in clinical trials of targeted therapy and immunotherapy of lung cancer in 2018. Transl Lung Cancer Res 2019;8:1091-106. [Crossref] [PubMed]
- Liao WY, Chen JH, Wu M, et al. Neoadjuvant chemotherapy with docetaxel-cisplatin in patients with stage III N2 non-small-cell lung cancer. Clin Lung Cancer 2013;14:418-24. [Crossref] [PubMed]
- Dasari S, Tchounwou PB. Cisplatin in cancer therapy: molecular mechanisms of action. Eur J Pharmacol 2014;740:364-78. [Crossref] [PubMed]
- Galluzzi L, Senovilla L, Vitale I, et al. Molecular mechanisms of cisplatin resistance. Oncogene 2012;31:1869-83. [Crossref] [PubMed]
- Liang J, Lu T, Chen Z, et al. Mechanisms of resistance to pemetrexed in non-small cell lung cancer. Transl Lung Cancer Res 2019;8:1107-18. [Crossref] [PubMed]
- Ganesh S, Iyer AK, Weiler J, et al. Combination of siRNA-directed Gene Silencing With Cisplatin Reverses Drug Resistance in Human Non-small Cell Lung Cancer. Mol Ther Nucleic Acids 2013;2:e110. [Crossref] [PubMed]
- Yousef M, Tsiani E. Metformin in Lung Cancer: Review of in Vitro and in Vivo Animal Studies. Cancers (Basel) 2017;9:45. [Crossref] [PubMed]
- Peng M, Darko KO, Tao T, et al. Combination of metformin with chemotherapeutic drugs via different molecular mechanisms. Cancer Treat Rev 2017;54:24-33. [Crossref] [PubMed]
- Lin CC, Yeh HH, Huang WL, et al. Metformin enhances cisplatin cytotoxicity by suppressing signal transducer and activator of transcription-3 activity independently of the liver kinase B1-AMP-activated protein kinase pathway. Am J Respir Cell Mol Biol 2013;49:241-50. [Crossref] [PubMed]
- Yasmeen A, Beauchamp MC, Piura E, et al. Induction of apoptosis by metformin in epithelial ovarian cancer: involvement of the Bcl-2 family proteins. Gynecol Oncol 2011;121:492-8. [Crossref] [PubMed]
- Zheng L, Yang W, Wu F, et al. Prognostic significance of AMPK activation and therapeutic effects of metformin in hepatocellular carcinoma. Clin Cancer Res 2013;19:5372-80. [Crossref] [PubMed]
- Sporn MB, Liby KT. NRF2 and cancer: the good, the bad and the importance of context. Nat Rev Cancer 2012;12:564-71. [Crossref] [PubMed]
- Ray PD, Huang BW, Tsuji Y. Reactive oxygen species (ROS) homeostasis and redox regulation in cellular signaling. Cell Signal 2012;24:981-90. [Crossref] [PubMed]
- Bryan HK, Olayanju A, Goldring CE, et al. The Nrf2 cell defence pathway: Keap1-dependent and -independent mechanisms of regulation. Biochem Pharmacol 2013;85:705-17. [Crossref] [PubMed]
- Best SA, Sutherland KD. "Keaping" a lid on lung cancer: the Keap1-Nrf2 pathway. Cell Cycle 2018;17:1696-1707. [Crossref] [PubMed]
- Hammad A, Namani A, Elshaer M, et al. "NRF2 addiction" in lung cancer cells and its impact on cancer therapy. Cancer Lett 2019;467:40-9. [Crossref] [PubMed]
- Furfaro AL, Traverso N, Domenicotti C, et al. The Nrf2/HO-1 Axis in Cancer Cell Growth and Chemoresistance. Oxid Med Cell Longev 2016;2016:1958174. [Crossref] [PubMed]
- Zhang J, Jiao K, Liu J, et al. Metformin reverses the resistance mechanism of lung adenocarcinoma cells that knocks down the Nrf2 gene. Oncology Letters 2018;16:6071-80. [Crossref] [PubMed]
- Do MT, Kim HG, Choi JH, et al. Metformin induces microRNA-34a to downregulate the Sirt1/Pgc-1α/Nrf2 pathway, leading to increased susceptibility of wild-type p53 cancer cells to oxidative stress and therapeutic agents. Free Radic Biol Med 2014;74:21-34. [Crossref] [PubMed]
- Yu C, Jiao Y, Xue J, et al. Metformin Sensitizes Non-small Cell Lung Cancer Cells to an Epigallocatechin-3-Gallate (EGCG) Treatment by Suppressing the Nrf2/HO-1 Signaling Pathway. Int J Biol Sci 2017;13:1560-9. [Crossref] [PubMed]
- Cloer EW, Goldfarb D, Schrank TP, et al. NRF2 Activation in Cancer: From DNA to Protein. Cancer Res 2019;79:889-98. [Crossref] [PubMed]
- Do MT, Kim HG, Khanal T, et al. Metformin inhibits heme oxygenase-1 expression in cancer cells through inactivation of Raf-ERK-Nrf2 signaling and AMPK-independent pathways. Toxicol Appl Pharmacol 2013;271:229-38. [Crossref] [PubMed]
- Yuan X, Xu C, Pan Z, et al. Butylated hydroxyanisole regulates ARE-mediated gene expression via Nrf2 coupled with ERK and JNK signaling pathway in HepG2 cells. Mol Carcinog 2006;45:841-50. [Crossref] [PubMed]
- Rojo AI, Medina-Campos ON, Rada P, et al. Signaling pathways activated by the phytochemical nordihydroguaiaretic acid contribute to a Keap1-independent regulation of Nrf2 stability: Role of glycogen synthase kinase-3. Free Radic Biol Med 2012;52:473-87. [Crossref] [PubMed]
- Ashinuma H, Takiguchi Y, Kitazono S, et al. Antiproliferative action of metformin in human lung cancer cell lines. Oncol Rep 2012;28:8-14. [PubMed]
- Mallik R, Chowdhury TA. Metformin in cancer. Diabetes Res Clin Pract 2018;143:409-19. [Crossref] [PubMed]
- Coyle C, Cafferty FH, Vale C, et al. Metformin as an adjuvant treatment for cancer: a systematic review and meta-analysis. Ann Oncol 2016;27:2184-95. [Crossref] [PubMed]
- Faria J, Negalha G, Azevedo A, et al. Metformin and Breast Cancer: Molecular Targets. J Mammary Gland Biol Neoplasia 2019;24:111-23. [Crossref] [PubMed]
- Ma Y, Guo FC, Wang W, et al. Kras gene mutation as a predictor of cancer cell responsiveness to metformin. Mol Med Rep 2013;8:763-8. [Crossref] [PubMed]
- Zhang J, Wu J, He Q, et al. The prognostic value of metformin for advanced non-small cell lung cancer: a systematic review and meta-analysis. Transl Lung Cancer Res 2018;7:389-96. [Crossref] [PubMed]
- Tseng SC, Huang YC, Chen HJ, et al. Metformin-mediated downregulation of p38 mitogen-activated protein kinase-dependent excision repair cross-complementing 1 decreases DNA repair capacity and sensitizes human lung cancer cells to paclitaxel. Biochem Pharmacol 2013;85:583-94. [Crossref] [PubMed]
- Kim SJ, Kim HS, Seo YR. Understanding of ROS-Inducing Strategy in Anticancer Therapy. Oxid Med Cell Longev 2019;2019:5381692. [Crossref] [PubMed]
- Marullo R, Werner E, Degtyareva N, et al. Cisplatin induces a mitochondrial-ROS response that contributes to cytotoxicity depending on mitochondrial redox status and bioenergetic functions. PLoS One 2013;8:e81162. [Crossref] [PubMed]
- Wang J, Yi J. Cancer cell killing via ROS: to increase or decrease, that is the question. Cancer Biol Ther 2008;7:1875-84. [Crossref] [PubMed]
- Trachootham D, Alexandre J, Huang P. Targeting cancer cells by ROS-mediated mechanisms: a radical therapeutic approach? Nat Rev Drug Discov 2009;8:579-91. [Crossref] [PubMed]
- Apopa PL, He X, Ma Q. Phosphorylation of Nrf2 in the Transcription Activation Domain by Casein Kinase 2 (CK2) Is Critical for the Nuclear Translocation and Transcription Activation Function of Nrf2 in IMR-32 Neuroblastoma Cells. J Biochem Mol Toxicol 2008;22:63-76. [Crossref] [PubMed]
- Huang HC, Nguyen T, Pickett CB. Phosphorylation of Nrf2 at Ser-40 by protein kinase C regulates antioxidant response element-mediated transcription. J Biol Chem 2002;277:42769-74. [Crossref] [PubMed]
- Wang X, Li R, Zhao X, et al. Metformin Promotes HaCaT Cell Apoptosis through Generation of Reactive Oxygen Species via Raf-1-ERK1/2-Nrf2 Inactivation. Inflammation 2018;41:948-58. [Crossref] [PubMed]
- Bloom DA, Jaiswal AK. Phosphorylation of Nrf2 at Ser40 by protein kinase C in response to antioxidants leads to the release of Nrf2 from INrf2, but is not required for Nrf2 stabilization/accumulation in the nucleus and transcriptional activation of antioxidant response element-mediated NAD(P)H: quinone oxidoreductase-1 gene expression. J Biol Chem 2003;278:44675-82. [Crossref] [PubMed]
- Rada P, Rojo AI, Chowdhry S, et al. SCF/{beta}-TrCP promotes glycogen synthase kinase 3-dependent degradation of the Nrf2 transcription factor in a Keap1-independent manner. Mol Cell Biol 2011;31:1121-33. [Crossref] [PubMed]
- Rada P, Rojo AI, Evrard-Todeschi N, et al. Structural and functional characterization of Nrf2 degradation by the glycogen synthase kinase 3/beta-TrCP axis. Mol Cell Biol 2012;32:3486-99. [Crossref] [PubMed]
- Yang X, Wang D, Ma Y, et al. Continuous activation of Nrf2 and its target antioxidant enzymes leads to arsenite-induced malignant transformation of human bronchial epithelial cells. Toxicol Appl Pharmacol 2015;289:231-9. [Crossref] [PubMed]
- Qian Z, Zhou T, Gurguis CI, et al. Nuclear factor, erythroid 2-like 2-associated molecular signature predicts lung cancer survival. Sci Rep 2015;5:16889. [Crossref] [PubMed]
- Yu S, Cheng C, Wang J, et al. Loss of Beclin1 Expression and Nrf2 Overexpression are Associated with Poor Survival of Patients with Non-Small Cell Lung Cancer. Anticancer Agents Med Chem 2018;18:1680-7. [Crossref] [PubMed]
- Solis LM, Behrens C, Dong W, et al. Nrf2 and Keap1 abnormalities in non-small cell lung carcinoma and association with clinicopathologic features. Clin Cancer Res 2010;16:3743-53. [Crossref] [PubMed]
- Cescon DW, She D, Sakashita S, et al. NRF2 Pathway Activation and Adjuvant Chemotherapy Benefit in Lung Squamous Cell Carcinoma. Clin Cancer Res 2015;21:2499-505. [Crossref] [PubMed]
- Tian Y, Liu Q, He X, et al. Emerging roles of Nrf2 signal in non-small cell lung cancer. J Hematol Oncol 2016;9:14. [Crossref] [PubMed]
- Zhang B, Ma Z, Tan B, et al. Targeting the cell signaling pathway Keap1-Nrf2 as a therapeutic strategy for adenocarcinomas of the lung. Expert Opin Ther Targets 2019;23:241-50. [Crossref] [PubMed]
(English Language Editor: J. Reynolds)


