Cholesterol synthesis disruption combined with a molecule-targeted drug is a promising metabolic therapy for EGFR mutant non-small cell lung cancer
Introduction
Lung cancer remains the most frequently diagnosed cancer and is the leading cause of cancer deaths worldwide. Non-small cell lung cancer (NSCLC) is the most common and accounts for 80% to 85% of all cases (1,2). Such patients are usually treated with platinum-doublet chemotherapy, which has helped to prolong overall survival, however, patients’ prognosis with advanced or metastatic NSCLC is poor.
Recently, developments in targeted therapy have provided new possibilities for NSCLC patients. Epidermal growth factor receptor (EGFR) mutations are the most common driver mutations in NSCLC and (3) first-generation, and second-generation EGFR-TKIs (EGFR-tyrosine kinase inhibitors) such as gefitinib, erlotinib, and afatinib have significantly extended progression-free survival (PFS) relative to chemotherapy, thus are considered the standard of care for patients with sensitizing EGFR mutations (4-6). As a result of the FLAURA study, the standard of care has shifted to third-generation EGFR-TKI osimertinib, as it markedly elongated PFS and overall survival compared with gefitinib, or erlotinib as a second-line treatment (7,8). Yet, acquired resistance inevitably develops against all these EGFR-TKIs due to secondary EGFR mutations, alternative pathway activation and or histology/phenotypic transformation (9). Therefore, novel treatment strategies are urgently needed to improve the outcomes of patients with sensitizing EGFR mutations.
Intriguingly, the therapeutic potential in targeting cholesterol in cancer development has gained attention (10). Low-density lipoprotein receptor (LDLR), is the key mediator of cholesterol uptake and has been identified as a novel metabolic target that limits the progression of several cancers, including pancreatic ductal adenocarcinoma (11), breast cancer (12), liver cancer (13) and bladder cancer (14). Of note, the targeting of LDLR potently promoted tumor cell death in an in vivo glioblastoma model (15). As such, we ask whether LDLR’s role in EGFR-mutant NSCLC might be a legitimate a therapeutic target.
Here, we investigate the association between EGFR mutations and LDLR, and the effect of cholesterol disruption on cell proliferation and growth in NSCLC cell lines. Lipid metabolism is a very complicated process in vivo (16). In vitro experiments can initially explore the sensitivity and reactivity of drugs to tumor cells. The therapeutic effects of drugs on tumors need to be reflected in the body. After all, we study tumor treatments that are ultimately used in humans. We used the characteristics of fast passage, fast growth, and similarity to humans in transplanted mice to simulate the real metabolism of drugs in the body to simulate drugs (atorvastatin and gefitinib/osimertinib) in patients with EGFR mutations. The animal xenograft model therapeutic effect of non-small cell lung cancer on human body is conducive to exploring related metabolic treatment models. We also evaluate the anti-tumor activity of a novel treatment strategy: the combination of an EGFR-TKI and a statin in EGFR mutant cell lines and in in vivo models.
We present the following article in accordance with the ARRIVE reporting checklist (available at http://dx.doi.org/10.21037/tlcr-20-812).
Methods
Cell lines and cell culture
H1993, A549, PC9, HCC827 and H1975 are human NSCLC cell lines, and Beas-2B an immortalized human lung bronchial epithelial cell line. We designed EGFR wild type cell lines (H1993, A549, Beas-2B) as control groups and EGFR mutant cell lines (PC9, HCC827, H1975) as experimental groups. Beas-2B Vector (17) is as control group while Beas-2B H is as experimental group. H1975 Mock is as control group and H1975 Gefitinib and 9291 are as experimental group. Repeating 3 times with the same method and in the same time period for each experiment to minimize the effects of subjective bias. Beas-2B-CAG-EGFR-Ex19del is a stable cell line and a CAG promoter that mimics the high expression of EGFR exon 19 deletion mutation, and Beas-2B-CAG-vector is a control. Beas-2B-EGFR-L858R is a cell line with an EGFR-L858R expression induced by doxycycline. The cell lines were cultured in DMEM supplemented with 10%, 1% fetal bovine serum (FBS) or 1% lipoprotein depleted fetal bovine serum (LPDS BS023201) and antibiotics (10,000 U/mL penicillin and 10 µg/mL streptomycin). All cells were maintained in a humidified incubator at 37 °C with 5% carbon dioxide.
Antibodies and reagents
Antibodies were used against the following: LDLR (Abcam, EP1553Y), SREBP-1 (Abcam, ab28481); phospho-EGFR (Tyr1068), phospho-p44/42MAPK (ERK1⁄2) (Thr202⁄Tyr204), p-Akt (Ser473), and β-actin (Sigma). Reagents used were EGF (recombinant human epidermal growth factor, cyt-217, PROSPEC Corporation, CA), low density lipoprotein (Abcam, LDL ab91115), atorvastatin calcium salt trihydrate (Sigma, PZ0001), osimertinib (Selleck, AZD9291 S7297), gefitinib (AZD1839 S1025), ERK1/2 inhibitor (SCH772984), and AKT1/2/3 inhibitor (MK-2206 2HCL), all were purchased from Selleckchem (Shanghai, China).
Western blot analysis
After harvesting, cells were suspended in a RIPA (Radio Immunoprecipitation Assay) lysis buffer (Thermo, Hercules, CA) containing the phosphatase inhibitor cocktail Phos STOP (Roche, Mannheim, Germany) and a protease inhibitor cocktail (Sigma-Aldrich Corporation, St Louis, MO) and incubated on ice for 30 minutes. The cell lysates were then centrifuged at 12,000 rpm, at 4 °C for 20 minutes. The protein supernatant was determined using a Thermo Protein Assay Reagent (Thermo, Hercules, CA). Equal amounts of protein extracts per well were separated by 8% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred into polyvinylidene difluoride membranes (Bio-Rad Laboratories, Hercules, CA). Blocked with 5% non-fat dried milk and diluted with TBST (Tris buffer solution with Tween, a mixture of Tris-HCl 20, NaCl 150 and 0.1% Tween-20, pH 7.5) at room temperature for 1 h, the membranes were probed with primary antibodies against LDLR (EP1553Y), phospho-EGFR (Tyr1068), β-actin (Sigma), phospho-p44/42MAPK (ERK1⁄2) (Thr202⁄Tyr204), phospho-Akt (Ser473), and SREBP-1 (ab28481) overnight at 4 °C. They were then washed three times, and the membranes further immunoblotted with horseradish peroxidase and conjugated donkey anti-rabbit secondary antibodies (1:2,000, Santa Cruz, CA) at room temperature for 1 h. Signals were detected using a super signal West Dura Luminol/Enhancer solution (Thermo, Hercules, CA).
MTT assays
Cells (2,000 cells/well) were seeded in 96 wells and were treated with different concentrations or types of serum and different concentrations of atorvastatin for 5 days. We tested the viability of cells with a CCK8 kit according to the manufacturer’s instructions (Cell Counting Kit-8, Dojindo Co., Japan). The viability of the control group was normalized to 100%. Cell viability rate (%) = (OD value of treatment group/OD value of control group) × 100%. Cell survival numbers were determined using Countess II (Life technologies). Dead cells were measured using trypan blue (EF T10282) exclusion assays.
SiRNA assay
Beas-2B and PC-9 cells (2.0×105 cells/well) were seeded in 25 cm2 well plates for 24 h, the growth of cells in the culture well plates reached approximately 80%, and were then replaced with a culture medium with fresh DMEM, and antibiotics (10,000 U/mL penicillin and 10 µg/mL streptomycin) but without FBS. Two Rnase-free, sterile EP tubes were used to add 250 µL Opti-MEM transfection optimized medium, respectively. One tube was added to 17 µL of 10 µM siRNA, and the other tube to 17 µL of Lipofectamine® RNAiMAX transfection reagent, and mixed gently. After that, the two EP tubes were mixed gently for 10 minutes and then the siRNA mixture was added to each well plate, and incubated at 37 °C for 6 h. The medium was then replaced with 10% FBS and incubated for 48 hours at 37 °C. After that, the cells were lysed for western blot analysis.
Pathological hematoxylin-eosin staining
The tissue sections from tumor resected animal xenograft models were deparaffinized. Tissue sections were stained with hematoxylin (BA4041 Baso) and eosin (BA4022 Baso). Images were captured using a photomicroscope and analyzed.
Immunohistochemical staining
Tissue sections from resected tumor animal xenograft models were deparaffinized. After citrate antigen repair and blocking, the samples were incubated with aKi67 antibody (1/400, ab16667 abcam) at 4 °C overnight in an incubator. The samples were then incubated with a goat anti-rabbit secondary antibody IgG H&L (1/400, HRP, ab6721 abcam) at 4 °C overnight in an incubator. Tissue sections were stained with DAB, and again with hematoxylin (BA4041 Baso). Images were captured using a photomicroscope and analyzed.
Animal xenograft model
Four-week-old, similar in weight (18±2g), female nude mice BALB/c-Foxn1nu/Nju (D000521) (Beijing Vital River Laboratory Animal Technology Co., Ltd.) received a subcutaneous injection of oncogenesis with 4×106 cells of T25 (HCC827 cells and H1975 cells, both were NSCLC cells) by the right armpit to study the effects on tumor growth (18). The 24 mice were numbered from 1 to 24 and randomly divided into eight groups: HCC827 normal saline group (control group), HCC827 atorvastatin group, HCC827 gefitinib group, HCC827 gefitinib with atorvastatin group (combination group), H1975 normal saline group (control group), H1975 atorvastatin group, H1975 osimertinib group, H1975 osimertinib with atorvastatin group, each with 6 mice in a cage. After 1 week of adaptive feeding, the mice received the subcutaneous injection on September 30th, and were tube fed at the same time (from 9:00 am to 10:00 am) every day with normal saline, atorvastatin (Y00000012187 Pfizer Pharmaceuticals Limited) (50 mg/kg QD) (19), gefitinib (AstraZeneca Pharmaceuticals) (5 mg/kg QD) (20), osimertinib (Y00000012800 AstraZeneca Pharmaceuticals) (4 mg/kg QD) (21) and the combination (gefitinib 5 mg/kg QD and atorvastatin 50 mg/kg QD or osimertinib 4 mg/kg QD and atorvastatin 50 mg/kg QD) from October 8th to the 28th, 2019. Mice were fed in plastic cage with sealed air filter device in SPF class housing of laboratory. Each cage is a unit by itself. This cage can be placed on a standard cage rack in a general laboratory. The filter material is in sheet form and is made of sparsely spun (but non-woven) polyester fiber. All cages were sterilized by high-pressure steam. When using it and feeding management, it was carried out in a protective cover, which contains positive pressure filtered air. And it was laminar (non-turbulent) and unidirectional. All items that came into contact with mice (normal SPF feed and other items) were sterilized. Drinking water was treated to remove bacteria. The full-time staff wore sterile surgical masks, hats, gloves, and back coats. When touching animals, they used rubber-covered tweezers which were immersed in denatured ethanol. The light cycle conditions are 12 hours bright/12 hours dark, without windows and artificial lighting, so that the light is constant throughout the year. The temperature is 26–28 °C (78–820 F), and the relative humidity is 40–60%. Ventilate 10–15 times per hour, and use a high-efficiency filter (99.9%) to ensure good air filtration and remove particles larger than 0.3 µm in the air. In the process of feeding, managing and using animals, effective measures have been taken to protect experimental animals from unnecessary harm, hunger, thirst, fright, torture, disease and pain, to ensure that animals can achieve natural behaviors and receive good management and care. It provides a clean and comfortable living environment, provides adequate and healthy food and drinking water, and avoids or reduces pain and suffering. The order in which animals in different experimental groups were processed was in accordance with the above grouping. All animals in the experimental group were processed and the tumor volumes were evaluated. The weight of the mice, tumor width and length were measured every two days (tumor volume was equal to the square of tumor width and then times tumor length and times π/6). On October 28th, 2019, the mice were placed on a wet ice slurry to reduce sensitivity prior to CO2 euthanasia (22). The tumors were then dissected and weighed. All these were carried out in SPF class housing of laboratory according to the Laboratory Guidelines for Animal Care. Pictures were taken to compare morphologic changes. Pathological slides were made for HE staining and Ki67 staining was used to analyze the malignancy of the tumors after treatment. This animal experiments were performed under a project license [SCXK(Shanghai) 20170011#] granted by the Veterinäramt des Kantons Zürich, Switzerland, in compliance with the Swiss Animal Protection Act (TSchG) and the Swiss Animal Protection Ordinance (TSchV).
Data analysis
Results are shown as mean ± SEM. Representative results shown are from triple independent experiments. Other comparisons in cell growth and proliferation assays and cell death were performed using two-tailed Student’s t test. A P value of <0.05 was considered statistically significant. SigmaPlot 10.0 was used to make the figures.
Results
EGFR promotes LDLR expression in non-small cell lung cancer cell lines
Highly proliferative cancer cells activate the endogenous synthesis of lipids and increase their uptake of lipids and lipoproteins showing strong lipid affinity (15). To study the relationship between EGFR and LDLR, we detected their protein expression in human NSCLC cell lines. Namely, H1993, A549, HCC827, PC9, H1975, and an immortalized human lung bronchial epithelial cell line, Beas-2B. Detection revealed that the protein level of LDLR in both the EGFR-wild type cell lines (A549 and H1993) and Beas-2B were significantly lower than in the EGFR mutant NSCLC cell lines (PC-9, HCC827, and H1975). Beta-actin was used to verify equal loading (Figure 1A). EGFR activation medium recombinant human epidermal growth factor (EGF) and doxycycline (23) (DOX) and a CAG promoter (24) were then used to stably express high levels of EGFR to analyse LDLR expression. Beas-2B cells were treated with 40 ng/mL of EGF for 48 and 72 hours. LDLR expression was upregulated at 48 and 72 hours, but not upregulated without EGF. After 72 hours, EGF stimulation was evident (Figure 1B). A plasmid CAG with higher promoter activity was then used to express the corresponding level of EGFR with exon 19 deletion(Ex19del), which significantly increased the protein expression level of LDLR, in contrast with the vector (Figure 1C). We employed a plasmid with a L858R mutation induced by doxycycline (2 µg/mL) for 0, 4, 8, and 12 hours to express the protein-escalation level of p-EGFR. The protein expressions of LDLR were as gradual and incremental as the p-EGFR (Figure 1D). We found that LDLR was highly expressed in NSCLC along with high expression of EGFR caused by the exon 19 deletion and the L858R mutation, including in the Beas-2B cells, that were stimulated by EGF. These results strongly show LDLR expression can be induced by different forms of EGFR activation, illustrating a relationship between EGFR mutation and low density lipoprotein cholesterol metabolism, and that EGFR upregulates LDLR in non-small cell lung cancer cell lines.

Suppressing EGFR reduces LDLR expression in non-small cell lung cancer cell lines
EGFR-TKIs have been shown to target EGFR mutations and lower protein expression. So, we applied gefitinib (a first-generation EGFR-TKI that targets the EGFR-Ex19del and EGFR-L858R mutations) and AZD 9291 (a third-generation EGFR-TKI that targets the T790M mutation) to assess whether they can reverse the up-regulation of LDLR and p-EGFR. In Figure 2A, both LDLR and p-EGFR up-regulation were reversed by gefitinib with EGF stimulation at 48 and 72 hours in Beas-2B cells. Figure 2B illustrates the same result, in which the up-regulation of LDLR and p-EGFR induced by 2 µg/mL doxycycline was reversed in Beas-2B-EGFR-L858R cells. In the other NSCLC EGFR-Ex19del mutation cell lines (PC-9 and HCC827) the protein expression of LDLR and p-EGFR clearly decreased using a dose escalation manner of 0 to 0.4 µM of gefitinib (Figure 2C,D). We tried the same protocol on the H1975 cell line containing the EGFR-T790M mutation that is resistant to gefitinib. Its p-EGFR expression level was the same pre- and post- gefitinib. Accordingly, the LDLR expression level remained unchanged due to the unchanged EGFR activity. By contrast, we used the AZD9291 to treat the H1975 cell line, here, the protein expression of p-EGFR was reversed, leading to a lower protein expression level of LDLR (Figure 2E). Together, these results demonstrate that suppressing EGFR activation can reduce LDLR expression of activated EGFR in NSCLC. Figures 1 and 2 show that the protein expression of LDLR is consistent with the expression of EGFR protein.

EGFR mutation cells count on lipids to survive and grow
Having ascertained that p-EGFR promotes LDLR expression, we explored what influences the proliferation and survival of EGFR-mutant cells when a low-density lipoprotein (LDL) was depleted from fetal bovine serum. We cultured EGFR-wild type and EGFR-mutant cells in 1% FBS and 1% LPDS (lipoprotein deficient serum). LDL was added to 1% LPDS as a rescue comparison. All cells were cultured for 5 days. In Figure 3A, the growth and proliferation of the H1993 cell line in 1% LPDS medium was marginal compared with the 1% FBS medium. However, for the PC-9 and H1975 cell lines, the growth and morphology were notable. The proliferation was significantly inhibited in the 1% LPDS medium. We found that LDL was required for EGFR-mutant cells to survive and grow and that it was able to return back to its baseline level in the 1% LPDS + LDL groups.

We measured the cell survival numbers in the 1% FBS, 1% LPDS and 1% LPDS + LDL media using a hemocytometer with a trypan blue assay. As shown in Figure 3B, the depletion of LDL caused EGFR-mutant cells to die and reduced the number of EGFR-mutant cells in the 1% LPDS medium. These results show that EGFR-mutant cells are dependent on LDL for their proliferation and survival, indicating that extracellular cholesterol is integral for EGFR mutation cells to grow. To further assess the importance of cholesterol synthesis on the growth and survival status of EGFR-mutant cells, we treated cells with a dose escalation of atorvastatin from 0 to 12.5 µM in 10% FBS medium for 5 days to suppress cholesterol synthesis. We seeded the cells in 96-well plates at a density of 2×103 cells/well in DMEM supplemented with non-fetal bovine serum for one night, and then transferred them into 10% FBS medium the next day and marked it as day 0. A cell Counting Kit-8 (Dojindo Co., Japan) assay was used to detect cell survival on day 5. The results are displayed in Figure 3C. The line chart shows the number of H1993 (EGFR-wild type) cells had slightly reduced, while PC-9, and H1975’s (EGFR-mutant type) had drastically decreased following a dose-dependent manner of atorvastatin. We seeded the cells in 6-well plates at a density of 4×104 cells/well to repeat the above phenomenon. The cell behavior is shown in Figure 3D. It demonstrates that NSCLC cells with an EGFR mutation can cause cell death in vitro using a certain dose of atorvastatin. Thus, we conclude significantly more lipids are needed for EGFR mutation cells to survive and grow.
Without extracellular cholesterol, atorvastatin treatment restrains NSCLC cells with EGFR mutations from proliferating and growing
The proliferation and growth of EGFR mutational cells ceases if the supply of cholesterol in the cells’ growth process is cut-off. We seeded the three cell lines, H1993, PC-9 and H1975 in serum-free DMEM at a density of 2×103 cells/well in 96-well plates overnight. They were then cultured in 1% FBS and 1% LPDS. The next day, we transferred the cells into 1% FBS and 1% LPDS media. We added atorvastatin with different doses to those media and marked them as day 0. We detected cell survival numbers with a CCKit8 (Cell Counting Kit-8, Dojindo Co., Japan) every day, for 5 consecutive days. Line charts shows the cells’ proliferation tendencies and histograms, the cell survival’s cumulative effects on the fifth day (Figure 4A,B,C). We discovered that a low dose of atorvastatin alone could inhibit EGFR-mutant cells’ proliferation and incite these cells to die over time. The effect was dose-dependent (Figure 4A). Contrasting Figure 4A with Figure 4B, shows that under the same concentration of atorvastatin treatment, the proliferation and growth of EGFR mutant cell lines in LPDS were more inhibited. In regards to LPDS combined with the inhibition of cholesterol synthesis treatment atorvastatin, it greatly suppressed proliferation and promoted the death of EGFR-mutant cells (Figure 4B). Figure 4C shows cells’ total survival status on the fifth day of cell culture in 1% FBS and 1% LPDS with different low doses of atorvastatin treatment. This validates that when there is a lack of extracellular cholesterol, atorvastatin treatment restrains NSCLC cells with an EGFR mutation from proliferating and growing.

EGFR signaling upregulates LDLR expression through the SREBP-1 dependent pathway
To discover the hidden molecular mechanism of LDLR upregulation via EGFR activation, we used western blot to test the downstream pathways of EGFR. Immunoblot showed LDLR expression was mediated by AKT, but not by ERK (Figure 5A,B,C), and that SREBP-1, being an upstream of LDLR protein expression, affects LDLR protein expression. In Figure 5D, the expression of LDLR was high without an AKT inhibitor but was low with an AKT inhibitor in PC-9 H (stable and high expression of SREBP-1 as experimental group), and PC-9 V (stable and low expression of SREBP-1 as control group). In Figure 5E,F, Beas-2B SREBP-1 group and PC-9 SREBP-1 group were as experimental groups that both were silenced by siRNA, and its protein expression decreased leading to low protein expression of LDLR, while Beas-2B Scramble group and PC-9 Scramble group were as control groups. In short, EGFR signaling upregulates LDLR expression through the AKT-mediated SREBP-1 dependent pathway.

A molecule-targeted drug and atorvastatin enhances the treatment effect in vitro
Based on the previous experiment, we wondered whether statins could enhance the effect of a molecule-targeted drug treatment and we did the following experiment. We observed the growth and survival of EGFR-wild type cells and EGFR-mutant type cells using a hemocytometer with a trypan blue assay to count the cell survival numbers in NSCLC with atorvastatin (2.5 µM), gefitinib (0.5 µM), osimertinib (AZD9291) (0.5 µM) and the combination of atorvastatin and a molecule-targeted drug (gefitinib/osimertinib). DMSO was used as a control. Atorvastatin did not affect the growth of EGFR wild type cells (Figure 6A), but helped to inhibit EGFR mutation cells’ survival and growth (Figure 6B,C). Thus, atorvastatin combined with molecule-targeted drugs could enhance the effect of a single treatment in vitro.

Atorvastatin mitigates the growth of NSCLC in vivo, especially when part of a combination treatment
We took BALB/c nude mice implanted with HCC827 cells, and H1975 cells (both NSCLC cells) to study the effects on tumor growth with monotherapy and combination therapy. We found that monotherapy, atorvastatin, held off HCC827 and H1975 cell growth compared with saline, but it was not as significant as gefitinib, or osimertinib. However, in combination groups, atorvastatin played a sensitizing role in retarding NSCLC growth (Figure 7A,B,C,D). We analyzed data and found that in HCC827 cell line, the average tumor shrinkage is about 68% in Gefitinib group, and about 49% in atorvastatin group, but about 89% in combination group. In H1975 cell line, the average tumor shrinkage is about 18% in Osimertinib group, and about 8% in atorvastatin group, but about 44% in combination group. In HCC827 control group, all mice were alive when processing euthanasia (6/6), in gefitinib group, one was dead before processing euthanasia (5/6), mice in atorvastatin group were all alive (6/6), mice in combination group were dead two (4/6). In H1975 control group, all mice were alive when processing euthanasia (6/6), in osimertinib group, two were dead before processing euthanasia (4/6), one died in atorvastatin group (5/6), mice in combination group were dead two (4/6). During the experiment, some mice died due to excessive tumor burden and did not reach the end of the study. Therefore, when inoculating tumor cells, the tumor cells should be strictly homogenized so that the tumor cells can be inoculated in the same part of the mouse to avoid tumors overloaded causing adverse events. Following dissection on the final day, the results further revealed that atorvastatin and a TKI have a synergistic effect on suppressing tumor growth in terms of tumor volume and pathological slide staining (Figure 7E,F,G,H). Thus, atorvastatin inhibits the growth of NSCLC especially when used as part of a combination treatment in vivo.

Summary of results
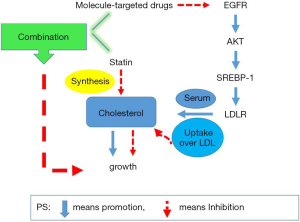
TKI can reduce the expression of LDLR by blocking the SREBP-1-dependent EGFR signaling pathway, which may reduce the utilization of cholesterol and lead to tumor growth inhibition. On the other hand, the application of atorvastatin drugs inhibits the synthesis of cholesterol and causes tumor growth restriction. These effects have a synergistic effect on TKI’s inhibition of tumor growth (Figure 8). This discovery may have clinical guiding significance for the treatment of NSCLC in the future.
Discussion
Here, we found that EGFR mutant lung cancer cell lines are more dependent on cholesterol than EGFR wild type lung cancer cell lines. Furthermore, that cholesterol-lowering treatment (statins) has an inhibitory effect on EGFR mutant lung cancer. At the level of EGFR mutant lung cancer cell lines and in an animal model, cholesterol-lowering therapy has a synergistic effect with EGFR-TKIs.
Targeted therapy for patients with EGFR mutations is initially effective but drug resistance inevitable emerges, therefore, new treatment strategies are urgently needed. Recent immunotherapy in the form of PD-1/PD-L1 has achieved great success in wild-type NSCLC, but the efficacy is poor in mutant lung cancer. Recent studies have shown that a combination of metformin and an EGFR-TKI can improve efficacy. This suggested that the treatment metabolism may be a new strategy for the treatment of EGFR mutant lung cancer and is worthy of exploration. In addition to anti-glucose metabolism, anti-lipid metabolism therapy has shown therapeutic potential in multiple tumor types, but it has not been studied in EGFR mutant lung cancer.
Advances in tumor metabolism have developed rapidly in recent years, especially in regards to the role of cholesterol (25,26). Studies have shown that an increase in de novo synthesis in lipid metabolism is a new sign of many aggressive cancers. For example, it was discovered that cholesterol could influence the proliferation and growth of pancreatic cancer (11,27). In the former research, it was discovered that an EGFR mutation enhances cholesterol uptake and promotes cell proliferation and growth in a glioblastoma EGFR-mutant model. It was also reported that LDLR expression was associated with EGFR mutational status (15). As is well-known, LDLR combines with LDL to transfer extracellular cholesterol intracellularly for metabolic growth (28). However, it is still unclear whether the different demand for cholesterol between EGFR-wild cell lines and EGFR-mutant cell lines is mediated by EGFR in NSCLC. In addition, further evidence is needed regarding the potential clinical significance of suppressing cholesterol synthesis in EGFR-TKI sensitive and EGFR-TKI resistant NSCLC with an EGFR mutation.
In this study, we investigated the association between EGFR and LDLR and the effect of cholesterol disruption on cell proliferation and growth in NSCLC cell lines. Our results show that the signaling pathway from EGFR to LDLR is through the AKT-mediated SREBP-1 dependent pathway and that downregulation of LDL receptors could decrease cholesterol uptake. As such, an EGFR inhibitor could be used to reduce cholesterol uptake to suppress EGFR-mutant cells’ proliferation and growth. Alternatively, a statin could be utilized to suppress cholesterol synthesis thereby repressing EGFR mutant cells proliferation and growth. This explains why the inhibition effect of tumor cell growth using a single EGFR-TKI is superior to statins alone, because EGFR-TKIs can inhibit the activator transcription pathway thereby increasing apoptosis (29) and inhibiting LDLR expression from cholesterol uptake over LDL. Moreover, statins inhibit cholesterol synthesis and indirectly reduce cholesterol uptake. Thus, taking advantage of the combination of a statin and a sensitive EGFR-TKI to treat EGFR-mutant cell lines may have better anti-tumor efficacy than a single drug. Statins may also be a good option for cells resistant to EGFR-TKIs.
The process of a tumor’s biological metabolism is complex, three metabolic pathways are transformed into each other for cells’ proliferation and growth (16). To better observe and test cells’ proliferation and lessen cell disturbances, we seeded cells in serum starved the media one night in advance (30). In the process of culturing cells, we observed through a microscope that the cell-to-cell line connections increased after statin treatment, but that the number of cells decreased, and that cell proliferation was suppressed. We suppose that a statin may make cells release an element that promotes them to die whilst the rest continue to survive. However, this is speculative and requires further study. Nevertheless, EGFR-mutant NSCLC cells did decrease following statin treatment. Based on this, molecule-targeted drugs might accelerate cell death. We anticipate that molecular targeted drugs may not only suppress EGFR and LDLR protein expression and reduce cholesterol uptake, but also cut down cell lines’ connections and isolate them resulting in cell death. Furthermore, we found that a combination treatment of a statin and an EGFR-TKI for sensitive EGFR-TKIs treatment cells has good efficacy, and for resistant EGFR-TKIs cells, a statin restrains its proliferation and growth. This is an exciting possibility that would help those who cannot afford expensive target therapy drugs or those who are resistant to target therapy drugs. We are currently planning clinical trials to advance our research in the hope to further understand cells’ communication after statin treatment.
Our study has some limitations. This study lacks research using lung cancer tissue specimens, and the relationship between EGFR mutations and LDLR in lung cancer patients is not yet clear. This study has not been tested in cell lines resistant to osimertinib (such as cells with cis-T790M and C797S mutations), and it is not clear whether this strategy can reverse resistance to osimertinib.
The results of this study show that EGFR mutant lung cancer cell lines are highly dependent on LDLR, and that anti-lipid metabolism therapy can improve the effect of EGFR-TKIs, and it is expected to reverse drug resistance, thus warranting clinical investigation.
Acknowledgments
We thank Mr. Christopher Lavender of Sun Yat-sen University Cancer Center’s in-house Editorial Office for the language editing of this article.
Funding: This work was supported by Chinese National Science Foundation project (Grant No. 81872499, Grant No. 81772476, Grant No. 81602011), Outstanding Young Talents Program of Sun Yat-sen University Cancer (16zxyc03) and Central Basic Scientific Research Fund for Colleges-Young Teacher Training Program of Sun Yat-sen University (17ykpy85).
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at http://dx.doi.org/10.21037/tlcr-20-812
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tlcr-20-812
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tlcr-20-812). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All these were carried out in SPF class housing of laboratory according to the Laboratory Guidelines for Animal Care. This animal experiments were performed under a project license [SCXK(Shanghai) 20170011#] granted by the Veterinäramt des Kantons Zürich, Switzerland, in compliance with the Swiss Animal Protection Act (TSchG) and the Swiss Animal Protection Ordinance (TSchV).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Mengoli MC, Longo FR, Fraggetta F, et al. The 2015 World Health Organization Classification of lung tumors: new entities since the 2004 Classification. Pathologica 2018;110:39-67. [PubMed]
- Shigematsu H, Lin L, Takahashi T, et al. Clinical and biological features associated with epidermal growth factor receptor gene mutations in lung cancers. J Natl Cancer Inst 2005;97:339-46. [Crossref] [PubMed]
- Rosell R, Carcereny E, Gervais R, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol 2012;13:239-46. [Crossref] [PubMed]
- Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med 2009;361:947-57. [Crossref] [PubMed]
- Sequist LV, Yang JC, Yamamoto N, et al. Phase III study of afatinib or cisplatin plus pemetrexed in patients with metastatic lung adenocarcinoma with EGFR mutations. J Clin Oncol 2013;31:3327-34. [Crossref] [PubMed]
- Soria JC, Ohe Y, Vansteenkiste J, et al. FLAURA Investigators. Osimertinib in Untreated EGFR-Mutated Advanced Non-Small-Cell Lung Cancer. N Engl J Med 2018;378:113-25. [Crossref] [PubMed]
- Ohe Y, Imamura F, Nogami N, et al. Osimertinib versus standard-of-care EGFR-TKI as first-line treatment for EGFRm advanced NSCLC: FLAURA Japanese subset. Jpn J Clin Oncol 2019;49:29-36. [Crossref] [PubMed]
- Wu SG, Shih JY. Management of acquired resistance to EGFR TKI-targeted therapy in advanced non-small cell lung cancer. Mol Cancer 2018;17:38. [Crossref] [PubMed]
- Kuzu OF, Noory MA, Robertson GP. The Role of Cholesterol in Cancer. Cancer Res 2016;76:2063-70. [Crossref] [PubMed]
- Guillaumond F, Bidaut G, Ouaissi M, et al. Cholesterol uptake disruption, in association with chemotherapy, is a promising combined metabolic therapy for pancreatic adenocarcinoma. Proc Natl Acad Sci U S A 2015;112:2473-8. [Crossref] [PubMed]
- Gallagher EJ, Zelenko Z, Neel BA, et al. Elevated tumor LDLR expression accelerates LDL cholesterol-mediated breast cancer growth in mouse models of hyperlipidemia. Oncogene 2017;36:6462-71. [Crossref] [PubMed]
- Pattanayak SP, Bose P, Sunita P, et al. Bergapten inhibits liver carcinogenesis by modulating LXR/PI3K/Akt and IDOL/LDLR pathways. Biomed Pharmacother 2018;108:297-308. [Crossref] [PubMed]
- Liang Z, Chen Y, Wang L, et al. CYP27A1 inhibits bladder cancer cells proliferation by regulating cholesterol homeostasis. Cell Cycle 2019;18:34-45. [Crossref] [PubMed]
- Guo D, Reinitz F, Youssef M, et al. An LXR agonist promotes glioblastoma cell death through inhibition of an EGFR/AKT/SREBP-1/LDLR-dependent pathway. Cancer Discov 2011;1:442-56. [Crossref] [PubMed]
- Akram M. Citric acid cycle and role of its intermediates in metabolism. Cell Biochem Biophys 2014;68:475-8. [Crossref] [PubMed]
- Niwa H, Yamamura K, Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 1991;108:193-9. [Crossref] [PubMed]
- Stachnik A, Yuen T, Iqbal J, et al. Repurposing of bisphosphonates for the prevention and therapy of nonsmall cell lung and breast cancer. Proc Natl Acad Sci U S A 2014;111:17995-8000. [Crossref] [PubMed]
- Peng P, Wei W, Long C, et al. Atorvastatin augments temozolomide’s efficacy in glioblastoma via prenylation-dependent inhibition of Ras signaling. Biochem Biophys Res Commun 2017;489:293-8. [Crossref] [PubMed]
- Chang TC, Chin YT, Nana AW, et al. Enhancement by Nano-Diamino-Tetrac of Antiproliferative Action of Gefitinib on Colorectal Cancer Cells: Mediation by EGFR Sialylation and PI3K Activation. Horm Cancer 2018;9:420-32. [Crossref] [PubMed]
- Ballard P, Yates JW, Yang Z, et al. Preclinical Comparison of Osimertinib with Other EGFR-TKIs in EGFR-Mutant NSCLC Brain Metastases Models, and Early Evidence of Clinical Brain Metastases Activity. Clin Cancer Res 2016;22:5130-40. [Crossref] [PubMed]
- Couto M, Cates C. Laboratory Guidelines for Animal Care. Methods Mol Biol 2019;1920:407-30. [Crossref] [PubMed]
- Li D, Shimamura T, Ji H, et al. Bronchial and peripheral murine lung carcinomas induced by T790M-L858R mutant EGFR respond to HKI-272 and rapamycin combination therapy. Cancer Cell 2007;12:81-93. [Crossref] [PubMed]
- Tokuzawa Y, Kaiho E, Maruyama M, et al. Fbx15 is a novel target of Oct3/4 but is dispensable for embryonic stem cell self-renewal and mouse development. Mol Cell Biol 2003;23:2699-708. [Crossref] [PubMed]
- Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat Rev Cancer 2011;11:85-95. [Crossref] [PubMed]
- Silvente-Poirot S, Poirot M. Cancer. Cholesterol and cancer, in the balance. Science 2014;343:1445-6. [Crossref] [PubMed]
- Merino Salvador M, Gómez de Cedrón M, Moreno Rubio J, et al. Lipid metabolism and lung cancer. Crit Rev Oncol Hematol 2017;112:31-40. [Crossref] [PubMed]
- Maxfield FR, van Meer G. Cholesterol, the central lipid of mammalian cells. Curr Opin Cell Biol 2010;22:422-9. [Crossref] [PubMed]
- da Cunha Santos G, Shepherd FA, Tsao MS. EGFR mutations and lung cancer. Annu Rev Pathol 2011;6:49-69. [Crossref] [PubMed]
- Chen N, Fang W, Zhan J, et al. Upregulation of PD-L1 by EGFR Activation Mediates the Immune Escape in EGFR-Driven NSCLC: Implication for Optional Immune Targeted Therapy for NSCLC Patients with EGFR Mutation. J Thorac Oncol 2015;10:910-23. [Crossref] [PubMed]